Abstract
Gene inactivation of the orphan G protein-coupled receptor LGR4, a paralogue of the epithelial-stem-cell marker LGR5, results in a 50% decrease in epithelial cell proliferation and an 80% reduction in terminal differentiation of Paneth cells in postnatal mouse intestinal crypts. When cultured ex vivo, LGR4-deficient crypts or progenitors, but not LGR5-deficient progenitors, die rapidly with marked downregulation of stem-cell markers and Wnt target genes, including Lgr5. Partial rescue of this phenotype is achieved by addition of LiCl to the culture medium, but not Wnt agonists. Our results identify LGR4 as a permissive factor in the Wnt pathway in the intestine and, as such, as a potential target for intestinal cancer therapy.
Keywords: GPCR, intestinal crypts, LGR, stem cells
Introduction
LGR4, 5 and 6 constitute a subfamily of orphan G protein-coupled receptors (GPCRs) that are evolutionarily related to the glycoprotein hormone receptors (Hsu et al, 1998). LGR5 and LGR6 have recently been identified as specific markers of stem cells in several epithelia (Barker et al, 2007; Barker & Clevers, 2010), and LGR5 has been shown to negatively control the Wnt pathway during intestinal development (Garcia et al, 2009). Whereas homozygous Lgr5-knockout mice die from ankyloglossia in the few hours after birth (Morita et al, 2004), homozygous or hypomorphic Lgr4-knockout mice display intrauterine growth retardation (Mazerbourg et al, 2004) and defects in the development of several organs (Kato et al, 2006, 2007; Mendive et al, 2006; Song et al, 2008; Weng et al, 2008; Luo et al, 2009; Yamashita et al, 2009; Li et al, 2010). No data are available about the possible functional relationship between LGR4 and LGR5 in epithelial stem cells or progenitors.
Intestinal crypts constitute a classical model in which to study the interaction and effects of various regulatory cascades on epithelial stem cells in physiology and diseases (Bjerknes & Cheng, 2005; Fodde & Brabletz, 2007; Scoville et al, 2008; Potten et al, 2009). We report here the expression pattern of Lgr4 in the mouse small intestine and demonstrate that it is required for terminal differentiation of Paneth cells in vivo, and maintenance of intestinal stem cells or progenitors in ex vivo cultures.
Results And Discussion
Lgr4 expression in the small intestine
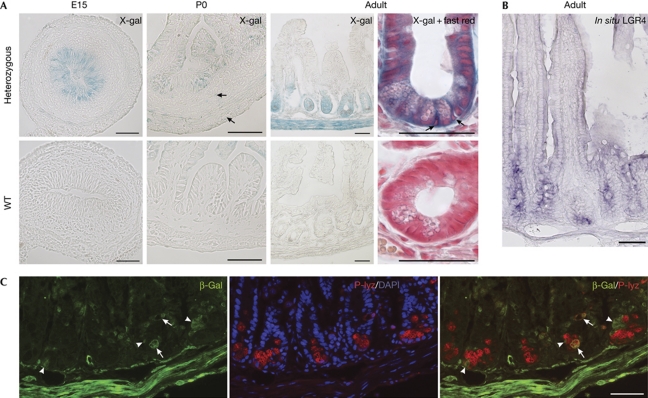
Heterozygous mice with a LacZ gene trap knocked in the Lgr4 locus (Leighton et al, 2001) were used to investigate Lgr4 gene expression in the ileum. At embryonic day (E) 15 and at birth, LacZ activity was detected in the pseudo-stratified epithelium and intervillus progenitors, respectively (Fig 1A), in a pattern similar to that of Lgr5 (Garcia et al, 2009). In adults, epithelial expression of Lgr4 was found all along the crypts, but not in the villi, using 5-bromo-4-chloro-3-indolyl-β-D-galactoside (X-gal) staining and in situ hybridization with an LGR4 riboprobe (Fig 1A,B). In the crypts, Lgr4 expression was found above the Paneth-cell zone, in the transit-amplifying cell region, in crypt basal columnar (CBC) cells, between Paneth cells (Fig 1A,C) and in rare Paneth cells (Fig 1C). Outside the epithelium, Lgr4 was expressed at low levels in the mesenchyme and smooth-muscle layers of embryo (E15) and newborn mice (Fig 1A) and in adults, more strongly in the smooth-muscle layers (Fig 1A,C), intestinal subepithelial myofibroblasts and enteric neurons (supplementary Fig S1A,B online). A similar expression pattern was found in the duodenum and colon (supplementary Fig S1A online).
Figure 1.
Lgr4 expression pattern in the ileum. (A) Lgr4/LacZ expression detected by X-gal staining of heterozygous or wild-type (WT) embryonic (E15), newborn (P0) and adult mice. Arrows in newborn heterozygous panel indicate faint but specific X-gal-positive mesenchymal or smooth muscle cells. In adults, arrows indicate CBC cells in an enlarged view of X-gal-stained crypt with fast red counterstaining. (B) In situ hybridization of an adult section showing Lgr4 expression all along the crypt. (C) Co-staining of β-gal and P-lyz antibodies with DAPI. In the bottom of the crypts, β-gal-expressing cells that do not express the P-lyz Paneth cell marker are shown (arrowheads), whereas few cells show double staining (arrows). Scale bars, 50 μm. β-gal, β-galactosidase; DAPI, 4,6-diamidino-2-phenylindole; E, embryonic day; P, postnatal day; P-lyz, P-lyzozome; WT, wild type; X-gal, 5-bromo-4-chloro-3-indolyl-D-galactoside.
LGR4 deficiency affects postnatal crypt development
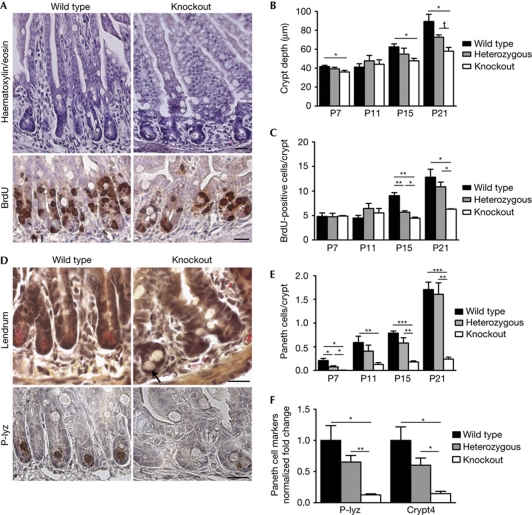
Mice homozygous for the gene trap LacZ knock-in allele, referred to as ‘Lgr4 knockout’, displayed a hypomorphic phenotype. Compared with wild-type ileum, expression of Lgr4 was reduced to 10% in Lgr4 knockout (supplementary Fig S2A online). Although the timing of crypt development was normal in Lgr4 knockout mice, a reduction in the crypt depth (25–35%) was obvious from postnatal day (P) 15 and was accompanied by a 50% reduction in epithelial-cell proliferation (Fig 2A–C). Differentiation of absorptive, enteroendocrine and goblet-cell lineages was not modified significantly (supplementary Fig S2B online). By contrast, a defect in Paneth-cell differentiation was observed at all postnatal stages tested, with an 85% reduction in their number at P21 (Fig 2E) and decreased expression of the terminal differentiation markers P-lyzozyme and cryptdin 4 (Fig 2D,F). Moreover, the degree of maturation of the rare Paneth-cells was decreased (supplementary Fig S2C online). Similar effects of LGR4 deficiency were observed in the duodenum (supplementary Fig S2D online). These data suggest a key role for LGR4 in normal postnatal epithelial cell proliferation and terminal Paneth-cell differentiation. This phenotype is similar to that observed in mice with a hypomorphic β-catenin allele (Andreu et al, 2008), suggesting that LGR4 might positively regulate the Wnt pathway. It is in contrast to the premature development of Paneth cells and upregulation of Wnt-target genes observed in Lgr5-null mice (Garcia et al, 2009). The apparently antagonistic effects of the two receptors agree with our surprising observation that Lgr4/Lgr5 double knockouts survive the neonatal period (supplementary Fig S2E online), whereas Lgr5 knockouts die at birth from ankyloglossia (Morita et al, 2004). However, similarly to Lgr4 knockouts, double knockout mice die before one month of age, probably from severe kidney lesions (supplementary Fig S2E online; Kato et al, 2006). These data indicate non-redundancy of LGR4 and LGR5, with a dominant effect of LGR4 deficiency.
Figure 2.
LGR4 deficiency affects postnatal crypt development in the small intestine. (A) Haematoxylin/eosin staining and immunohistochemical detection of BrdU of ileal sections of P15 wild-type and knockout mice that were killed 90 min after injection. (B) Measurement of postnatal ileal crypt-depth; a total of 20–50 well-oriented crypt–villus units were assessed per mouse; n=2–5 per group. *P<0.03, tP=0.074. (C) Quantification of BrdU staining; a total of 20 crypts per mouse; n=2–7 per group; **P<0.003, *P<0.03. (D) Paneth cell lineage differentiation assessed by Lendrum's staining and immunohistochemical detection of P-lyz performed on ileal sections of P15 mice. Arrow indicates an immature Paneth cell in the knockout section. (E) Quantification of Paneth-cell differentiation by Lendrum's staining (30–60 crypts per mouse; n=2–5 per group); ***P⩽0.0002, **P<0.003, *P<0.03. (F) Quantitative real-time–PCR analysis of RNA from P15 mice (n=6 for wild type and heterozygous, n=4 for knockout). P-Lyz and Crypt4 transcripts are normalized to wild-type levels; *P<0.02; **P<0.005. (A,D) scale bars, 20 μm. (B,C,E,F) values are means±s.e.m. Significance computed from unpaired t-test. BrdU, 5-bromodeoxyuridine; P, postnatal day; P-lyz, P-lyzozyme.
LGR4 is required for ex vivo maintenance of crypts
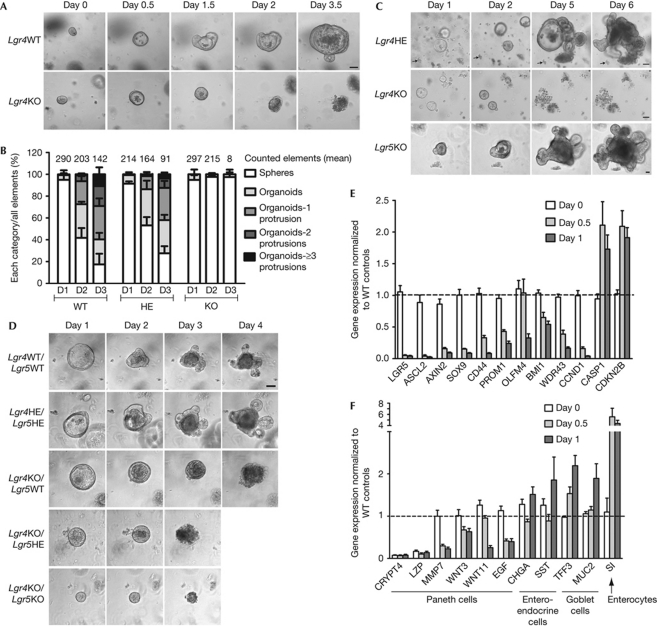
To determine the role of LGR4 in crypt development without the influence of the mesenchyme, Lgr4-knockout crypts were studied in the ‘minigut’ culture system (Sato et al, 2009). In agreement with the original description, crypts cultured from P15 wild-type or heterozygous mice differentiated into multi-fingered organoids after 3 days in culture (Fig 3A,B). By contrast, after generation of hollow spheres containing mainly stem and transit-amplifying cells (day 0.5–1), structures grown from P15 Lgr4-knockout intestine did not increase further in size and became filled with cellular material (days 1.5–2; Fig 3A). Lgr4-knockout organoids started to disaggregate between days 2 and 3, and died before day 7 (Fig 3A). The same phenotype was observed in Lgr4-knockout progenitors isolated from newborn mice (Fig 3C), when fully differentiated Paneth cells are not yet present, despite detection of Crypt4 transcripts in both wild-type and knockout tissues (supplementary Fig S3B online).
Figure 3.
LGR4 is required for ex vivo development of organoids. (A) Organoid formation of crypts from P15 mice. (B) Quantification of organoid complexity during the first 3 days of culture (D1, D2 and D3) of crypts from P15 mice (complexity classes are defined in supplementary Fig S3A online). Numbers above the columns represent the average of elements counted per genotype (n=3 wild-type (WT), 3 heterozygous (HE) and 4 knockout (KO) animals). (C) Organoid formation from intervillus progenitors of P0 mice. ‘Remarkable’ debris are used as tracking landmarks (arrows). (D) Organoid formation of crypts collected from P11 mice generated from crosses between double heterozygous Lgr4/Lgr5 mating pairs. Scale bars, 50 μm. (E,F) Quantitative real-time–PCR analysis of transcripts from P15-derived crypt cultures. Transcripts in Lgr4 KO samples were normalized to WT levels at each time point. Statistical analyses compare day 1 with day 0 in KO samples; values are means±s.e.m. Significance was calculated from unpaired t-test: P⩽0.0006 in (E); P<0.04 (Wnt3, Muc2), P⩽0.0003 (Mmp7, Wnt11, Egf, Tff3, Si) in (F). P, postnatal day.
Due to neonatal lethality, the ex vivo phenotype of Lgr5 knockout could only be studied from the culture of P0 progenitors. In contrast to Lgr4-knockout, Lgr5-knockout progenitors survived and evolved into complex organoids within 5 days (Fig 3C). Lgr5 deficiency did not rescue the Lgr4-knockout phenotype in Lgr4/Lgr5 double knockout crypts (Fig 3D). Thus, unlike LGR5, LGR4 is required for ex vivo survival of both fetal progenitors and adult stem cells, and the effect of LGR4 deficiency is dominant in double knockouts.
To investigate the mechanism leading to death of Lgr4-knockout crypts, we compared gene expression profiles of Lgr4-knockout and wild-type crypts during culture, and validated these data by quantitative real-time (qRT)–PCR on a selection of gene markers (Fig 3E,F; supplementary Table S1 online). At the time of seeding, the most strongly downregulated transcripts in P15-knockout crypt preparations were terminal differentiation markers of Paneth cells (Crypt4 and Lzp). Stem-cell and other differentiation markers were at wild-type levels (Fig 3E,F). Other Paneth-specific genes (Wnt3, Wnt11, Mmp7 and Egf) were not substantially affected (Fig 3F). These data are consistent with a blockade in terminal differentiation of Paneth cells in Lgr4-knockout crypts in vivo. After 12 h in culture—when the morphology of Lgr4-knockout and wild-type crypts was similar—a marked decrease in expression (80–90%) of CBC cell markers (Gracz et al, 2010) and Wnt target genes (Ascl2, Lgr5, Axin2, Sox9 and Cyclin D1) was observed (Fig 3E). This effect was stronger after 24 h. Downregulation of other stem-cell markers (Bmi1 and Prom1; Sangiorgi & Capecchi, 2008; Zhu et al, 2009) and transit-amplifying cells (Wdr43; Barker et al, 2007) also occurred, but to a lesser extent and at a later stage (Fig 3E). Among the genes that were upregulated during culture of Lgr4-knockout crypts were the CDK inhibitor Cdkn2b, the proapoptotic gene Casp1 and differentiation markers of all lineages, except Paneth cells (Fig 3E,F). The latter probably reflects the decrease in Wnt signalling tone and the progressive exhaustion of stem and transit-amplifying cells during culture of Lgr4-knockout crypts, with relative increase in the proportion of differentiated cells. In agreement with this view, Lgr4-knockout spheres showed only rare Ki67-positive cells at day one, compared with wild-type (supplementary Fig 3C online).
These findings indicate that Lgr4 deficiency causes a blockade in terminal differentiation of Paneth cells in vivo and leads to acute loss of stem cells in crypts cultured ex vivo. The wild-type levels of Wnt3 and Wnt11 transcripts in Lgr4 knockout at the start of the culture argue against the hypothesis that death of CBC cells ex vivo would result from the loss of Wnt signals normally produced by Paneth cells (Sato et al, 2011).
Partial rescue of Lgr4 knockout crypts in culture by LiCl
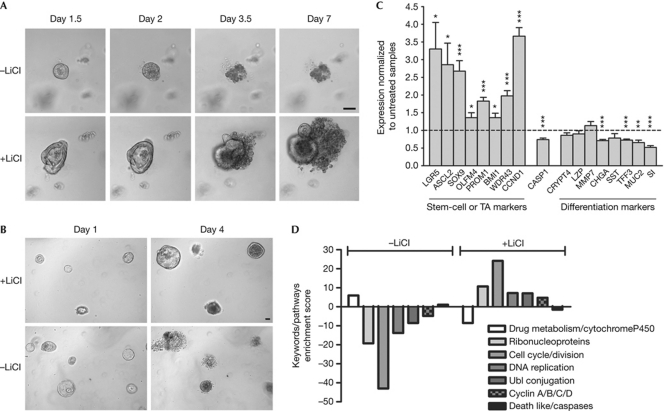
Death of Lgr4-knockout crypts ex vivo suggests that in wild-type organoids, LGR4 receptor is activated by its agonist—either in the culture medium or produced by the organoids themselves—or is endowed with constitutive activity. Accordingly, we sought to rescue Lgr4-knockout crypts by activating various GPCR signalling pathways. Although LGR4 has been reported to activate cyclic AMP production through Gs (Li et al, 2010), forced activation of this cascade by forskolin was ineffective. Activation of several GPCRs known to be expressed in intestinal stem cells and coupled to different cascades did not rescue the phenotype Lgr4-knockout crypts (supplementary Table S3 online; van der Flier et al, 2009). As Wnt target genes were strongly and rapidly downregulated in cultured Lgr4-knockout crypts, we tested several agents known to stimulate the Wnt pathway, including Wnt3a and inhibitors of gsk3β (supplementary Table S3 online). Only LiCl (1–10 mM) was effective; 6% of Lgr4-knockout crypts survived after 7 days of culture in the presence of 5-mM LiCl, compared with 0% when cultured in control medium (Fig 4A). In comparison, 60% of wild-type crypts survived in control medium, showing that rescue by LiCl was only partial. Lgr4-knockout organoids rescued by LiCl could be replated several times over a period of more than one month and remained dependent on LiCl for survival (Fig 4B).
Figure 4.
LGR4-deficient phenotype ex vivo is partly rescued by LiCl. (A) Crypts from P15 Lgr4 knockout cultured in the presence or absence of 7.5-mM LiCl. One representative experiment out of 11 is shown. (B) Dependence of LGR4-deficient organoids on LiCl treatment. After dissociation, Lgr4-knockout-LiCl rescued organoids were replated in the presence (+LiCl) or absence (−LiCl) of LiCl. Scale bars, 50 μm. (C) Quantitative real-time–PCR analysis at day 1 of transcripts from P15-derived Lgr4-knockout crypts cultured with or without 5-mM LiCl. Transcripts were normalized to untreated samples. Statistical analyses: values are means±s.e.m. Significance was computed from paired t-test: *P<0.03, **P=0.0024, ***P⩽0.0005. (D) Ontogeny analysis of microarray results with the David software, comparing gene expression at day 1 of Lgr4 knockout with wild-type crypts cultured in control medium (−LiCl), and knockout crypts treated with or without LiCl.
The effect of LiCl on the expression of a panel of markers was investigated in Lgr4-knockout crypts by qRT–PCR after one day in culture. LiCl induced upregulation of Wnt-dependent and -independent stem cell and progenitor markers and—with the exception of Paneth cells—downregulation of terminal lineage differentiation markers (Fig 4C). Global transcriptome analysis by the DAVID software (Huang et al, 2009) confirmed and extended the observation that LiCl counteracts the effects of culture on the gene expression pattern in Lgr4-knockout crypts (Fig 4D).
In line with the almost normal levels of Wnt transcripts in Lgr4-knockout crypts at day 0, the inability of exogenous Wnt3a to rescue Lgr4-knockout crypts suggests that the death of CBC cells or P0 progenitors is cell-autonomous, with LGR4 functioning as a survival factor, probably through a permissive effect on the Wnt signalling cascade. Accurate analyses of the connections between LGR4 and Wnt regulatory pathways will only be possible when LGR4 agonists become available. Failure of gsk3β inhibitors to rescue Lgr4-knockout crypts ex vivo suggests that LiCl functions, at least partly, independently of its known inhibitory effect on this enzyme (Phiel & Klein, 2001). In this regard, LiCl has recently been shown to control gsk3β by dissociating β-arrestin–PP2a–Akt complexes (Beaulieu et al, 2009). This provides an interesting link to GPCRs and indicates that the action of LiCl—and possibly LGR4—could involve interaction between the Wnt and PI3K/Akt pathways.
Our observations in vivo suggest a role for LGR4 in allowing normal cell proliferation in intestinal crypts and terminal differentiation of Paneth cells. Ex vivo, they demonstrate that LGR4 is required for the maintenance of crypt stem cells, in contrast to the survival and development into organoids of Lgr5-knockout progenitors. The milder phenotype of Lgr4 knockout observed in vivo implies that extra-epithelial signals of systemic or, more likely, mesenchymal or muscular origin, (Powell et al, 2011) partly compensate for LGR4 deficiency in vivo. Given the role of Wnt in stem-cell survival and proliferation, as well as in Paneth-cell differentiation, we hypothesize that LGR4 permissively controls the Wnt pathway in CBC cells. Our results indicate non-redundancy between Lgr4 and Lgr5 genes and identify LGR4 as a potential target for the development of antagonists with therapeutic use in intestinal cancers.
Methods
Animal experiments and tissue processing. Animal procedures complied with the guidelines of the European Union and were approved by the local ethics committee. The Lgr4/Gpr48^Gt mice (Leighton et al, 2001) were crossed with Lgr5/LacZ–NeoR knock-in mice (Morita et al, 2004) to generate double Lgr4/Lgr5 knockouts.
Tissue processing, histological protocols, immunohistochemistry and in situ hybridization experiments were carried out as described previously (Garcia et al, 2009). The following primary antibodies were used for tissue and organoid staining: goat villin (SantaCruz Biotechnology); rabbit chromogranin A (Immunostar); P-lyzozyme (Dako) and Ki67 (Abcam); mouse vimentin (Dako) and β-catenin (BD Transduction Lab.); rat BrdU (Abcam); and chicken β-galactosidase (Abcam).
Ex vivo culture. Samples were cultured according to Sato et al (2009). Crypt medium was supplemented with 2-mM L-glutamine, penicillin–streptomycin cocktail and 2% fetal bovine serum. After 6–10 days in culture, organoids were collected, dissociated with 2.5-mg ml−1 dispase II (Roche), and replated in fresh Matrigel. In experiments with P0 progenitors, 1-μM JAG-1 (Anaspec) was added to the Matrigel and the culture medium was supplemented with 10-μM Y-27632 (Sigma). Wnt3a (Milteny, Germany) was added to Matrigel and medium (100 ng ml−1), and its bioactivity was checked using the C2C12 myoblast differentiation and TOPflash reporter assays.
qRT–PCR. qRT–PCR was performed on total RNA from ileum or ex vivo crypt cultures, as reported previously (Garcia et al, 2009). Expression levels were normalized to that of the Gapdh and Ywhaz genes. Each sample was run in duplicate. Primer sequences are listed in supplementary Table S2. For qRT–PCR data, depending on the gene analysed, the number of animals used was: 10–11 for wild type and 14–16 for knockout at day 0; 5–7 for wild type and 10 for knockout at day 0.5; and 5–7 for wild type and 8–12 for knockout at day 1 (Fig 3E), and 3 for wild type and 6–9 for knockout (Figs 3F; 4C).
Microarray experiments. Two-channel microarray experiments, with dye-swap, were performed from a single wild-type/Lgr4 knockout P15 crypt pair on MEEBO slides (38,784 seventy-mer probes; Stanford Functional Genomics Facility, CA, USA), as described previously (Garcia et al, 2009). For the effect of LiCl on Lgr4-knockout crypts at day one, genes with mean duplicate ratios of >1.5 or <0.66 were selected. These data, together with those from Lgr4 knockout compared with wild-type at day one, were independently uploaded into the DAVID Bioinformatics Resources 6.7 database (NIAID), and functional annotation clustering was determined by using the default setting (Huang et al, 2009). Complete microarray data sets were deposited in Gene Expression Omnibus (GEO; accession number: GSE27904).
Statistical evaluation. Statistical analyses were performed with Graph Pad Prism. All experimental data are expressed as mean±s.e.m. Significance of differences between genotypes was determined by unpaired t-test analysis. In Fig 4C, a paired t-test was used to compare untreated with LiCl-treated knockout crypts.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We are grateful to William C. Skarnes and Hans Clevers for providing us with gene trap LacZ knock-in (LST20) and Lgr5 knockout mice, respectively. We thank David Communi for critical reading of the manuscript and Gabriela Vasile for help with Wnt assays. This work was supported by the Interuniversity Attraction Poles Programme–Belgian State–Belgian Science Policy (6/14), the Fonds de la Recherche Scientifique Médicale of Belgium, the Walloon Region (programme ‘Cibles’) and the Francqui Foundation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Andreu P, Peignon G, Slomianny C, Taketo MM, Colnot S, Robine S, Lamarque D, Laurent-Puig P, Perret C, Romagnolo B (2008) A genetic study of the role of the wnt/β-catenin signalling in paneth cell differentiation. Dev Biol 324: 288–296 [DOI] [PubMed] [Google Scholar]
- Barker N, Clevers H (2010) Leucine-rich repeat-containing G-Protein-coupled receptors as markers of adult stem cells. Gastroenterology 138: 1681–1696 [DOI] [PubMed] [Google Scholar]
- Barker N et al. (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007 [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, Gainetdinov RR, Caron MG (2009) Akt/Gsk3 signaling in the action of psychotropic drugs. Annu Rev Pharmacol Toxicol 49: 327–347 [DOI] [PubMed] [Google Scholar]
- Bjerknes M, Cheng H (2005) Gastrointestinal stem cells. ii. intestinal stem cells. Am J Physiol Gastrointest Liver Physiol 289: G381–G387 [DOI] [PubMed] [Google Scholar]
- Fodde R, Brabletz T (2007) Wnt/β-Catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol 19: 150–158 [DOI] [PubMed] [Google Scholar]
- Garcia MI, Ghiani M, Lefort A, Libert F, Strollo S, Vassart G (2009) Lgr5 deficiency deregulates wnt signaling and leads to precocious paneth cell differentiation in the fetal intestine. Dev Biol 331: 58–67 [DOI] [PubMed] [Google Scholar]
- Gracz AD, Ramalingam S, Magness ST (2010) Sox9 expression marks a subset of Cd24-expressing small intestine epithelial stem cells that form organoids in vitro. Am J Physiol Gastrointest Liver Physiol 298: G590–G600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SY, Liang SG, Hsueh AJ (1998) Characterization of two lgr genes homologous to gonadotropin and thyrotropin receptors with extracellular leucine-rich repeats and a G protein-coupled, seven-transmembrane region. Mol Endocrinol 12: 1830–1845 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Kato S, Matsubara M, Matsuo T, Mohri Y, Kazama I, Hatano R, Umezawa A, Nishimori K (2006) Leucine-rich repeat-containing G protein-coupled receptor-4 (Lgr4, Gpr48) is essential for renal development in mice. Nephron Exp Nephrol 104: E63–E75 [DOI] [PubMed] [Google Scholar]
- Kato S, Mohri Y, Matsuo T, Ogawa E, Umezawa A, Okuyama R, Nishimori K (2007) Eye-open at birth phenotype with reduced keratinocyte motility in Lgr4 null mice. FEBS Lett 581: 4685–4690 [DOI] [PubMed] [Google Scholar]
- Leighton PA, Mitchell KJ, Goodrich LV, Lu X, Pinson K, Scherz P, Skarnes WC, Tessier-Lavigne M (2001) Defining brain wiring patterns and mechanisms through gene trapping in mice. Nature 410: 174–179 [DOI] [PubMed] [Google Scholar]
- Li XY et al. (2010) G protein-coupled receptor 48 upregulates estrogen receptor alpha expression via Camp/Pka signaling in the male reproductive tract. Development 137: 151–157 [DOI] [PubMed] [Google Scholar]
- Luo J et al. (2009) Regulation of bone formation and remodeling by G-protein-coupled receptor 48. Development 136: 2747–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazerbourg S, Bouley DM, Sudo S, Klein CA, Zhang JV, Kawamura K, Goodrich LV, Rayburn H, Tessier-Lavigne M, Hsueh AJ (2004) Leucine-rich repeat-containing, G protein-coupled receptor 4 null mice exhibit intrauterine growth retardation associated with embryonic and perinatal lethality. Mol Endocrinol 18: 2241–2254 [DOI] [PubMed] [Google Scholar]
- Mendive F, Laurent P, Van SG, Skarnes W, Pochet R, Vassart G (2006) Defective postnatal development of the male reproductive tract in Lgr4 knockout mice. Dev Biol 290: 421–434 [DOI] [PubMed] [Google Scholar]
- Morita H, Mazerbourg S, Bouley DM, Luo CW, Kawamura K, Kuwabara Y, Baribault H, Tian H, Hsueh AJ (2004) Neonatal lethality of Lgr5 null mice is associated with ankyloglossia and gastrointestinal distension. Mol Cell Biol 24: 9736–9743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phiel CJ, Klein PS (2001) Molecular targets of lithium action. Annu Rev Pharmacol Toxicol 41: 789–813 [DOI] [PubMed] [Google Scholar]
- Potten CS, Gandara R, Mahida YR, Loeffler M, Wright NA (2009) The stem cells of small intestinal crypts: where are they? Cell Prolif 42: 731–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell DW, Pinchuk IV, Saada JI, Chen X, Mifflin RC (2011) Mesenchymal cells of the intestinal lamina propria. Annu Rev Physiol 73: 213–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangiorgi E, Capecchi MR (2008) Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 40: 915–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T et al. (2009) Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459: 262–265 [DOI] [PubMed] [Google Scholar]
- Sato T, Van Es JH, Snippert HJ, Stange DE, Vries RG, Van Den Born M, Barker N, Shroyer NF, Van De Wetering M, Clevers H (2011) Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469: 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoville DH, Sato T, He XC, Li L (2008) Current view: intestinal stem cells and signaling. Gastroenterology 134: 849–864 [DOI] [PubMed] [Google Scholar]
- Song H, Luo J, Luo W, Weng J, Wang Z, Li B, Li D, Liu M (2008) Inactivation of G-Protein-coupled receptor 48 (Gpr48/Lgr4) impairs definitive erythropoiesis at midgestation through down-regulation of the Atf4 signaling pathway. J Biol Chem 283: 36687–36697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier LG et al. (2009) Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell 136: 903–912 [DOI] [PubMed] [Google Scholar]
- Weng J et al. (2008) Deletion of g protein-coupled receptor 48 leads to ocular anterior segment dysgenesis (Asd) through down-regulation Of Pitx2. Proc Natl Acad Sci USA 105: 6081–6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita R, Takegawa Y, Sakumoto M, Nakahara M, Kawazu H, Hoshii T, Araki K, Yokouchi Y, Yamamura K (2009) Defective development of the gall bladder and cystic duct in Lgr4- hypomorphic mice. Dev Dyn 238: 993–1000 [DOI] [PubMed] [Google Scholar]
- Zhu L, Gibson P, Currle DS, Tong Y, Richardson RJ, Bayazitov IT, Poppleton H, Zakharenko S, Ellison DW, Gilbertson RJ (2009) Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature 457: 603–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.