Abstract
By using mass spectrometry, we have identified Ser 402 as a new phosphorylation site within the catalytic domain of human slingshot 1 (SSH1). Phosphorylation at this site inhibits substrate binding and, thus, phosphatase activity in vitro, resulting in enrichment of phosphorylated cofilin in monolayer cell culture. We further demonstrate that protein kinase D (PKD) is upstream from Ser 402 phosphorylation. Accordingly, expression of active PKD in Drosophila phenotypically mimics the loss of SSH activity by inducing accumulation of phosphorylated cofilin and filamentous actin. We thus identify a universal mechanism by which PKD controls SSH1 phosphatase activity.
Keywords: cofilin, protein kinase D, slingshot, focal adhesion
Introduction
Cofilin is an actin-binding protein that depolymerizes filaments through its actin-severing activity and creates new ends for elongation, thereby adjusting the direction of cell movement (overview in Desmarais et al, 2005). Cofilin activity is inhibited by phosphorylation on Ser 3. The family of slingshot (SSH) phosphatases comprises three members in mammals—SSH1, SSH2 and SSH3—which reactivate phosphorylated cofilin (P-cofilin) by dephosphorylation (Niwa et al, 2002). SSH1 has an amino-terminal, noncatalytic (SSH-N) domain and is activated by binding to filamentous actin (F-actin). Both the noncatalytic and the phosphatase domains are required for SSH1 binding to P-cofilin (Kurita et al, 2008). In the absence of SSH1, cells accumulate P-cofilin and F-actin (Niwa et al, 2002). It was shown that the protein kinase D (PKD) family of serine/threonine kinases negatively regulates SSH1 by direct phosphorylation at Ser 937 and 978, which are located within the S domain of the carboxy-terminus (Eiseler et al, 2009; Peterburs et al, 2009). Phosphorylation of these residues induces the binding of 14-3-3 scaffolding proteins leading to the sequestration of SSH1 in the cytoplasm, thereby limiting the amount of active SSH1 at the lamellipodia (Nagata-Ohashi et al, 2004). However, the S domain is present in SSH1 and SSH2, but not in SSH3 or Drosophila SSH (Ohta et al, 2003). Here, we identify Ser 402 in SSH1 as a new phosphorylation site, that is conserved in SSH2 and across species. We provide evidence that PKD-mediated phosphorylation at Ser 402 blocks the phosphatase activity of SSH1, resulting in enrichment of P-cofilin. Accordingly, SSH1 and Drosophila SSH peptides that overlap with this site are in vitro targets for PKD. Similarly, on loss of SSH activity, overexpression of active PKD in Drosophila tissues induces enrichment of F-actin and P-cofilin. We identify here a universal mechanism by which PKD controls SSH1 phosphatase activity.
Results And Discussion
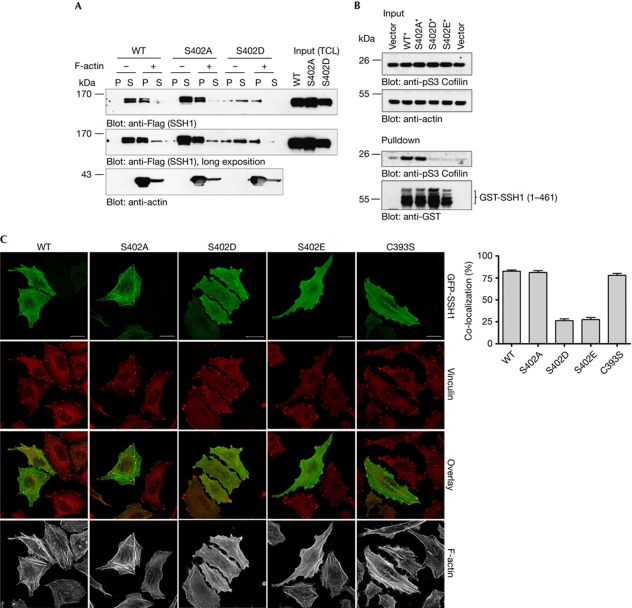
To identify new phosphorylation sites within SSH1, nano liquid chromatography–mass spectrometry analysis of trypsin-digested Flag-tagged SSH1 ectopically expressed in H2O2-stimulated human embryonic kidney (HEK)293T cells was performed. A singly phosphorylated, doubly charged peptide detected at m/z 611.29, corresponding to amino acids 400–410 of SSH1, was fragmented. The resulting fragmentation range showed a good sequence coverage by y- and b-ions, enabling unambiguous localization of the phospho-site to Ser 402 (Fig 1A). Ser 402 is next to the phosphatase domain of SSH1, raising the possibility that phosphorylation at this site might be involved in P-cofilin recognition and thereby regulation of phosphatase activity. To test whether in vitro phosphatase activity was altered by Ser 402 phosphorylation, we generated a GST-tagged SSH1 protein comprising amino acids 1–461, which has full phosphatase activity in an F-actin-dependent manner (Kurita et al, 2008). Furthermore, we exchanged Ser 402 with an alanine (S402A), glutamate (S402E) and aspartate (S402D) to generate phospho-deficient and phospho-mimic mutants, respectively. SSH1 wild type and the mutants were pulled down from HEK293T cell lysates and subjected to in vitro phosphatase assays together with Flag-tagged P-cofilin and F-actin. In this assay, the wild-type and the S402A protein showed similar phosphatase activity resulting in a decrease of P-cofilin levels in comparison to control (Fig 1B), whereas the S402D and S402E proteins had no effect on P-cofilin. Therefore, we next tested whether phosphorylation at Ser 402 affects SSH1 activity in cells. We transiently expressed the truncated and full-length wild-type SSH1 and the S402A, S402D and S402E mutants in HEK293T cells and analysed the level of P-cofilin. Expression of the wild-type and the S402A protein decreased, whereas S402D and S402E mutants enriched P-cofilin levels compared with control, respectively, indicating that Ser 402 phosphorylation affects the phosphatase activity of SSH1 in vitro and in cells (Fig 1C,D). Moreover, introduction of S402D and E mutations into a non-phosphorylatable SSH1 S937/978A protein with higher phosphatase activity (Nagata-Ohashi et al, 2004) restored P-cofilin to the control level (Fig 1D), showing that Ser 402 phosphorylation is a crucial mechanism in the control of phosphatase activity.
Figure 1.
Phosphorylation at Ser 402 inhibits SSH1-phosphatase activity. (A) Mass spectra of the SSH1 phosphopeptide SASpTVIAYAMK were measured with a mass deviation of −0.1 p.p.m. A good coverage of the whole sequence by fragment (b and y) ions is shown. The fragment ions y9 and y10 with corresponding neutral losses of phosphoric acid indicate that Ser 402 is a modification site. (B) Purified GST–SSH1 (1–461) proteins were subjected to in vitro phosphatase assay using cofilin-Flag as a substrate and filamentous actin as cofactor. Proteins were detected with specific antibodies as indicated. One of three independent experiments is shown. (C,D) HEK293T cells were transfected with plasmids encoding the indicated (C) GST-tagged SSH1 (1–461) and (D) Flag-tagged SSH1 full-length proteins. Cells were lysed and SSH1 and phospho-cofilin (P-cofilin) were analysed using specific antibodies. Detection of tubulin verified equal protein levels. One of three independent experiments is shown (B–D). GST, glutathione S-transferase; HEK, human embryonic kidney; SSH1, slingshot1; WT, wild type.
To determine the mechanism by which Ser 402 phosphorylation inhibits phosphatase activity, we analysed the capacity of SSH1 to interact with F-actin. Try 458, next to the phosphatase domain, is essential to enhance SSH1 activity on F-actin binding (Nishita et al, 2005). However, F-actin co-sedimentation assays demonstrate that the interaction between F-actin and full-length SSH1 is not affected by Ser 402 phosphorylation (Fig 2A). Ser 402 is located within the phosphatase domain, which is important for binding of P-cofilin (Kurita et al, 2008). It is possible that binding of the phosphate group of P-cofilin to the active site in the catalytic domain is essential to form a stable enzyme–substrate complex. Increase of local negative charge next to the active site could thus negatively affect P-cofilin binding. Notably, replacement of Cys 393 by serine (CS) creates a phosphatase-inactive SSH1 that forms a stable complex with P-cofilin (Kurita et al, 2008). We thus constructed CS mutants of glutathione S-transferase (GST)–SSH1 (1–461) wild type, S402A, S402D and S402E and analysed their ability to bind to P-cofilin (Fig 2B). P-cofilin coprecipitated with wild type and S402A, but not with the S402D and E mutant SSH1 proteins, indicating that Ser 402 phosphorylation blocks substrate binding. SSH1 has been shown to localize to the leading edge and to vinculin-positive focal adhesions (Soosairajah et al, 2005; Marshall et al, 2009). In HeLa cells, wild-type and S402 mutant proteins colocalized with F-actin at lamellipodia in RacQ61L-expressing cells independently of Ser 402 phosphorylation (data not shown). Instead, only 25% of focal adhesions were positive for green fluorescent protein (GFP)-tagged SSH1 S402D and E proteins, whereas almost 80% of focal adhesions colocalized with SSH1 wild-type and S402A proteins. The average number of focal adhesions per cell, however, was independent of SSH1 mutant expression (data not shown).
Figure 2.
Phosphorylation at Ser 402 blocks binding of SSH1 to P-cofilin. (A) F-actin cosedimentation assay using lysates from HEK293T cells transiently transfected with the indicated Flag-tagged SSH1 proteins. Samples without F-actin were included as controls. SSH1 proteins and actin were detected by western blotting with Flag- and actin-specific antibodies, respectively. (B) HEK293T cells expressing the indicated GST-tagged SSH1 proteins were lysed, incubated with recombinant Flag-cofilin, and precipitated with glutathione sepharose 4B. Proteins were detected with specific antibodies as indicated. Asterisk indicates C393S mutation. (C) HeLa cells expressing the indicated GFP-tagged SSH1 proteins (shown in green) were serum-starved, fixed and stained for vinculin (red) and F-actin (grey). Scale bar, 20 μm. The diagram shows the mean percentage (±s.e.m.) of focal adhesions positive for GFP-SSH1 per cell. A total of 36 cells were analysed per sample. GFP, green fluorescent protein; HEK, human embryonic kidney; P, pellet fraction; S, supernatant; SSH, slingshot; WT, wild type.
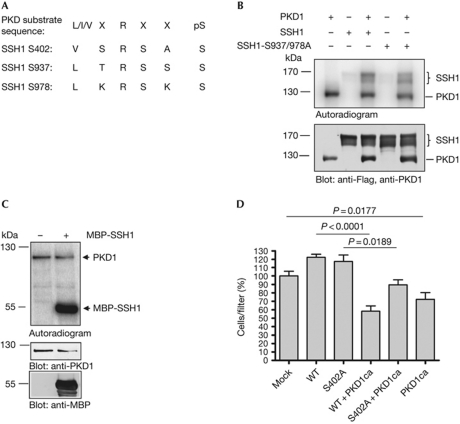
Ser 402 is in the consensus motif of PKD (L/I/VxRxxS/T; Fig 3A; Hutti et al, 2004; Doppler et al, 2005), a kinase that phosphorylates SSH1 on Ser 937 and 978 (Peterburs et al, 2009). To investigate whether PKD phosphorylates further sites within SSH1, we performed an in vitro kinase assay using purified PKD1 together with purified Flag-SSH1 wild type and the S937/978A protein. Wild-type SSH1 and the S937/978A mutant were phosphorylated by PKD in a similar manner, suggesting that more PKD sites are present (Fig 3B). Indeed, a truncated SSH1 protein, comprising the amino acids 327–474 and thus lacking the C-terminal PKD sites, is a substrate for PKD1 in vitro (Fig 3C), suggesting that PKD1 is upstream from Ser 402.
Figure 3.
Phosphorylation of Ser402 occurs downstream from protein kinase D. (A) Alignment of the PKD consensus phosphorylation motif and the PKD motifs in SSH1. (B) Flag-SSH1 WT and S937/978A mutant fusion proteins were incubated with [32P]-γ-ATP in the absence (−) and presence (+) of purified PKD1. Incorporation of radioactive phosphate was analysed using a PhosphoImager (top), followed by immunoblotting with Flag- and PKD1-specific antibodies to verify equal loading of the SSH1 and PKD1 proteins, respectively. (C) In vitro kinase assay with purified PKD1 and recombinant MBP-SSH1 protein (amino acids 327–474) as described in (B). Immunoblotting was performed with MBP- and PKD1-specific antibodies. (D) HeLa cells were transfected with empty vector, SSH1 WT, and SSH1 S402A alone or together with active PKD1. Transwell chemotaxis assays were performed for 4 h. Significance was calculated by an unpaired two-tailed Student's t-test. P<0.05 was accepted as statistically significant. The mean of three independent experiments±s.e.m is shown. ca, constitutive active; GST, glutathione S-transferase; MBP, maltose binding protein; PKD, protein kinase D; SSH1, slingshot 1; WT, wild type.
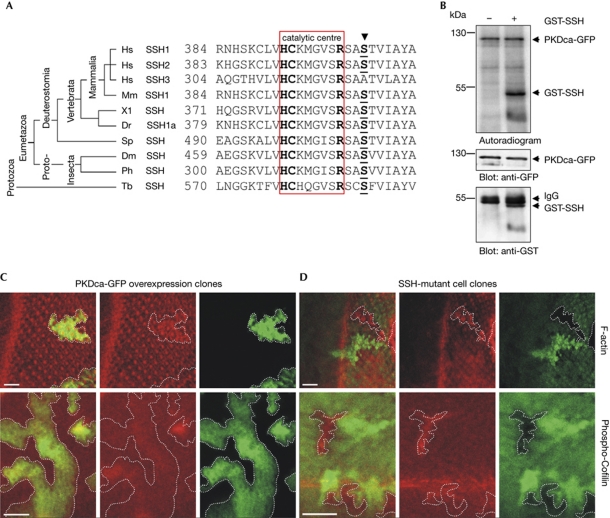
PKD inhibits directed cell migration through phosphorylation of SSH1 at Ser 937 and 978 (Eiseler et al, 2009). The co-expression of constitutive active (ca) PKD1 with SSH1 wild type significantly decreased cell migration in comparison to SSH1 wild type alone. However, this was partly rescued by the SSH1 S402A mutant (Fig 3D), indicating that PKD-controlled phosphorylation of this site substantially contributes to the regulation of SSH1-dependent directed cell migration. The sequences surrounding Ser 402 are highly conserved across species, suggesting that the mechanism of negative regulation through phosphorylation at Ser 402 could be universal (Fig 4A). We used Drosophila melanogaster as a model organism for further in vivo studies. Here, the C-terminus of SSH lacks the two PKD phospho-sites, whereas the previously identified phosphorylation site at Ser 477 (Ser 402 in mammals) is conserved (Fig 4A; Ohta et al, 2003). Similarly to the human PKD1ca protein, Drosophila PKDca-GFP (Maier et al, 2007) displayed constitutive activity (supplementary Fig S1 online). In in vitro kinase assays, Drosophila PKDca was able to directly phosphorylate a truncated Drosophila SSH–GST-fusion protein comprising the amino acids 461–671 (Fig 4B). These results show that Drosophila SSH is a PKD substrate in vitro. Loss of Drosophila SSH results in an accumulation of both P-cofilin and F-actin (Niwa et al, 2002). If PKD is a negative regulator of SSH in Drosophila, we also expect an accumulation of P-cofilin and of F-actin, when active PKD is expressed in Drosophila tissues. We thus induced cell clones overexpressing PKDca-GFP that are surrounded by wild-type tissue in developing wing and eye imaginal anlagen. There was a significant accumulation of F-actin and P-cofilin that was detectable in cells overexpressing the active PKD protein, compared with the neighbouring wild-type sister cells (Fig 4C). Both F-actin and P-cofilin were also enriched in cells that lack the SSH protein (Fig 4D). These data indicate that PKD negatively regulates SSH activity in Drosophila tissue, probably by phosphorylation of Ser 477.
Figure 4.
Drosophila PKDca induces the accumulation of F-actin and P-cofilin. (A) Phylogenetic comparison of SSH proteins. The catalytic centre is highlighted (HCxxGxxR; Keyse, 1995). The arrow indicates Ser 402 in human SSH1; homologous sites are shown in bold and are underlined. Note the high conservation of this region in the annotated sequences of vertebrates and invertebrates. Human SSH3 has an alanine instead of the serine at this position, as do other SSH3 orthologues. Only one isoform is shown for the vertebrates; however, orthologues to SSH1 and SSH2 have sequences identical to the ones shown here. The alignment shows the human SSH proteins (Hs SSH1, Hs SSH2, Hs SSH3), SSH from mouse (Mm), frog (Xenopus laevis, Xl), zebra fish (Danio rerio, Dr), sea urchin (Strongylocentrotus purpuratus, Sp), Drosophila melanogaster (Dm), body louse (Pediculus humanus capitis, Ph) and from a protozoan parasitic trypanosome (Trypanosoma brucei gambiense, Tb). (B) In vitro kinase assay using GST-tagged fly SSH protein (amino acids 461–671) as a substrate for constitutive active Drosophila PKDca-GFP. The blots show the presence of the respective proteins. (C) PKDca was overexpressed in cell clones marked by GFP (green, white outlines). (D) Cell clones mutant for Ssh are marked by the absence of GFP (white outlines); the wild-type twin clones are recognized by enhanced GFP staining. P-cofilin was detected with a specific antibody and filamentous actin was visualized with Rhodamine-phalloidin (red). Overlays are shown in the first panel each. Scale bar, 20 μm. Enrichment of P-cofilin on PKDca induction is highly significant (n=16, P<0.0001, Student's t-test). GFP, green fluorescent protein; GST, glutathione S-transferase; IgG, immunoglobulin G; PKD, protein kinase D; SSH, slingshot.
Ser 402 is not present in SSH3, suggesting that this phosphatase is regulated in a different manner. Accordingly, SSH3 interacts with 14-3-3 proteins through a different domain than SSH1 and SSH2, which remains to be identified (Kligys et al, 2007). It was reported that PAK4 directly phosphorylates SSH1 in the N-terminal region in vitro (Soosairajah et al, 2005). The phosphorylation negatively regulates phosphatase activity towards P-cofilin and LIMK1. The authors therefore proposed that PAK4 regulates SSH1 by phosphorylation on sites other than Ser 937 and 978. On the basis of these data, one could speculate that phosphorylation at Ser 402 is mediated by PAK4; however, the consensus phosphorylation motif of PAK4 does not support a valine in the −5 position (Rennefahrt et al, 2007). Accordingly, by mass spectrometry, no increase of Ser 402 phosphorylation was detected when SSH1 was incubated with active PAK4 (data not shown). Instead, our data suggest that PKD is the responsible kinase, because it is upstream from Drosophila SSH, which lacks the two C-terminal PKD target sites. We therefore conclude that SSH activity is regulated downstream of PKD by phosphorylation of Ser 477 and 402 in Drosophila and humans, respectively. Mammalian SSH1 seems to require further regulatory mechanisms. PKD-regulated 14-3-3 binding to the phosphorylated Ser 937 and 978 reduces the interaction of SSH1 with F-actin and sequesters SSH1 in the cytosol, but has no direct effect on the ability of SSH1 to dephosphorylate cofilin (Soosairajah et al, 2005). This is corroborated by studies showing that a C-terminal-truncated SSH1 protein has full phosphatase activity in an F-actin-dependent manner (Yamamoto et al, 2006; Kurita et al, 2008). We thus conclude that PKD determines local and cellular SSH1-phosphatase activity by phosphorylation of Ser 937/978 and 402, respectively, thereby fine-tuning cofilin activation. SSH1 has been demonstrated to dephosphorylate and thereby inactivate LIMK1 (Soosairajah et al, 2005). One could therefore speculate that 14-3-3 protein-bound SSH1 is able to dephosphorylate LIMK1 in the cytosol, whereas local cofilin activation at the lamellipodium is blocked. This is in line with a model showing that active cofilin is exclusively present in the actin compartment of migrating cells (Oser & Condeelis, 2009). We further show that phosphorylation at Ser 402 impedes binding of P-cofilin and focal adhesion localization. On the basis of our data, we suggest that SSH1 is recruited to focal adhesions by interaction with P-cofilin. This hypothesis is strengthened by the fact that, similarly to the SSH1 wild-type protein, the C393S mutant is enriched in focal adhesions (Fig 2C). Interestingly, expression of active SSH1 increases focal adhesion disassembly, a process that is central in directed cell migration (Marshall et al, 2009). Whether phosphorylation of SSH1 at Ser 402 controls directed cell migration by regulating focal adhesion turnover will be addressed in the future. Taken together, the results of our study identify a universal mechanism by which PKD regulates SSH1 activity and, thus, cofilin activation.
Methods
Reagents, plasmids and antibodies. See supplementary information online.
Cell culture. HEK293T and HeLa cells were maintained in RPMI supplemented with 10% FCS and transfected with expression plasmids using TransIT 293 and TransIT Monster (both Mirus, USA), respectively.
Phosphatase and kinase assay. Phosphatase assays were performed as described previously (Niwa et al, 2002; Yamamoto et al, 2006). Details can be found in the supplementary information online. In vitro kinase assays were done as described previously (Hausser et al, 2002; Peterburs et al, 2009).
F-actin co-sedimentation and P-cofilin binding assay. F-actin co-sedimentation assays were performed as described previously (Kurita et al, 2008). Details can be found in the supplementary information online. For the P-cofilin binding assay, HEK293T cells expressing GST–SSH1 (1–461) proteins were lysed, and purified Flag-cofilin was added. GST-tagged proteins were precipitated using glutathione sepharose 4B beads, washed three times and sampled for SDS–PAGE. GST-tagged proteins and bound P-cofilin were analysed by immunoblotting using GST- and P-cofilin-specific antibodies, respectively.
Transwell chemotaxis assay. Cell migration assays were performed as described previously (Peterburs et al, 2009) with the following modifications: HeLa cells were seeded at a density of 20,000 cells per insert. Mean migration of control cells was set as 100%.
Immunofluorescence and microscopy. Immunofluorescence was performed as described previously (Peterburs et al, 2009). Details are in the supplementary information online.
Nano liquid chromatography–mass spectrometry analysis of purified Flag-SSH1, fly stocks and clonal analysis. See supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank I. Wech for technical help, J. Schmid for generating GST–SSH fusion protein, and K. Basler, O. Bernard, the Bloomington Stock Centre, the Developmental Studies Hybridoma Bank (DSHB) and Drosophila Genomics Resource Center (DGRC) for flies, DNA and antibodies. This work was supported by grants to D.M. (DFG MA 1328/8-1) and to A.H. (DFG HA 3557/4-1 and WIN-Kolleg, HAW).
Footnotes
The authors declare that they have no conflict of interest.
References
- Desmarais V, Ghosh M, Eddy R, Condeelis J (2005) Cofilin takes the lead. J Cell Sci 118: 19–26 [DOI] [PubMed] [Google Scholar]
- Doppler H, Storz P, Li J, Comb MJ, Toker A (2005) A phosphorylation state-specific antibody recognizes Hsp27, a novel substrate of protein kinase D. J Biol Chem 280: 15013–15019 [DOI] [PubMed] [Google Scholar]
- Eiseler T, Doppler H, Yan IK, Kitatani K, Mizuno K, Storz P (2009) Protein kinase D1 regulates cofilin-mediated F-actin reorganization and cell motility through slingshot. Nat Cell Biol 11: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausser A, Link G, Bamberg L, Burzlaff A, Lutz S, Pfizenmaier K, Johannes FJ (2002) Structural requirements for localization and activation of protein kinase C mu (PKC mu) at the Golgi compartment. J Cell Biol 156: 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, Cantley LC, Turk BE (2004) A rapid method for determining protein kinase phosphorylation specificity. Nat Methods 1: 27–29 [DOI] [PubMed] [Google Scholar]
- Keyse SM (1995) An emerging family of dual specificity MAP kinase phosphatases. Biochim Biophys Acta 1265: 152–160 [DOI] [PubMed] [Google Scholar]
- Kligys K, Claiborne JN, DeBiase PJ, Hopkinson SB, Wu Y, Mizuno K, Jones JC (2007) The slingshot family of phosphatases mediates Rac1 regulation of cofilin phosphorylation, laminin-332 organization, and motility behavior of keratinocytes. J Biol Chem 282: 32520–32528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita S, Watanabe Y, Gunji E, Ohashi K, Mizuno K (2008) Molecular dissection of the mechanisms of substrate recognition and F-actin-mediated activation of cofilin-phosphatase Slingshot-1. J Biol Chem 283: 32542–32552 [DOI] [PubMed] [Google Scholar]
- Maier D, Nagel AC, Gloc H, Hausser A, Kugler SJ, Wech I, Preiss A (2007) Protein kinase D regulates several aspects of development in Drosophila melanogaster. BMC Dev Biol 7: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall TW, Aloor HL, Bear JE (2009) Coronin 2A regulates a subset of focal-adhesion-turnover events through the cofilin pathway. J Cell Sci 122: 3061–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata-Ohashi K et al. (2004) A pathway of neuregulin-induced activation of cofilin-phosphatase Slingshot and cofilin in lamellipodia. J Cell Biol 165: 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishita M, Tomizawa C, Yamamoto M, Horita Y, Ohashi K, Mizuno K (2005) Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration. J Cell Biol 171: 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T (2002) Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell 108: 233–246 [DOI] [PubMed] [Google Scholar]
- Ohta Y, Kousaka K, Nagata-Ohashi K, Ohashi K, Muramoto A, Shima Y, Niwa R, Uemura T, Mizuno K (2003) Differential activities, subcellular distribution and tissue expression patterns of three members of Slingshot family phosphatases that dephosphorylate cofilin. Genes Cells 8: 811–824 [DOI] [PubMed] [Google Scholar]
- Oser M, Condeelis J (2009) The cofilin activity cycle in lamellipodia and invadopodia. J Cell Biochem 108: 1252–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterburs P, Heering J, Link G, Pfizenmaier K, Olayioye MA, Hausser A (2009) Protein kinase D regulates cell migration by direct phosphorylation of the cofilin phosphatase slingshot 1 like. Cancer Res 69: 5634–5638 [DOI] [PubMed] [Google Scholar]
- Rennefahrt UE, Deacon SW, Parker SA, Devarajan K, Beeser A, Chernoff J, Knapp S, Turk BE, Peterson JR (2007) Specificity profiling of Pak kinases allows identification of novel phosphorylation sites. J Biol Chem 282: 15667–15678 [DOI] [PubMed] [Google Scholar]
- Soosairajah J, Maiti S, Wiggan O, Sarmiere P, Moussi N, Sarcevic B, Sampath R, Bamburg JR, Bernard O (2005) Interplay between components of a novel LIM kinase-slingshot phosphatase complex regulates cofilin. EMBO J 24: 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Nagata-Ohashi K, Ohta Y, Ohashi K, Mizuno K (2006) Identification of multiple actin-binding sites in cofilin-phosphatase Slingshot-1L. FEBS Lett 580: 1789–1794 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.