Abstract
Mitochondria form intricate networks through fission and fusion events. Here, we identify mitochondrial dynamics proteins of 49 and 51 kDa (MiD49 and MiD51, respectively) anchored in the mitochondrial outer membrane. MiD49/51 form foci and rings around mitochondria similar to the fission mediator dynamin-related protein 1 (Drp1). MiD49/51 directly recruit Drp1 to the mitochondrial surface, whereas their knockdown reduces Drp1 association, leading to unopposed fusion. Overexpression of MiD49/51 seems to sequester Drp1 from functioning at mitochondria and cause fused tubules to associate with actin. Thus, MiD49/51 are new mediators of mitochondrial division affecting Drp1 action at mitochondria.
Keywords: fission, fusion, Drp1, mitochondria, morphology
Introduction
Mitochondria are dynamic organelles that undergo fission and fusion events and are distributed along cytoskeletal elements (Frazier et al, 2006; Liesa et al, 2009; Westermann, 2010). As mitochondria cannot be created de novo, an active fission apparatus is required to promote transmission of mitochondria to dividing cells. Division also takes place in cells undergoing differentiation and growth, and has a role in removing damaged and aging organelles through mitophagy (Twig et al, 2008; Westermann, 2010). Two fission mediators conserved between yeast and mammals are the mainly cytosolic GTPase dynamin-related protein 1 (Drp1; Dnm1 in yeast) and the mitochondrial outer-membrane protein Fis1 (Otsuga et al, 1998; Smirnova et al, 1998; Mozdy et al, 2000; Yoon et al, 2003). Overexpression of Fis1 accelerates mitochondrial division, whereas its inactivation blocks fission, resulting in fused and elongated mitochondria (Yoon et al, 2003; Stojanovski et al, 2004). Drp1, and Dnm1, polymerize into spirals and show a mitochondrial population of foci or rings at constriction sites (Bleazard et al, 1999; Labrousse et al, 1999; Smirnova et al, 2001). In yeast, Dnm1 is recruited to mitochondria by Fis1 and the effector proteins Mdv1 and Caf4 to drive mitochondrial fission (Mozdy et al, 2000; Tieu & Nunnari, 2000; Griffin et al, 2005; Koirala et al, 2010). However, in mammals, Mdv1 and Caf4 homologues are absent and the role of Fis1 in fission has recently been disputed (Otera et al, 2010). Other proteins have been implicated in mitochondrial fission, including endophilin BI (Karbowski et al, 2004), MARCH5 (Karbowski et al, 2007) and Mff (Gandre-Babbe & van der Bliek, 2008; Otera et al, 2010). Nevertheless, understanding of the precise mechanisms that coordinate the complex process of mitochondrial fission and fusion remains limited. Further understanding of the machinery involved in mitochondrial morphogenesis, including the regulation of Drp1 function, is therefore required.
Results And Discussion
MiD49/51 alter mitochondrial morphology
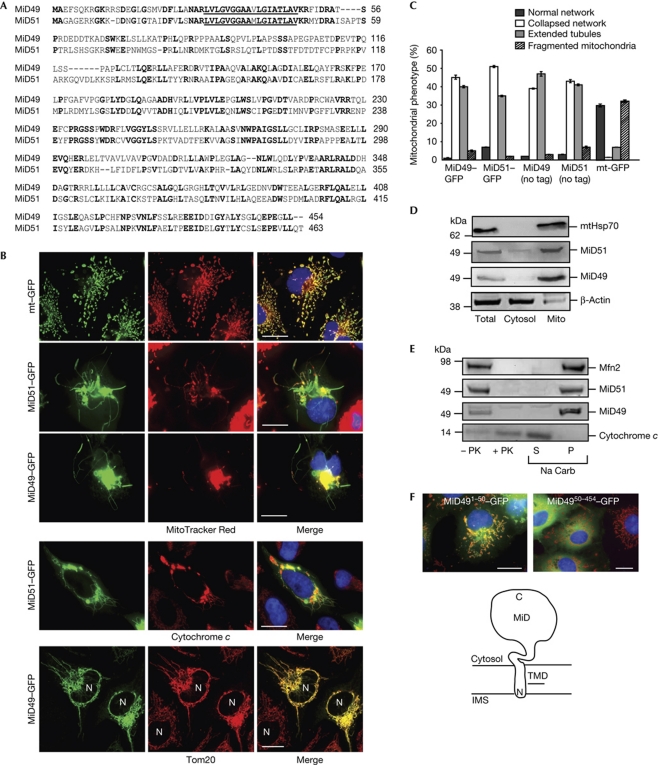
As part of a random cellular localization screen of uncharacterized human proteins (Simpson et al, 2000), it was found that expression of SMCR7L caused unique changes in mitochondrial distribution and resulted in the appearance of fused mitochondrial tubules. The SMCR7L protein shares 45% sequence identity with the human Smith–Magenis syndrome chromosome region candidate gene 7 protein (SMCR7; Fig 1A). SMCR7 is not responsible for Smith–Magenis syndrome (Slager et al, 2003), and here we rename the SMCR7 and SMCR7L gene products mitochondrial dynamics proteins of 49 kDa (MiD49) and 51 kDa (MiD51), respectively. When MiD49/51 were expressed as carboxy-terminal green fluorescent protein (GFP) fusions, they both localized to mitochondria. The GFP-fluorescent tubules counterstain with MitoTracker Red, or mitochondrial proteins cytochrome c and Tom20, confirming their mitochondrial origin (Fig 1B). Moreover, their expression caused the formation of long mitochondrial tubules projecting out of a perinuclear collapsed network (Fig 1C). These mitochondrial tubules appeared to be morphologically distinct from those reported following unregulated fusion (Karbowski et al, 2004; Stojanovski et al, 2004).
Figure 1.
MiD49/51 are mitochondrial outer-membrane proteins that affect mitochondrial morphology. (A) Sequence alignment with conserved residues shown in bold and predicted transmembrane domains underlined. (B) COS7 cells expressing GFP fusion constructs and stained with MitoTracker Red (red) and Hoechst (blue; upper panels). HeLa cells expressing MiD–GFP fusion constructs and immunostained for cytochrome c or Tom20 (lower panels). (C) COS7 cells expressing different constructs were blind-counted for mitochondrial morphology (mean±s.e.m., n=3, 100 cells counted per experiment). (D) Fractionated COS7 cells were analysed by immunoblotting using antibodies as indicated. (E) COS7 mitochondria were subjected to proteinase K (PK) or sodium carbonate (Na Carb) extraction, followed by western blot analysis using antibodies as indicated. (F) MiD491−50–GFP and MiD4950−454–GFP were expressed in COS7 cells and stained with MitoTracker Red and Hoechst. Scale bars, 20 μm. GFP, green fluorescent protein; IMS, intermembrane space; MiD49/51, mitochondrial dynamics proteins of 49 and 51 kDa; Mito, mitochondria; P, pellet; S, supernatant; TMD, transmembrane domain.
Mitochondrial localization of endogenous MiD proteins was confirmed by western blot analysis (Fig 1D). Both MiD49/51 were susceptible to protease treatment of mitochondria, such as the integral outer-membrane protein mitofusin 2 (Mfn2), whereas cytochrome c was protected (Fig 1E). MiD49/51 were also found in the pellet fraction following alkaline extraction of peripheral membrane proteins, whereas cytochrome c was in the supernatant fraction, as expected. On the basis of these results, together with sequence information (Fig 1A), we conclude that MiD49/51 are amino-terminally anchored in the mitochondrial outer membrane with C-terminal cytosolic domains. Indeed, residues 1–50 of MiD49 directed GFP to the mitochondrial outer membrane, whereas loss of the predicted transmembrane domain (MiD4950−454–GFP) was cytosolic (Fig 1F). These results were also obtained for MiD51 (data not shown). Expression of MiD4950−454–GFP did not alter mitochondrial morphology, indicating that the cytosolic domain must be at the mitochondrial surface in order for the MiD proteins to be active.
Mitochondrial extensions are attached to F-actin
Mammalian mitochondria are usually transported along microtubules, although they are also known to interact with microfilaments and intermediate filaments (Anesti & Scorrano, 2006). Confocal time-lapse imaging showed mitochondria in cells expressing MiD51–GFP to be dynamic, with the tubules showing considerable movement and a lack of apparent fission events (Fig 2A; supplementary Movie S1 online). These mitochondrial tubules associated with F-actin (Fig 2B) and disruption of actin filaments with cytochalasin D led to retraction of the extended mitochondrial tubules and collapsed networks (Fig 2C; supplementary Movie S2 online). By contrast, disruption of microtubules with nocodazole did not reduce the presence of fused mitochondrial tubules (Fig 2D). It is not clear whether MiD49/51 influence actin polymerization at the surface of mitochondria or facilitate myosin-directed transportation events. Analysis of Arp2/3 complex localization showed no obvious changes (data not shown), suggesting that actin nucleation is not markedly affected.
Figure 2.
Cells expressing MiD51–GFP contain motile mitochondrial tubules that colocalize with F-actin. (A) Confocal time-lapse imaging of COS7 cells expressing MiD51–GFP (supplementary Movie S1 online). Representative images are shown. Solid line denotes cell boundary, dotted line denotes nucleus (N). (B) Confocal images of COS7 cells expressing MiD51– or MiD49–GFP and stained for F-actin. Insets are magnified in the lower panels. (C) Time-lapse images of a COS7 cell coexpressing MiD51–GFP following addition of 10-μM cytochalasin D at t=0. Arrows indicate mitochondrial tubules that retract following cytochalasin D treatment (supplementary Movie S2 online). Solid line denotes cell boundary. (D) Mitochondrial morphology was blind-counted in cells expressing MiD51–GFP following various treatments (mean±s.e.m., n=3, 100 cells counted per experiment). Scale bars, 20 μm. GFP, green fluorescent protein; MiD49/51, mitochondrial dynamics proteins of 49 and 51 kDa.
Although overexpression of the MiD proteins induces mitochondrial attachment to actin, we note that such attachments might not be physiological. However, we cannot exclude the idea that the MiD proteins are involved in making transient attachments between mitochondria and actin. Indeed, De Vos et al (2005) reported that actin is important for recruiting Drp1 to the mitochondrial surface to facilitate fission in mammalian cells. It is therefore plausible that the MiD proteins might be involved in such an event under physiological conditions.
MiD49/MiD51 recruit Drp1 to the mitochondrial surface
The appearance of fused mitochondrial tubules following overexpression of MiD49/51 indicates that mitochondrial fission is blocked and/or fusion is enhanced. However, at low-level MiD51–GFP expression (Fig 3A; supplementary Fig S1 online), mitochondrial morphology was relatively normal, whereas MiD51–GFP was found at discrete foci on the mitochondrial surface (Fig 3A). Similar results were obtained for MiD49 (data not shown). The presence of endogenous foci formed by MiD49/51 could not be analysed because of cross-reacting immune sera. Nevertheless, this fluorescence pattern differed from the even fluorescence seen in cells expressing GFP fused to mitochondrial outer-membrane proteins Tom7 (Translocase of outer membrane 7kDa) or Miro1 (Mitochondrial Rho GTPase 1; Fig 3A). Confocal analysis of the MiD51–GFP foci showed the presence of rings around potential mitochondrial constriction sites (Fig 3B; supplementary Movie S3 online), similar to those reported for the mitochondrial-fission mediator Drp1 (Smirnova et al, 2001; Legesse-Miller et al, 2003). Indeed, it was found that in cells showing both high and low levels of MiD49–GFP fluorescence, most Drp1 was found at the mitochondrial surface (Fig 3C). Pearson's correlational analysis of cells expressing MiD49/51–GFP showed a clear increase in Drp1 colocalization with mitochondria, in contrast to cells expressing GFP–Miro1 or GFP–Tom7 (Fig 3C,D). The MiD–GFP foci were not dependent on the presence of Drp1 as they were still present following Drp1 knockdown (Fig 3E; supplementary Movie S4 online). Our results indicate that MiD49/51 have a direct role in the recruitment or stabilization of Drp1 at mitochondria.
Figure 3.
MiD49/51 form foci at mitochondria and alter Drp1 localization. (A) COS7 cells expressing GFP constructs were stained with MitoTracker Red (red) and Hoechst (blue). Insets are magnified in the right panels. All images were obtained at the same exposure. Scale bar, 20 μm. (B) Reconstruction of mitochondria containing MiD49–GFP and stained with MitoTracker Red. Arrows indicate a MiD49–GFP ring at a potential mitochondrial constriction site (supplementary Movie S3 online). Scale bar, 1 μm. (C) COS7 cells expressing MiD49–GFP (low and high fluorescence) or GFP–Miro1 were immunostained for Drp1 (red) and stained with Hoechst. (D) Linescans of cell regions were analysed for Drp1 and GFP fluorescence. Relative fluorescence intensities were measured (n>45), Drp1 colocalization with mitochondria was analysed and the degree of association is shown as Pearson's correlation units (r, bottom panel; mean±s.e.m., n=3). (E) HeLa cells depleted of Drp1 were transfected with a construct expressing MiD51–GFP and stained with MitoTracker Red. MiD51–GFP foci are enlarged in the inset (supplementary Movie S4 online). Scale bars, 20 μm. Western blot analysis confirmed knockdown of Drp1, mitochondrial matrix B17.2L was used as a control. Drp1, dynamin-related protein 1; GFP, green fluorescent protein; MiD49/51, mitochondrial dynamics proteins of 49 and 51 kDa; Miro1, Mitochondrial Rho GTPase 1; Tom7, Translocase of outer membrane 7kDa.
Next, we assessed whether Drp1 recruitment to mitochondria could be saturated. When expressed alone or with the outer-membrane protein human Milt1 (Koutsopoulos et al, 2010), GFP–Drp1 was predominantly cytosolic. However, in most cells overexpressing MiD49 or MiD51, GFP–Drp1 was enriched at mitochondria (Fig 4A). Furthermore, we generated mouse embryonic fibroblasts expressing MiD51 under 4-hydroxytamoxifen-inducible conditions (Dunning et al, 2007). Induction of MiD51 led to the appearance of swollen and fused mitochondrial extensions (Fig 4B) and recruitment of GFP–Drp1 to mitochondria (Fig 4C). A dominant-negative GTPase mutant GFP–Drp1K38A was also recruited to mitochondria (Fig 4C; Yoon et al, 2001). Similarly, GFP–Drp1S600D, which shows decreased GTPase activity and mitochondrial recruitment (Chang & Blackstone, 2007), was also found at mitochondria following MiD51 expression (Fig 4C). However, expression of Drp1 dominant-negative mutants containing the middle domain mutations A395D or G363D—which have defects in higher-ordered assembly (Chang et al, 2010)—were not detected at mitochondria (Fig 4C). Middle domain mutants of Dnm1/Drp1 maintain dimer/trimer formation, but multimeric scission complex formation is impaired (Bhar et al, 2006; Chang et al, 2010). Drp1 dimer/trimer formation was evident after overexpression and knockdown of MiD49/51, as seen after chemical crosslinking (data not shown). These data, in conjunction with the Drp1 mutant analysis, indicate that for Drp1 to be retained at mitochondria following MiD49/51 expression, Drp1 must have the ability to assemble into a higher-ordered species. Live-cell imaging after the addition of 4-hydroxytamoxifen showed that GFP–Drp1 was recruited to mitochondria, and fission events in these cells were observed (supplementary Movie S5 online), whereas uninduced and wild-type cells did not recruit GFP–Drp1 (supplementary Movies S5 and S6 online). As cells remained viable after increased fission events, this indicates that Drp1 was functional and apoptosis was not induced. Immunoblot analysis also revealed that Drp1 was enriched in mitochondria isolated from cells following MiD51 expression (Fig 4D). Complete enrichment of Drp1 with mitochondria was not seen, presumably because of mechanical treatment when isolating mitochondria. To determine a direct interaction between MiD49 and Drp1, a yeast two-hybrid assay was carried out using a truncated form of MiD49 lacking the transmembrane domain and a predicted unstructured region (residues 1–120). By using high-stringency selection, a direct interaction between MiD49ΔN120 and Drp1 was confirmed (Fig 4E). Furthermore, coimmunoprecipitation analysis of MiD49–GFP following crosslinking showed a direct association with Drp1 (Fig 4F). The control Tom40 was not coprecipitated, and GFP–Miro1 and GFP–Fis1 did not coprecipitate with Drp1 (Fig 4F).
Figure 4.
MiD49/51 recruit GFP–Drp1 to the mitochondrial surface. (A) COS7 cells expressing GFP–Drp1 alone or with MiD49, MiD51 or huMilt1 were blind-counted for GFP–Drp1 localization (mean±s.e.m., n=3, 100 cells counted per experiment). (B) Mouse embryonic fibroblasts (MEFs) before (−4HT) and 24 h after (+4HT) induction of MiD51 were stained with MitoTracker Red (red) and visualized by epifluorescence. Western blot analysis confirmed MiD51 induction, with mitochondrial matrix B17.2L as a control. Scale bars, 20 μm. (C) MEFs transfected with GFP constructs were visualized by epifluorescence before (−4HT) and 24 h after (+4HT) MiD51 induction. (D) Western blot analysis of whole-cell (W), mitochondria (M) and cytosolic (C) fractions from MEFs before (−4HT) and after (+4HT) MiD51 expression. Cytosolic Hsc70 and mitochondrial matrix B17.2L acted as controls. (E) By using a yeast two-hybrid assay, a direct interaction between truncated MiD49ΔN120 and Drp1 was observed, similar to the positive control p53 and large T-antigen. MiD49ΔN120 did not interact with the soluble domain of hFis1 or the large T-antigen, similar to the negative control Lamin and T-antigen. (F) Coimmunoprecipitation of endogenous Drp1 with MiD49–GFP following 20-μM bis(maleimido)hexane crosslinking of HeLa cells. Western blot analysis was conducted with antibodies as indicated. Arrow indicates crosslinked species. Drp1, dynamin-related protein 1; GFP, green fluorescent protein; MiD49/51, mitochondrial dynamics proteins of 49 and 51 kDa; Miro1, Mitochondrial Rho GTPase 1; WB, western blot; 4HT, 4-hydroxytamoxifen.
Loss of MiD49/51 alters Drp1 recruitment
To determine the role of endogenous MiD49/51 in the regulation of mitochondrial morphology and distribution, we used RNA interference. After knockdown of both MiD proteins (approximately 80% reduction), the levels of other morphology and control proteins were similar, except for Drp1, which was reduced at mitochondria (Fig 5A). Knockdown of both MiD49 and 51 resulted in an irregular distribution of the network and fused mitochondria (Fig 5B,C). Changes in mitochondrial morphology were only clearly observed following knockdown of both MiD proteins, suggesting that they are functionally redundant. Drp1 association with mitochondria was significantly reduced following knockdown of MiD49/51, in comparison to the control Hsc70 (Fig 5D; supplementary Fig S2 online).
Figure 5.
Effect of MiD49/51 RNA interference on mitochondrial morphology and Drp1 distribution. (A) COS7 cells were transfected with either scrambled or MiD-specific RNAi constructs (separately or together). Mitochondrial fractions were isolated 48 h post-transfection and analysed by immunoblotting. (B) Confocal microscopy of COS7 cells co-transfected with mt-Dendra2 and MiD49/MiD51 RNAi constructs. Epifluorescent microscopy of HeLa cells co-transfected with both MiD RNAi constructs then immunostained for cytochrome c. Inset is magnified in the lower right panel. (C) COS7 mitochondrial phenotypes were blind-counted and scored following knockdown (mean±s.e.m., n=3, 100 cells counted per experiment). (D) Confocal imaging of COS7 cells co-transfected with either scrambled or MiD-specific RNAi constructs along with mt-Dendra2 to visualize mitochondria. Cells were fixed and immunostained for Drp1 or Hsc70. Relative fluorescence intensities of 30-μm linescans were measured (n>37). Drp1 or Hsc70 colocalization with mitochondria was analysed and the degree of association was shown as Pearson's correlation units (r; mean±s.e.m., n=3). (E) HeLa cells transfected with scrambled, MiD49/MiD51 RNAi or Drp1 RNAi constructs were incubated for 60 min with either 20-μM CCCP or DMSO. Cells were fixed and probed for cytochrome c and then mitochondrial morphology was blind-counted (mean±s.e.m., n=3, 100 cells counted per experiment). Scale bars, 20 μm. CCCP, carbonyl cyanide m-chlorophenylhydrozone; DMSO, dimethylsulphoxide; Drp1, dynamin-related protein 1; GFP, green fluorescent protein; Mfn, mitofusin 2; MiD49/51, mitochondrial dynamics proteins of 49 and 51 kDa; mt, matrix; RNAi, RNA interference.
Addition of CCCP (carbonyl cyanide m-chlorophenylhydrozone)—a proton ionophore that uncouples the mitochondrial membrane potential—blocks fusion leading to unopposed Drp1-dependent mitochondrial fission (Legros et al, 2002). If MiD49/51 are involved in regulating Drp1 activity, it is expected that their knockdown would also block CCCP-induced mitochondrial fragmentation. As can be seen (Fig 5E), knockdown of MiD49 and 51 led to an approximate fourfold reduction in CCCP-induced fragmentation. Similar effects have also been observed following reduction of Mff levels (Gandre-Babbe & van der Bliek, 2008).
The process of mitochondrial fission is complex and requires several regulatory steps involving Drp1. This includes mitochondrial recruitment, assembly at the scission complex, division and Drp1 disassembly. At which step might the MiD proteins function? As MiD49/51 can form foci and rings around mitochondria, it is possible that they act as scaffolds for Drp1. Further evidence supporting this comes from the fact that MiD proteins were found at the foci and apparent constriction sites, independently of Drp1 expression. At higher expression levels, MiD–GFP fluorescence seems to be more uniformly distributed along the mitochondrial outer membrane. Following MiD51-induced expression, GFP–Drp1 also seems to be more uniformly distributed at the mitochondrial outer membrane. Under these conditions, we suggest that Drp1 still binds to mitochondria, but is unable to organize into active scission complexes, blocking fission and resulting in unbiased fusion. Following knockdown of both MiD49 and 51, we propose that Drp1 is unable to form higher-order complexes at mitochondria, resulting in unbiased fusion. Drp1 middle domain mutants were not recruited to mitochondria on induction of MiD51 expression, also suggesting that MiD49 and 51 function downstream from the initial recruitment of Drp1. Coimmunoprecipitation and yeast two-hybrid results confirmed a direct interaction between MiD49 and Drp1; however, we cannot exclude the idea that the MiD proteins might regulate other proteins such as Mff or Fis1, which also influence Drp1 action. In summary, MiD49/51 are new members of a growing family of proteins that regulate mitochondrial morphology in higher eukaryotes. The challenge for the future is to understand the way in which these proteins regulate and coordinate their activity.
Methods
Plasmids. Complementary DNA encoding human MiD49 (SMCR7; Accession number: Q96C03) and MiD51 (SMCR7 L; Accession number: Q9NQG6) were cloned into the pE-GFP-N1 (Clontech, CA, USA) vector. RNA interference oligonucleotides were cloned into the pSilencer 3.0-H1 (Ambion, TX, USA) vector. The GFP and untagged constructs and all mutants were generated by PCR.
Cell culture and imaging. Generation, selection and induction of stable mouse embryonic fibroblast cell lines were performed as reported previously (Dunning et al, 2007). Transfections were performed using Lipofectamine 2000 (Invitrogen, CA, USA). For immunofluorescence studies, cells were fixed with 4% (wt/vol) paraformaldehyde or 100% ice-cold acetone. Nuclei were stained with 10-μg/ml Hoechst 33258 and F-actin with Phalloidin Alexa Fluor 568 (Molecular Probes, CA, USA). The final concentration of nocodazole (Sigma, MO, USA) was 5 μM.
Antibodies. Antibodies were raised in rabbits using a peptide against residues 438–454 of MiD49 or recombinant MiD51 as the antigen. Western blots were probed with in-house antibodies against matrix-Hsp70 and Tom40 (Johnston et al, 2002), mitofusin-2, Hsc70 and B17.2L. Commercial antibodies used were cytochrome c, Drp1 and OPA1 (BD Biosciences, MD, USA), β-actin (Sigma, MO, USA) and Tom20 (Santa Cruz, CA, USA).
Mitochondrial treatments and western blotting. Mitochondrial isolation, fractionation, proteinase K digestions and sodium carbonate extractions were performed using standard methods (Johnston et al, 2002).
Coimmunoprecipitation. HeLa cells transiently transfected with GFP–Fis1, MiD49–GFP or GFP–Miro1 constructs were crosslinked with 20-μM bis(maleimido)hexane (Thermo Scientific, IL, USA). Transfected cells were immunoprecipitated using GFP-Trap beads (Chromotek, Martinsried, Germany), according to the manufacturer's instructions.
Protein–protein interaction assay. The MiD49 soluble domain (amino acids 121–463, MiD49ΔN120) was cloned into the pGBKT7 vector (Clontech). Drp1 and the Fis1 soluble domain were cloned into the pGADT7 vector. The two-hybrid analysis, with controls, was performed according to the manufacturer's instructions (Clontech). Cotransformants were selected on high selection media.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank D. Chudakov, J. Shaw, L. Scorrano, A. van der Bliek, R. Youle, E. Hanssen, Q. Zhao and X. Wang for various constructs, reagents and assistance. This work was supported by grants from the National Health and Medical Research Council, Australian Research Council, the William Buckland Foundation and the Human Frontier Science Program Organization.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anesti V, Scorrano L (2006) The relationship between mitochondrial shape and function and the cytoskeleton. Biochim Biophys Acta 1757: 692–699 [DOI] [PubMed] [Google Scholar]
- Bhar D, Karren MA, Babst M, Shaw JM (2006) Dimeric Dnm1–G385D interacts with Mdv1 on mitochondria and can be stimulated to assemble into fission complexes containing Mdv1 and Fis1. J Biol Chem 281: 17312–17320 [DOI] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM (1999) The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol 1: 298–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CR, Blackstone C (2007) Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem 282: 21583–21587 [DOI] [PubMed] [Google Scholar]
- Chang CR, Manlandro CM, Arnoult D, Stadler J, Posey AE, Hill RB, Blackstone C (2010) A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division. J Biol Chem 285: 32494–32503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos KJ, Allan VJ, Grierson AJ, Sheetz MP (2005) Mitochondrial function and actin regulate dynamin-related protein 1-dependent mitochondrial fission. Curr Biol 15: 678–683 [DOI] [PubMed] [Google Scholar]
- Dunning CJ, McKenzie M, Sugiana C, Lazarou M, Silke J, Connelly A, Fletcher JM, Kirby DM, Thorburn DR, Ryan MT (2007) Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J 26: 3227–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier AE, Kiu C, Stojanovski D, Hoogenraad NJ, Ryan MT (2006) Mitochondrial morphology and distribution in mammalian cells. Biol Chem 387: 1551–1155 [DOI] [PubMed] [Google Scholar]
- Gandre-Babbe S, van der Bliek AM (2008) The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell 19: 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin EE, Graumann J, Chan DC (2005) The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J Cell Biol 170: 237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AJ, Hoogenraad J, Dougan DA, Truscott KN, Yano M, Mori M, Hoogenraad NJ, Ryan MT (2002) Insertion and assembly of human tom7 into the preprotein translocase complex of the outer mitochondrial membrane. J Biol Chem 277: 42197–42204 [DOI] [PubMed] [Google Scholar]
- Karbowski M, Jeong SY, Youle RJ (2004) Endophilin B1 is required for the maintenance of mitochondrial morphology. J Cell Biol 166: 1027–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Neutzner A, Youle RJ (2007) The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol 178: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koirala S, Bui HT, Schubert HL, Eckert DM, Hill CP, Kay MS, Shaw JM (2010) Molecular architecture of a dynamin adaptor: implications for assembly of mitochondrial fission complexes. J Cell Biol 191: 1127–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsopoulos OS, Laine D, Osellame L, Chudakov DM, Parton RG, Frazier AE, Ryan MT (2010) Human Miltons associate with mitochondria and induce microtubule-dependent remodeling of mitochondrial networks. Biochim Biophys Acta 1803: 564–574 [DOI] [PubMed] [Google Scholar]
- Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM (1999) C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell 4: 815–826 [DOI] [PubMed] [Google Scholar]
- Legesse-Miller A, Massol RH, Kirchhausen T (2003) Constriction and Dnm1p recruitment are distinct processes in mitochondrial fission. Mol Biol Cell 14: 1953–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros F, Lombes A, Frachon P, Rojo M (2002) Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol Biol Cell 13: 4343–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesa M, Palacin M, Zorzano A (2009) Mitochondrial dynamics in mammalian health and disease. Physiol Rev 89: 799–845 [DOI] [PubMed] [Google Scholar]
- Mozdy AD, McCaffery JM, Shaw JM (2000) Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol 151: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K (2010) Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol 191: 1141–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D, Keegan BR, Brisch E, Thatcher JW, Hermann GJ, Bleazard W, Shaw JM (1998) The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J Cell Biol 143: 333–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JC, Wellenreuther R, Poustka A, Pepperkok R, Wiemann S (2000) Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep 1: 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slager RE, Newton TL, Vlangos CN, Finucane B, Elsea SH (2003) Mutations in RAI1 associated with Smith-Magenis syndrome. Nat Genet 33: 466–468 [DOI] [PubMed] [Google Scholar]
- Smirnova E, Shurland D, Ryazantsev SN, van der Bliek AM (1998) A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol 143: 351–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM (2001) Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell 12: 2245–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D, Koutsopoulos OS, Okamoto K, Ryan MT (2004) Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J Cell Sci 117: 1201–1210 [DOI] [PubMed] [Google Scholar]
- Tieu Q, Nunnari J (2000) Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J Cell Biol 151: 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G et al. (2008) Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27: 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B (2010) Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol 11: 872–884 [DOI] [PubMed] [Google Scholar]
- Yoon Y, Pitts KR, McNiven MA (2001) Mammalian dynamin-like protein DLP1 tubulates membranes. Mol Biol Cell 12: 2894–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Krueger EW, Oswald BJ, McNiven MA (2003) The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol 23: 5409–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.