Abstract
In the Drosophila circadian clock, the CLOCK/CYCLE complex activates the period and timeless genes that negatively feedback on CLOCK/CYCLE activity. The 24-h pace of this cycle depends on the stability of the clock proteins. RING-domain E3 ubiquitin ligases have been shown to destabilize PERIOD or TIMELESS. Here we identify a clock function for the circadian trip (ctrip) gene, which encodes a HECT-domain E3 ubiquitin ligase. ctrip expression in the brain is mostly restricted to clock neurons and its downregulation leads to long-period activity rhythms in constant darkness. This altered behaviour is associated with high CLOCK levels and persistence of phosphorylated PERIOD during the subjective day. The control of CLOCK protein levels does not require PERIOD. Thus, CTRIP seems to regulate the pace of the oscillator by controlling the stability of both the activator and the repressor of the feedback loop.
Keywords: circadian, CLOCK, PERIOD, ubiquitination, UFD pathway
Introduction
Circadian clocks rely on transcriptional negative feedback loops. In Drosophila, the basic helix–loop–helix PAS domain proteins CLOCK (CLK) and CYCLE (CYC) activate the transcription of period (per) and timeless (tim) in the evening (Weber, 2009; Allada & Chung, 2010). PER and TIM progressively accumulate and enter the nucleus, in which they repress CLK/CYC-dependent transcription late in the night. PER and TIM are subject to complex post-translational controls that involve several kinases such as DOUBLE TIME CK1e, CK2 and SHAGGY GSK3, as well as the PP1 and PP2A phosphatases. CLK phosphorylation cycles in a similar manner, with mildly phosphorylated CLK activating transcription in the evening (Kim & Edery, 2006; Yu et al, 2006). PER represses CLK activity by triggering its hyperphosphorylation and release from DNA (Kim & Edery, 2006; Yu et al, 2006, 2009; Menet et al, 2010). However, CLK protein turnover remains unclear as the contributions of protein-level cycling and phosphorylation cycling are still debated (Houl et al, 2006; Yu et al, 2006; Hung et al, 2009).
The stability of phosphorylated PER relies on a proteasome-dependent pathway that requires the SUPERNUMERARY LIMBS E3 ubiquitin ligase (Chiu et al, 2008). SUPERNUMERARY LIMBS is part of a CULLIN 1-based SCF complex that belongs to the RING family of E3 ubiquitin ligases (Nakayama & Nakayama, 2005). Another family of E3 ubiquitin ligases has been characterized, whose members contain a HECT (homologous to E6AP C-terminus) domain (Rotin & Kumar, 2009). The mammalian protein TRIP12 (thyroid hormone receptor-interacting protein 12; Lee et al, 1995) is one of them and has been proposed to be involved in amino-terminal ubiquitination (Park et al, 2009; Chen et al, 2010).
In an enhancer trap screen, two Gal4-encoding P-elements secreting the pigment-dispersing factor (PDF) neuropeptide were found that are strongly expressed in the neurons which are key pacemaker cells for the control of Drosophila rest–activity rhythms (Dubruille & Emery, 2008). Here, we show that the target gene is the orthologue of trip12, and we named the Drosophila gene circadian trip (ctrip) because its downregulation lengthens the period of behavioural rhythms. Molecular analysis indicates that CTRIP destabilizes CLK protein in a PER-independent manner and helps degradation of phosphorylated PER and TIM in the morning.
Results and Discussion
ctrip is strongly expressed in the PDF clock neurons
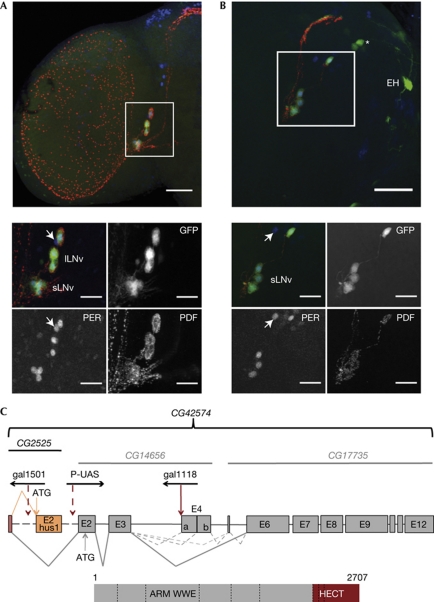
The expression of the gal1118 Gal4 enhancer trap has been described as essentially restricted to the PDF-expressing ventral lateral neurons of the Drosophila brain, with weaker expression in other clock cells as well as a few non-clock cells (Blanchardon et al, 2001). Another insertion from the same collection of P-element Gal4 lines (Boquet et al, 1999), gal1501, showed a similar expression pattern in the adult (Fig 1A) and larval (Fig 1B) brain, with an even higher specificity for the PDF neurons. The two P-elements are located in a region that includes the DNA repair gene Hus1-like (CG2525; Abdu et al, 2007) and a large gene (CG42574; http://flybase.org/) that is homologous to the human trip12 gene (Fig 1C). CG42574 encodes a putative E3 ubiquitin ligase that contains Armadillo repeats, a WWE protein–protein interaction domain (Aravind, 2001) and a carboxy-terminal HECT domain.
Figure 1.
Brain expression and genomic organization of ctrip. (A, B) GFP labelling of adult (A) or larval (B) brain of w; UAS-gfp; gal1501/+ flies at zeitgeber time (ZT) 0 in light–dark conditions (ZT 0 is lights-ON). gal1501-driven GFP fluorescence is combined with PER and PDF antibodies. All images are projections of apotome stacks. The white squares on the half-brain images correspond to the small images. Colours in the merge pictures are green for GFP, blue for PER and red for PDF. Arrow points to the fifth PDF-negative SLNV. GFP can be seen in at least three PER-negative cells in each dorsal hemisphere of the larval brain: the EH-synthesizing neurons and a pair of dorsal neurons (asterisk) that seem to be the prothoracic-gland-innervating neurons of the lateral protocerebrum (PG–LP; Blanchardon et al, 2001). Weak expression in other clock neuron subsets can be detected in flies homozygous for the gal1501 and UAS-gfp transgenes (not shown). Scale bars represent 50 (half brains) or 25 (small images) μm. (C) Schematic representation of the ctrip gene. Top: current (black) and previous (grey) Flybase nomenclature. Middle: exon/intron structure with common (red), Hus1-like-specific (orange) and ctrip-specific (grey) exons as boxes. Dotted lines represent out-of-scale regions (see http://flybase.org/). Splicing events are shown as grey lines for the most abundant transcripts and dotted grey lines for the less abundant ones. E4a/b boundary is defined by a splicing acceptor site located 630 bp downstream of the beginning of E4. Putative translation start sites (ATG) are indicated for both Hus1-like and ctrip. P-element insertions are shown with the transcription orientation of the gal4 gene (gal1118 and gal1501) or UAS sequence (P-UAS). gal1501 is inserted 3208 bp upstream from Hus1-like coding exon and gal1118 is inserted 78 bp upstream from E4. P-UAS refers to P-UAS 11430 that is inserted 1673 bp upstream from E2. Bottom: predicted protein encoded by the most abundant transcript. Exon boundaries are represented by dotted lines. ARM and WWE domains are encoded by E6, whereas the HECT domain is encoded by E11–12. ARM, Armadillo; ctrip, circadian trip; EH, eclosion hormone; GFP, green fluorescent protein; HECT, homologous to E6AP C-terminus; lLNv, large ventral lateral neuron; PDF, pigment-dispersing factor; PER, PERIOD; P-UAS, P-element-carrying Upstream Activating Sequence; RNAi, RNA interference; sLNv, small ventral lateral neuron.
The localization of gal1118 supported the idea that CG42574 is responsible for the brain expression pattern and CG42574 was named ctrip. Reverse transcription (RT)–PCR and sequence analysis of head transcripts indicated that Hus1-like and ctrip messenger RNAs (mRNAs) share a non-coding first exon, and that ctrip contained 12 exons with alternative splicing in the E3–E6 region (Fig 1C; supplementary Fig S1 online). In the adult head, ctrip mRNA levels were approximately tenfold more than per mRNA levels (not shown), suggesting a broad expression outside the brain. The most abundant ctrip head transcript contained 10 exons (lacking E4–E5) and encoded a 2707-amino-acid putative protein, but transcripts including E4–E5 or E4b–E5 or only E5 could be detected at levels 10–15 times lower (Fig 1C; supplementary Figs S1,2 online). None of the ctrip mRNAs showed oscillations of its abundance in the head in light–dark cycles (supplementary Fig S2 online).
The CG17735 transcript was recently identified as 5–15-fold enriched in the PDF neurons compared with all brain neurons (Kula-Eversole et al, 2010), validating the expression pattern of the two ctrip P-gal4 insertions. In contrast to ctrip transcripts in whole-head extracts, CG17735 transcripts cycled in the PDF cells, reaching peak levels in the first half of the night (Kula-Eversole et al, 2010). The strong expression of ctrip in the head suggests that it includes both clock cells and non-clock cells, which could mask oscillations in specific subsets.
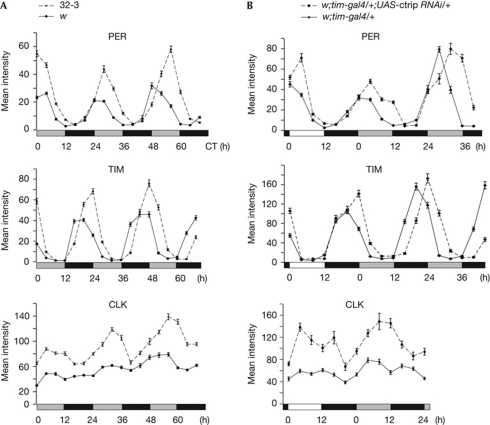
Loss of ctrip lengthens the clock period in larval neurons
Deletions were generated by P-element excision. The Y4 deletion removed the Hus1-like coding exon and induced female sterility, similarly to the Hus1-like37-null mutant (Abdu et al, 2007). Homozygous Y4 or Hus1-like37 flies showed normal activity rhythms, indicating that Hus1-like is not involved in the behavioural clock (Table 1). The 19F1 deletion removed the ctrip alternative exons E4–E5, leading to an mRNA species that is identical to the most abundant wild-type transcript. 19F1 homozygous flies were viable and behaved similarly to wild-type (Table 1), indicating that the glutamine-rich region encoded by E4–E5 was not required for clock function. The 32.3 deletion removed both Hus1-like and exons 1–5 of ctrip, and was homozygously lethal at the pupal stage. ctrip larvae were analysed for clock protein oscillations in the lateral neurons (Fig 2A; supplementary Fig S3 online). High-amplitude PER and TIM oscillations were observed in the 32.3 mutant, with an increasing delay over 2.5 days in dark–dark conditions, revealing a long period. As reported previously (Hung et al, 2009), the wild-type larvae displayed low-amplitude CLK oscillations, but higher CLK levels and higher cycling amplitude were observed in 32.3 larvae (Fig 2A; supplementary Fig S3 online). A similar CLK and PER increase was observed in the dorsal neurons, indicating that ctrip was also acting in PDF-negative cells (supplementary Fig S4 online). To verify that the phenotype was due to ctrip loss, we analysed larvae carrying 32.3 over a deficiency that did or did not encompass ctrip region. At the beginning of the first day in dark–dark conditions, CLK and PER levels were increased only in larvae homozygous for the loss of the ctrip locus (supplementary Fig S4 online). In agreement with their wild-type behaviour, Hus1-like37 adults had normal CLK and PER levels (supplementary Fig S4 online). The absence of ctrip thus increases CLK levels and slows clock-protein cycling in the clock neurons.
Table 1. Locomotor activity rhythms in dark–dark conditions.
| Genotype | Number of flies | Rhythmic flies (%) | Tau (s.e.m.) | Power (s.e.m.) |
|---|---|---|---|---|
| w | 26 | 96 | 23.4 (0.1) | 108 (9) |
| w;pdf-gal4/+ | 8 | 100 | 24.0 (0.1) | 68 (2) |
| w;tim-gal4/+ | 29 | 97 | 24.1 (0.1) | 129 (4) |
| w;;17R3/TM6 | 8 | 100 | 23.7 (0.1) | 135 (6) |
| w;;17R3 | 7 | 100 | 24.0 (0.2) | 117 (9) |
| w;;14R3/TM6 | 16 | 88 | 24.0 (0.0) | 53 (3) |
| w;;14R3 | 16 | 94 | 24.2 (0.2) | 80 (10) |
| w;;Y4 | 20 | 90 | 23.7 (0.1) | 86 (9) |
| w;;hus1-like 37 | 30 | 100 | 24.3 (0.1) | 104 (4) |
| w;;19F1 | 17 | 94 | 24.2 (0.1) | 121 (4) |
| w;pdf-gal4/+;17R3/+ | 32 | 81 | 25.3 (0.1) | 56 (4) |
| w;pdf-gal4/+;14R3/+ | 46 | 89 | 25.1 (0.1) | 66 (2) |
| w;pdf-gal4/+;17R3,14R3/+ | 16 | 100 | 26.2 (0.1) | 59 (3) |
| w;pdf-gal4;17R3,14R3/+ | 26 | 96 | 27.0 (0.2) | 61 (4) |
| w;pdf-gal4;17R3 | 29 | 83 | 27.8 (0.2) | 84 (7) |
| w;pdf-gal4;14R3 | 34 | 91 | 26.0 (0.1) | 84 (7) |
| w;pdf-gal4;17R3,14R3 | 16 | 87 | 28.2 (0.6) | 71 (8) |
| w;tim-gal4/+;17R3/+ | 52 | 94 | 26.2 (0.1) | 125 (6) |
| w;tim-gal4/+;14R3/+ | 75 | 88 | 25.3 (0.1) | 88 (6) |
| w;tim-gal4/+;17R3,14R3/+ | 30 | 100 | 26.4 (0.1) | 146 (4) |
| 14R3 and 17R3 are UAS-ctrip RNAi transgenes targeting ctrip exons 2 and 6, respectively. | ||||
| DD, dark–dark conditions; Tau, period in hours; s.e.m., standard error of the mean. | ||||
Figure 2.
Clock-protein cycling in ctrip mutant larvae and ctrip RNAi adults. CLK, PER and TIM immunofluorescence in the larval (A) or adult small ventral lateral neurons (B). Error bars indicate the s.e.m. (n=20–25 brain hemispheres). White, grey and black bars represent day, subjective day and night, respectively. Time is indicated in hours. (A) Larvae were grown in 12:12 LD cycles and transferred to DD conditions. Experiments were done at 20°C to slow development, so that larval brains could be dissected up to 3 days after transfer to DD conditions. Larvae were collected every 4 h from the first day in DD conditions for brain dissection. (B) Flies were entrained in 12:12 LD cycles and transferred to DD conditions, at 25°C. Brains were dissected every 4 h on the last day of LD conditions, followed by 1 or 2 days in DD conditions. CLK, CLOCK; CT, circadian time (CT0 is the beginning of the first day in DD); ctrip, circadian trip; DD, dark–dark; LD, light–dark; PER, PERIOD; RNAi, RNA interference; TIM, TIMELESS.
ctrip RNA interference slows the behavioural clock
RNA interference (RNAi) constructs directed against ctrip exons 2 (14R3) or 6 (17R3) were expressed in either the PDF-expressing cells or all clock cells (tim-expressing cells), and the transgenic flies were analysed for locomotor activity rhythms in dark–dark conditions. A lengthening of the period was observed with both pdf-gal4 and tim-gal4 drivers (Table 1), indicating that ctrip is required for 24-h behavioural rhythms. A P-UAS insertion (see Fig 1B) was used to overexpress ctrip under tim-gal4 control, but this did not alter behaviour (supplementary Table S1 online), possibly because CTRIP acts with a partner that is present in limiting amounts. However, 24-h behavioural rhythms were rescued in flies co-expressing ctrip RNAi and the P-UAS-induced transcript, further supporting the idea that ctrip downregulation is responsible for the behavioural defect found in ctrip RNAi flies (supplementary Table S1 online). A large-scale RNAi screen has revealed that flies downregulated for CG17735 (see Fig 1) had long-period behavioural rhythms (Sathyanarayanan et al, 2008). Although we could not detect the reported inhibition of light-induced CRY degradation with our ctrip RNAi flies (Peschel et al, 2009), it provides an independent confirmation of our behavioural results.
Clock-protein oscillations were analysed in the small ventral lateral neurons of w;tim-gal4/+;17R3,14R3/+ flies, hereafter referred to as tim-gal4 UAS-ctrip RNAi (Fig 2B; supplementary Figs S3,5 online). A progressive delay of clock-protein oscillations was observed in dark–dark conditions, in agreement with the behavioural results. The increase at night of PER and TIM was not delayed in light–dark conditions (Fig 2B) and no difference in the nuclear entry of the proteins was observed (not shown). By contrast, the decrease of protein levels was shifted during the first subjective day of dark–dark conditions, suggesting that lengthening of the period is due to delayed protein degradation. As seen in the lateral neurons of the 32.3 mutant larvae, high CLK levels were observed at all time points. However, a stronger effect seemed to occur late in the day, suggesting that CTRIP levels (see also Kula-Eversole et al, 2010) and/or function might be regulated. Clock-protein oscillations were also analysed in the small ventral lateral neurons of flies expressing ctrip RNAi under pdf-gal4 control, with similar results (supplementary Fig S5 online).
CTRIP controls CLK protein stability
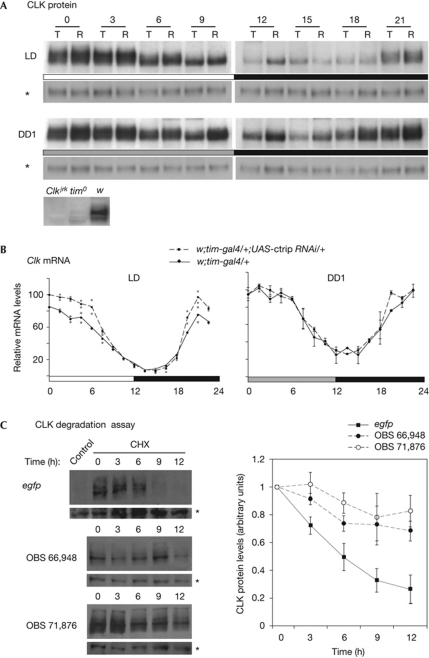
CLK protein and Clk mRNA levels were analysed in tim-gal4 UAS-ctrip RNAi head extracts. Slightly higher CLK levels were observed in light–dark conditions, and a stronger effect was observed in dark–dark conditions (Fig 3A; supplementary Fig S6 online). The increase of CLK in ctrip RNAi flies was less important in head extracts than in clock neurons (see Fig 2), suggesting that RNAi expression in the eye was weaker. Although a slight increase in Clk mRNA peak levels was observed in light–dark conditions, ctrip downregulation did not affect Clk mRNA levels in dark–dark conditions (Fig 3B). This indicated that the higher protein levels were a consequence of post-transcriptional control. As ctrip encodes a ubiquitin ligase, the simplest interpretation is that ctrip induces CLK protein degradation.
Figure 3.
CTRIP inhibition stabilizes CLK protein. Flies were entrained in 12:12 LD cycles and transferred to DD conditions, at 25°C. Head extracts were prepared from time points collected every 3 h (A) or 1.5 h (B) during the last LD day and the first DD day. (A) Western blot analysis of CLK protein. T, w;tim-gal4/+ controls; R, w;tim-gal4;UAS-ctrip RNAi. Asterisks indicate a PER antibody unspecific band on the same gel that was used as a loading control. Antibody specificity is shown by the absence of signal (CT3) in Clkjrk extracts and the very weak signal in tim0 extracts under the same conditions. (B) Quantitative RT–PCR of Clk mRNA. The results are the average of two (LD) or three (DD) independent experiments. Grey dots indicate the maximum and minimum values. Error bars indicate the s.e.m. (n=3 experiments). (C) RNAi-mediated inhibition of CTRIP reduces CLK degradation in Drosophila S2 cells. Degradation kinetics of CLK were analysed after blocking new protein synthesis by the addition of cycloheximide in the presence of dsRNA targeting ctrip exons 3 (OBS 71874) or 6 (OBS 66948) or egfp. Left panel: anti-CLK western blots of S2 cell lysates harvested at 0, 3, 6, 9, and 12 h after cycloheximide treatment. The egfp-dsRNA control is from untransfected and untreated cells. The CLK antibody used in this experiment was described in Hung et al (2007) and recognizes an unspecific band (asterisk) that was used as a loading control. Right panel: average degradation kinetics of CLK protein from five cycloheximide chase experiments (as in the left panel), with the amount of CLK protein prior to the addition of cycloheximide (time 0) set to 1. Error bars indicate the s.e.m. CHX, cycloheximide; CLK, CLOCK; ctrip, circadian trip; DD, dark–dark; dsRNA, double-stranded RNA; LD, light–dark; mRNA, messenger RNA; OBS, Open BioSystem; PER, PERIOD; RNAi, RNA interference; tim, timeless.
CLK protein stability was thus tested in S2 cell culture. Cycloheximide chase experiments showed reduced CLK degradation kinetics in the presence of either of two ctrip double-stranded RNAs directed against exons 3 and 6 of the gene, compared with an unspecific egfp control double-stranded RNA (Fig 3C). This indicated that CTRIP destabilizes CLK in S2 cells, supporting the idea of such a function in the clock neurons.
ctrip downregulation decreases PER/TIM degradation
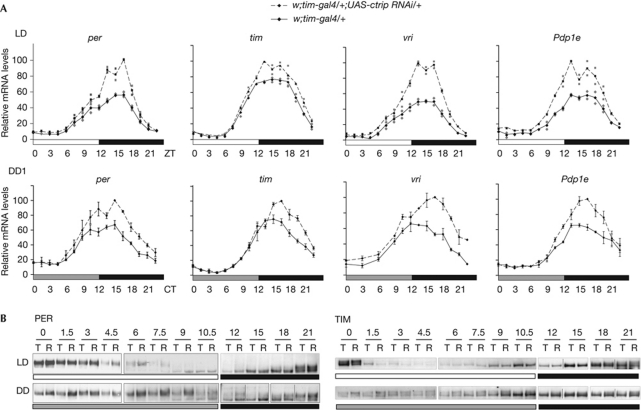
Transcripts of CLK direct targets (per, tim, vri and Pdp1e) were quantified in tim-gal4 UAS-ctrip RNAi flies (Fig 4A). In light–dark conditions, peak levels of the four transcripts were approximately 1.5-fold higher in the RNAi flies, but decreased to basal levels at the end of the night. On the first day of dark–dark conditions, the RNAi flies did not show a delayed start of mRNA increase but rather a longer increase of mRNA levels, resulting in a higher peak with a slight delay. This indicates higher CLK transcriptional activity of the RNAi flies in the middle of the night. PER and TIM protein cycling was tested by western blots of head extracts (Fig 4; supplementary Fig S7 online). In light–dark conditions, only slightly more phosphorylated PER was detected in the morning in ctrip RNAi flies. In dark–dark conditions, phosphorylated PER was more abundant throughout the subjective day, supporting the idea of deficient protein degradation in RNAi flies in the absence of light. A similar but milder effect was observed for the TIM protein.
Figure 4.
Levels of per and tim RNA and PER and TIM protein in ctrip RNAi flies. (A) Relative mRNA levels of per, tim, vri and Pdp1e in head extracts were determined by quantitative RT–PCR in LD conditions and during the first day of DD conditions. The results are the average of two (LD) or three (DD) independent experiments. Grey dots indicate the maximum and minimum values. Error bars indicate the s.e.m. (B) Western blot analysis of PER and TIM protein in head extracts during LD conditions and the first day of DD conditions. Flies were collected every 1.5 h during day or subjective day and every 3 h during night or subjective night. T, w;tim-gal4/+; R, w;tim-gal4/+;UAS-ctrip RNAi/+. Boxes are different gels and vertical lines indicate that the lanes were not adjacent on the gel. ctrip, circadian trip; CT, circadian time; DD, dark–dark; LD, light–dark; mRNA, messenger RNA; Pdp1e, Par domain protein1; per, period; RNAi, RNA interference; RT, reverse transcription; tim, timeless; vri, vrille; ZT, zeitgeber time.
Transgenic flies with increased CLK levels have been shown to display either no period change (Kim et al, 2002) or a mild shortening of the period, which might result from a premature nuclear entry of PER (Kadener et al, 2008). By contrast, the persistence of phosphorylated PER and TIM during the subjective day has been associated with a long behavioural period in various mutants (Rothenfluh et al, 2000; Suri et al, 2000). It therefore seems likely that the long-period phenotype of ctrip RNAi flies is a consequence of long-lasting PER and TIM during the day, in the absence of light-induced degradation.
The higher peak levels of CLK target transcripts in the RNAi flies indicates that CLK-dependent transcription is more active when ctrip is downregulated. However, PER and TIM proteins do not seem to accumulate faster, but show a persistence of their phosphorylated forms during the subjective day. By contrast, per and tim mRNAs reach trough levels early in the day, as in the wild type. Increased PER and TIM levels in the morning are thus not a consequence of elevated per and tim mRNA levels, but seem to be due to a post-translational action of CTRIP on PER and TIM. By destabilizing both the CLK activator and the PER repressor, CTRIP might have a buffering role in the transcriptional feedback loop.
Although PER and TIM persist during the subjective day, the evening accumulation of their mRNAs is not delayed in ctrip RNAi flies. Higher hypophosphorylated CLK levels probably counteract the repressing effect of higher PER levels. In the evening, the normal kinetics of mRNA accumulation might thus be the consequence of elevated CLK and PER levels. In the middle of the night, PER and TIM levels do not seem to be higher and CLK target transcripts continue to increase for a longer time, indicating that the repression phase is delayed in RNAi flies. A simple interpretation of this is that elevated CLK levels are responsible for extending the time of active transcription. Alternatively, downregulation of ctrip might delay repression by destabilizing the PER/CLK complex or reducing its repressing function.
CTRIP-mediated CLK regulation is PER-independent
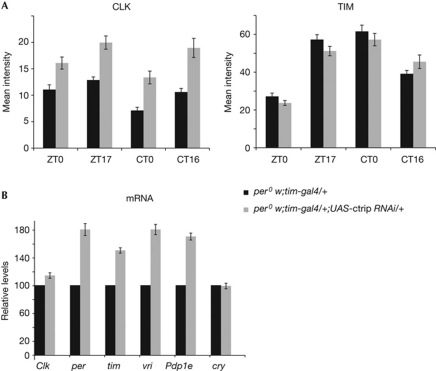
We asked whether increased CLK levels might be a consequence of abnormal PER degradation in the ctrip RNAi flies. per0; tim-gal4 UAS-ctrip RNAi flies were thus analysed for CLK and TIM levels in the PDF neurons (Fig 5A). In the absence of PER, TIM levels were not affected, but ctrip RNAi induced a 1.5-fold increase of CLK levels. In agreement with their increased CLK levels, per0; tim-gal4 UAS-ctrip RNAi flies displayed higher transcript levels for the CLK target genes, whereas Clk and cry were not affected (Fig 5B). This indicates that the persistence of phosphorylated TIM in the morning requires PER, whereas the control of CLK levels by CTRIP is a PER-independent mechanism.
Figure 5.
Effect of ctrip RNAi in per0 background. (A) Immunofluorescence quantification of CLK and TIM protein levels in the PDF-expressing small ventral lateral neurons of adult flies in LD (ZT) and DD (CT). Error bars indicate the s.e.m. (n=20–25 brain hemispheres). (B) Quantitative RT–PCR analysis of Clk, cry and CLK direct targets per, tim, vri and Pdp1e in LD conditions (ZT14). Each experiment has been normalized to the control value (black bar). Error bars indicate the s.e.m. (n=3 experiments). CLK, clock; ctrip, circadian trip; CT, circadian time; DD, dark–dark; LD, light–dark; Pdp1e, Par domain protein1; per, period; RNAi, RNA interference; tim, timeless; vri, vrille; ZT, zeitgeber time.
CLK and PER thus seem to be the main targets of CTRIP. The ubiquitin ligase might act independently on CLK and PER, with the two proteins possibly competing for CTRIP binding. Alternatively, a CTRIP-mediated effect on CLK could affect PER stability. Such a mechanism might provide an efficient way to counterbalance changes in CLK levels. For example, it could help to keep the pace of the oscillator more resistant to variations in CLK levels, which might be induced by physiological stress or environmental changes.
In mammals, TRIP12 has recently been shown to be part of the ubiquitin fusion degradation (UFD) pathway, in which poly-ubiquitin is added to the N-terminus of the target protein as a degradation signal (Ciechanover & Ben-Saadon, 2004; Park et al, 2009; Chen et al, 2010). Putative UFD pathway components are present in Drosophila (Lundgren et al, 2005), but no role for N-terminal ubiquitination has been shown. Our results raise the possibility that the UFD pathway is involved in tuning the speed of the circadian oscillator by controlling the stability of both CLK and PER.
Methods
Fly strains and behavioural analysis. pdf-gal4 (Park et al, 2000), tim-gal4 (Kaneko, 1998), gal1118 (Blanchardon et al, 2001) and hus1-like37 (Abdu et al, 2007) have been previously reported. The P-UAS 11430 line is described at http://gsdb.biol.metro-u.ac.jp/~dclust/index.html. The ctrip RNAi lines 14656R3 (named 14R3 here) and 17735R3 (named 17R3 here) are described at http://www.shigen.nig.ac.jp/fly/nigfly/index.jsp, and target E2 and E6 ctrip exons, respectively. A recombinant chromosome carrying both 14R3 and 17R3 was used for all molecular analyses. The Y4 and 32–3 deletions were obtained by imprecise excision of the gal1501 insertion and 19F1 deletion by imprecise excision of gal1118, through standard P-element excision procedures. The gal1501, gal1118, 19F1 and 32.3 stocks were outcrossed for five generations against a w control stock (Canton S genetic background) before further analyses. Behavioural assays were carried out with 1–7-day-old males at 25°C in Drosophila activity monitors (TriKinetics), as described previously (Klarsfeld et al, 2003). For dark–dark conditions analysis, flies were first entrained in 12 h:12 h light–dark conditions cycles during 2–4 days before transfer to dark–dark conditions, and data were analysed for 6 days from the second day in dark–dark conditions. Data analysis was done with the FaasX software, which is derived from the Brandeis Rhythm Package and is available on request. All behavioural experiments were performed two or three times with similar results.
Immunolabellings. In situ immunolabellings were done on whole-mounted brains of larvae or adult males, as described previously (Blanchardon et al, 2001). For quantifications, 20–25 brain hemispheres were used for each time point. Primary antibodies used were rabbit PER (Stanewsky et al, 1997) at 1:15,000 dilution, rat TIM (Grima et al, 2002) at 1:10,000, guinea-pig CLK GP47 (Houl et al, 2006) at 1:10,000 and mouse PDF (Developmental Studies Hybridoma Bank) at 1:50,000. Secondary goat antibodies (Invitrogen) were FP546-conjugated rabbit (1:2,000), Alexa 647-conjugated rat (1:5,000), Alexa 594-conjugated mouse (1:2,000) and Alexa 488-conjugated guinea-pig (1:2,000).
Western blots. Protein extracts were made from 30–40 male heads for each time point. Frozen heads were homogenized in 150 μl ice-cold RBS buffer (Yu et al, 2006) plus protease inhibitors (complete mini Roche), phosphatase inhibitors cocktails 1 (0.5%) and 2 (1%; Sigma), and 2-mM β-glycerophosphate. For SDS–PAGE, 50 μg of protein extracts were separated on 3–8% Tris–acetate gels (Invitrogen) and transferred to PVDF membranes. Primary antibodies used were rabbit PER (Stanewsky et al, 1997; 1:10,000), rat TIM (Grima et al, 2002; 1:2,000) and goat CLK sc-27070 (Santa Cruz Biotechnology; 1:1,000). HRP-conjugated secondary antibodies (Santa Cruz Biotechnology) used were rabbit goat (1:10,000), rat goat (1:15,000) and goat donkey (1:20,000). All western blots were reproduced two or three times with similar results. Degradation assays were performed as described in Hung et al (2009).
Genomic DNA and complementary DNA analysis. Genomic DNA surrounding the gal1118 and gal1501 insertions was isolated by standard cloning procedures to localize the P-elements. RT–PCR and 5′-rapid amplification of cloned ends (RACE) analysis of ctrip head complementary DNAs (cDNAs) were used to characterize the different ctrip transcripts from a w stock (Canton S background). For quantitative RT–PCR, cDNAs were synthesized from 1 μg of male head total RNA (Promega SV total RNA isolation system), and PCR was performed with a LightCycler (Roche) using the SYBR green detection protocol, as described previously (Grima et al, 2002). mRNA levels were normalized to the levels of tubulin mRNA and expressed as a percentage of the maximum value set to 100. The results were then averaged from two or three independent experiments.
Additional methods are available as supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank T. Préat for providing the gal1501 line, T. Aigaki for the P-UAS 11430 line, R. Stanewsky, P. Hardin and P. Emery for antibodies, and E. Nagoshi and M. Rosbash for helpful discussions. We also thank M. Boudinot for the FaasX software, L. Collet for artwork, and S. Bouleau, B. Grima, A. Klarsfeld, C. Michard-Vanhée and A. Szabo for their comments on the manuscript. This work was supported by the Agence Nationale de la Recherche ‘DrosoClock’ programme and the European Union Sixth Framework Programme ‘EUCLOCK’. A.L. was supported by Région Ile de France and Association pour la Recherche sur le Cancer and F.R. by the Institut National de la Santé et des Etudes et Recherches Médicales. F.W. was supported by grant WE2608/2-1 from the German Research Foundation (DFG).
Footnotes
The authors declare that they have no conflict of interest.
References
- Abdu U, Klovstad M, Butin-Israeli V, Bakhrat A, Schupbach T (2007) An essential role for Drosophila hus1 in somatic and meiotic DNA damage responses. J Cell Sci 120: 1042–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, Chung BY (2010) Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol 72: 605–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L (2001) The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem Sci 26: 273–275 [DOI] [PubMed] [Google Scholar]
- Blanchardon E, Grima B, Klarsfeld A, Chélot E, Hardin PE, Préat T, Rouyer F (2001) Defining the role of Drosophila lateral neurons in the control of circadian activity and eclosion rhythms by targeted genetic ablation and PERIOD protein overexpression. Eur J Neurosci 13: 871–888 [DOI] [PubMed] [Google Scholar]
- Boquet I, Hitier R, Dumas M, Chaminade M, Préat T (1999) Central brain postembryonic development in Drosophila: implication of genes expressed at the interhemispheric junction. J Neurobiol 42: 33–48 [PubMed] [Google Scholar]
- Chen D, Shan J, Zhu WG, Qin J, Gu W (2010) Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature 464: 624–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu JC, Vanselow JT, Kramer A, Edery I (2008) The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev 22: 1758–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Ben-Saadon R (2004) N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol 14: 103–106 [DOI] [PubMed] [Google Scholar]
- Dubruille R, Emery P (2008) A plastic clock: how circadian rhythms respond to environmental cues in Drosophila. Mol Neurobiol 38: 129–145 [DOI] [PubMed] [Google Scholar]
- Grima B, Lamouroux A, Chélot E, Papin C, Limbourg-Bouchon B, Rouyer F (2002) The F-box protein SLIMB controls the levels of clock proteins PERIOD and TIMELESS. Nature 429: 178–182 [DOI] [PubMed] [Google Scholar]
- Houl JH, Yu W, Dudek SM, Hardin PE (2006) Drosophila CLOCK is constitutively expressed in circadian oscillator and non-oscillator cells. J Biol Rhythms 21: 93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung HC, Maurer C, Kay SA, Weber F (2007) Circadian transcription depends on limiting amounts of the transcription co-activator nejire/CBP. J Biol Chem 282: 31349–31357 [DOI] [PubMed] [Google Scholar]
- Hung HC, Maurer C, Zorn D, Chang WL, Weber F (2009) Sequential and compartment-specific phosphorylation controls the life cycle of the circadian CLOCK protein. J Biol Chem 284: 23734–23742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Schoer R, Rosbash M (2008) Circadian transcription contributes to core period determination in Drosophila. PLoS Biol 6: e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M (1998) Neural substrates of Drosophila rhythms revealed by mutants and molecular manipulations. Curr Opin Neurobiol 8: 652–658 [DOI] [PubMed] [Google Scholar]
- Kim EY, Edery I (2006) Balance between DBT/CKIε kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc Natl Acad Sci USA 103: 6178–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Bae K, Ng FS, Glossop NR, Hardin PE, Edery I (2002) Drosophila CLOCK protein is under posttranscriptional control and influences light-induced activity. Neuron 34: 69–81 [DOI] [PubMed] [Google Scholar]
- Klarsfeld A, Leloup JC, Rouyer F (2003) Circadian rhythms of locomotor activity in Drosophila. Behav Processes 64: 161–175 [DOI] [PubMed] [Google Scholar]
- Kula-Eversole E, Nagoshi E, Shang Y, Rodriguez J, Allada R, Rosbash M (2010) Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc Natl Acad Sci USA 107: 13497–13502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Choi HS, Gyuris J, Brent R, Moore DD (1995) Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol 9: 243–254 [DOI] [PubMed] [Google Scholar]
- Lundgren J, Masson P, Mirzaei Z, Young P (2005) Identification and characterization of a Drosophila proteasome regulatory network. Mol Cell Biol 25: 4662–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M (2010) Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev 24: 358–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama KI, Nakayama K (2005) Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol 16: 323–333 [DOI] [PubMed] [Google Scholar]
- Park JH, Helfrich-Förster C, Lee G, Liu L, Rosbash M, Hall JC (2000) Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci USA 97: 3608–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Yoon SK, Yoon JB (2009) The HECT domain of TRIP12 ubiquitinates substrates of the ubiquitin fusion degradation pathway. J Biol Chem 284: 1540–1549 [DOI] [PubMed] [Google Scholar]
- Peschel N, Chen KF, Szabo G, Stanewsky R (2009) Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr Biol 19: 241–247 [DOI] [PubMed] [Google Scholar]
- Rothenfluh A, Young MW, Saez L (2000) A TIMELESS-independent function for PERIOD proteins in the Drosophila clock. Neuron 26: 505–514 [DOI] [PubMed] [Google Scholar]
- Rotin D, Kumar S (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 10: 398–409 [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan S, Zheng X, Kumar S, Chen CH, Chen D, Hay B, Sehgal A (2008) Identification of novel genes involved in light-dependent CRY degradation through a genome-wide RNAi screen. Genes Dev 22: 1522–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R, Frisch B, Brandes C, Hamblen-Coyle MJ, Rosbash M, Hall JC (1997) Temporal and spatial expression patterns of transgenes containing increasing amounts of the Drosophila clock gene period and a lacZ reporter: mapping elements of the PER protein involved in circadian cycling. J Neurosci 17: 676–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri V, Hall JC, Rosbash M (2000) Two novel doubletime mutants alter circadian properties and eliminate the delay between RNA and protein in Drosophila. J Neurosci 20: 7547–7755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F (2009) Remodeling the clock: coactivators and signal transduction in the circadian clockworks. Naturwissenschaften 96: 321–337 [DOI] [PubMed] [Google Scholar]
- Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE (2006) PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev 20: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zheng H, Price JL, Hardin PE (2009) DOUBLETIME plays a noncatalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock. Mol Cell Biol 29: 1452–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.