Abstract
Mitochondria manganese superoxide dismutase (SOD2) is an important antioxidant enzyme, deficiency of which is associated with various human diseases. The known primary regulation of SOD2 is through transcriptional activation. Here, we report that SOD2 is acetylated at Lys 68 and that this acetylation decreases SOD2 activity. Mitochondrial deacetylase SIRT3 binds to, deacetylates and activates SOD2. Increase of reactive oxygen species (ROS) levels stimulates SIRT3 transcription, leading to SOD2 deacetylation and activation. SOD2-mediated ROS reduction is synergistically increased by SIRT3 co-expression, but is cancelled by SIRT3 depletion. These results reveal a new post-translational regulation of SOD2 by means of acetylation and SIRT3-dependent deacetylation in response to oxidative stress.
Keywords: acetylation, ROS, SIRT3, SOD2
Introduction
Superoxide dismutases (SODs) are a class of enzymes that catalyse the detoxification of superoxide into oxygen and hydrogen peroxide, which is then converted to oxygen and water by catalase. SOD is believed to be present in all oxygen-metabolizing organisms, and the physiological role of SOD is to balance the level of intracellular reactive oxygen species (ROS), a product of aerobic metabolism normally produced in the mitochondria. Although low levels of ROS contribute to cell signalling and cell proliferation, excess of ROS, because of its highly reactive nature, causes damage to a range of cellular constituents, including proteins, lipids and, in particular, DNA (Finkel & Holbrook, 2000).
Mammalian cells express three forms of SOD that, although catalysing the same reaction, differ in metal co-factor binding and subcellular localization. SOD1 binds to copper and zinc and localizes in the cytosol; SOD2 binds to manganese and localizes in the mitochondria; SOD3 binds to copper and zinc and is secreted to extracellular fluid. Of these three forms, mitochondrial SOD2 is thought to have a crucial role in controlling the level of ROS, as mitochondria consume over 90% of intracellular oxygen and produce a large flux of ROS. Mutations in SOD2 are associated with ageing and various human diseases including idiopathic cardiomyopathy, sporadic motor neuron disease and cancer (Miao & St Clair, 2009).
Regulation of SOD2 level has so far been mostly reported at the transcriptional level. Oxidative stress-induced SOD2 expression is believed to be an important cellular defence mechanism (Miao & St Clair, 2009). By contrast, little is known about whether the level or function of SOD2 is also regulated post-translationally. Acetylation of the lysine side chain is a post-translational modification that is used extensively in regulation of histones and transcription regulators (Yang & Seto, 2008). Several recent acetylome proteomic studies have identified more than 2,000 potential acetylation substrates, many of which are localized in the cytotosol and mitochondria (Kim et al, 2006; Choudhary et al, 2009; Zhao et al, 2010), markedly expanding the scope of acetylation in cell regulation (Guan & Xiong, 2010). Among these newly identified acetylated proteins is SOD2, implicating a new regulation of SOD2 at the post-translational level. In this study, we investigate the role of acetylation in the regulation of SOD2 activity and in the cellular anti-oxidative response.
Results And Discussion
K 68 is an important acetylation site of SOD2
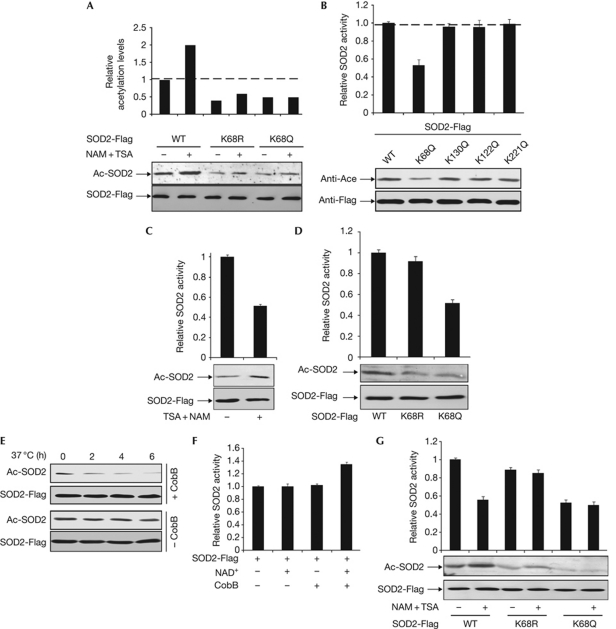
Of the many acetylated peptides identified from mitochondrial fraction, HHAAYVNNLNVTEEKAcYQEALAK is one that matched the human SOD2 sequence with acetylation at K68 (supplementary Fig S1A,B online). The notion that K68 can be acetylated in vivo was substantiated by deacetylase inhibitor treatment experiments. Addition of nicotinamide (NAM) and trichostatin A (TSA), two inhibitors that would inhibit most known deacetylases, caused a more than twofold increase in the acetylation level of ectopically expressed SOD2 (Fig 1A). Consistently, when K68 was changed to either glutamine (SOD2K68Q) or arginine (SOD2K68R), the acetylation levels of the mutant SOD2s decreased by about 50%, and they did not show a significant increase of acetylation after TSA and NAM treatment (Fig 1A). As well as K68, three additional lysine residues, K122, K130 and K221, were also identified by mass spectrometry studies. We generated the site-specific mutants individually targeting these three lysines and determined the effect of these mutations on SOD2 acetylation. Whereas K130Q substitution slightly reduced the level of SOD2 acetylation, both K122Q and K221Q mutations had little effect on SOD2 acetylation (Fig 1B). These results suggest that K68 is the main acetylation site in SOD2 under these assay conditions.
Figure 1.
Acetylation at K68 reduces SOD2 activity. (A) Inhibition of deacetylases increased the acetylation levels of wild-type SOD2, but not SOD2K68Q and SOD2K68R mutants. Flag-tagged SOD2, SOD2K68Q and SOD2K68R were expressed in HEK293T cells with or without TSA (0.5 μM) and NAM (10 mM) treatment and affinity purified. Acetylation levels and enzyme activity of SOD2 in this and other figures were determined by western blot analysis and enzyme assay, respectively. The average value of duplicate and s.d. is shown. (B) K68 is the main site of SOD2 acetylation. SOD2 was expressed in HEK293T and affinity purified, followed by western blotting and enzyme assay. (C) Inhibition of deacetylases decreased SOD2 specific activity. SOD2 was expressed in HEK293T cells, which were treated with or without NAM+TSA. (D) Acetylation mimetic mutation at K68 decreases SOD2 activity. (E,F) SOD2 is activated by in vitro deacetylation. SOD2 was expressed in HEK293T cells, purified by affinity purification and incubated with recombinant CobB. Samples were taken at different time points and analysed for acetylation levels (E) and specific SOD2 activities (F). (G) Inhibition of deacetylases in vivo decreased the activity of wild-type, but not K68 mutant SOD2. SOD2 proteins were expressed in HEK293T cells, followed by treatment with deacetylase inhibitors. HEK, human embryonic kidney; NAM, nicotinamide; SOD; superoxide dismutase; TSA, trichostatin A; WT, wild type.
Acetylation at K68 inhibits SOD2 catalytic activity
Newly synthesized human SOD2 proteins contain 223 amino acids, and cleavage of the amino-terminal 24-signal sequences occurs during import into mitochondria. In mature SOD2, K68 is found in the middle of the N-terminal helical hairpin domain that is responsible for coordinating Mn3+, the metal ion that is required for SOD2 catalytic activity (Borgstahl et al, 1992), suggesting a potential interference with SOD2 catalytic activity by K68 acetylation. To test this hypothesis, we immunopurified ectopically expressed Flag-tagged SOD2 and found that SOD2 purified from cells treated with TSA and NAM showed an approximately 50% decrease in specific activity compared with the enzyme isolated from the control cells, whereas acetylation of SOD2 was increased in the TSA- and NAM-treated samples (Fig 1C). The notion that acetylation inhibits SOD2 activity was supported by the analysis, which showed that acetylation mimetic mutant SOD2K68Q had an approximately 60% stronger decrease of specific activity than the wild-type SOD2. Conversely, substitution of Lys 68 to Arg (SOD2K68R), had a slight, but insignificant, effect on the enzyme activity of SOD2 (Fig 1D).
We then incubated purified SOD2 with bacterial deacetylase CobB. The level of SOD2 acetylation decreased in a time-dependent manner and became barely detectable after 6 h (Fig 1E), whereas the specific activity of SOD2 increased by as much as 35% (Fig 1F). Incubation of purified SOD2 with either NAD+-containing deacetylation buffer without CobB or CobB in the absence of NAD+ had little effect on SOD2 enzyme activity (Fig 1F). These results demonstrate that the in vitro deacetylation contributed to SOD2 activation, suggesting that acetylation negatively regulates SOD2 catalytic activity.
Next, we immunopurified SOD2, SOD2K68Q and SOD2K68R from human embryonic kidney (HEK)293T cells either untreated or treated with deacetylase inhibitors, and determined the specific activity of SOD2. Treatment of cells with TSA and NAM reduced the specific activity of wild-type SOD2 by 44%, but had almost no effect on the activity of either SOD2K68Q or SOD2K68R mutants (Fig 1G). Together, these results support the conclusion that acetylation negatively regulates SOD2 enzyme activity and that K68 is an important regulatory acetylation site.
SOD2 selectively binds to SIRT3
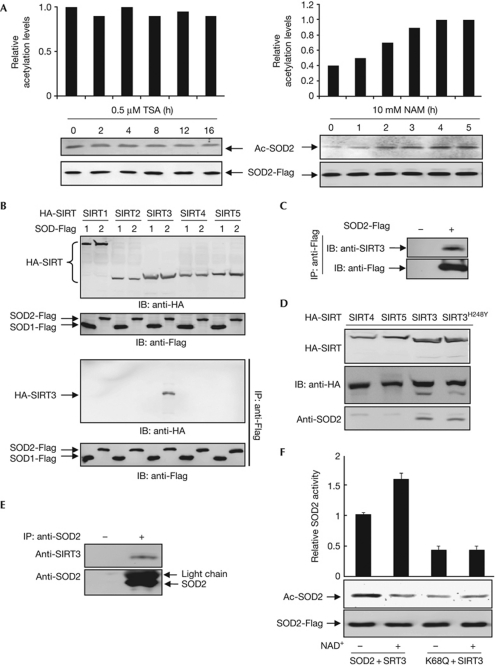
Reversible acetylation is achieved by the action of acetyltransferases and deacetylases. Four classes of 18 lysine deacetylases and more than 20 lysine acetyltransferases have been identified in mammalian cells (Blander & Guarente, 2004; Yang & Seto, 2008). Classes I, II and IV are referred to as ‘classical’ histone deacetylases and are sensitive to inhibition by TSA, whereas the class III NAD+-dependent sirtuin 3 (SIRT) can be inhibited by NAM (Smith et al, 2008). We treated HEK293T cells expressing Flag-tagged SOD2 with either TSA or NAM separately, and found that levels of SOD2 acetylation were unchanged with time in TSA-treated cells, but were increased in a time-dependent manner when cells were treated with NAM (Fig 2A), suggesting that deacetylation of SOD2 is probably catalysed by an NAD+-dependent sirtuin family deacetylase.
Figure 2.
SOD2 interacts with SIRT3. (A) NAM, but not TSA, increases SOD2 acetylation level. HEK293T cells were transfected with plasmids expressing Flag-tagged SOD2 and treated with either NAM (10 mM) or TSA (5 μM), followed by immunoblotting with acetyl lysine antibody and quantified by measuring the relative band density. (B) SOD2 coimmunoprecipitated with transfected SIRT3. SOD2 was expressed in HEK293T cells together with individual HA-tagged SIRTs, followed by IP–western blot analysis. (C) Interaction between transfected SOD2 and endogenous SIRT3. SOD2 was expressed in HEK293T cells, immunoprecipitated and analysed for association with endogenous SIRT3 by western blot. (D) Interaction between ectopically expressed SIRT3 and endogenous SOD2. (E) SOD2 associates with SIRT3 in vivo. Endogenous SOD2 protein was immunoprecipitated from HEK293T cells, resolved by SDS–PAGE, followed by immunoblotting with antibodies to SOD2 and SIRT3. (F) SIRT3 deacetylates SOD2 in vitro. HEK293T cells transfected with plasmid expressing SOD2 were treated with NAM (10 mM). HA-SIRT3 was expressed in HEK293T cells. Both proteins were immunopurified separately and then incubated together in the presence or absence of NAD+, followed by the measurement of catalytic activities of SOD2. HA, haemagglutinin; HEK, human embryonic kidney; IB, immunoblotting; IP, immunoprecipitation; NAM, nicotinamide; PAGE, polyacrylamide gel electrophoresis; SOD2; superoxide dismutase; TSA, trichostatin A.
Given that SOD2 is synthesized in the cytosol and translocates to and exerts its catalytic activity in mitochondria, we determined the interactions between SOD2 and three mitochondrially localized SIRTs—SIRT3, SIRT4 and SIRT5—and two cytoplasm SIRTs—SIRT1 and SIRT2. Reciprocal immunoprecipitation and western blot analyses demonstrate that SIRT3, but not the other SIRTs tested, binds to SOD2 (Fig 2B–D). Furthermore, endogenous SIRT3 can be readily detected in an SOD2 immunocomplex derived from HEK293T cells (Fig 2E), demonstrating a physical association between SOD2 and SIRT3 in vivo. Lastly, incubation of immunopurified SOD2 with separately purified SIRT3 in vitro demonstrates that SIRT3 deacetylates and activates wild-type SOD2 in an NAD+-dependent manner, but has little effect on the SOD2K68Q mutant (Fig 2F), supporting the notion that SIRT3 activates SOD2, at least in part through deacetylating K68.
SIRT3 deacetylates and activates SOD2 to scavenge ROS
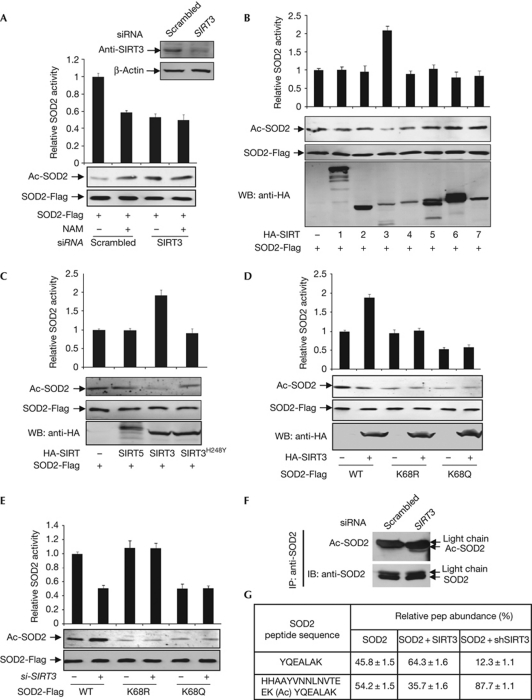
To determine the functional significance of SOD2–SIRT3 interaction, we knocked down SIRT3 in HEK293T cells and determined the enzyme activity of ectopically expressed SOD2 in the presence or absence of NAM. Knocking down SIRT3 reduced SOD2 activity by 46% (Fig 3A), providing additional evidence that acetylation negatively regulates the activity of SOD2. This reduction of SOD2 activity by SIRT3 knockdown was similar to that caused by NAM treatment, which reduced the SOD2 activity by 41%. Moreover, NAM treatment could not further inhibit SOD2 in SIRT3 knockdown cells. Conversely, we found that the co-expression of SIRT3 resulted in a decrease of SOD2 acetylation and a 2.3-fold increase of SOD2 activity, although expression of the other six SIRTs had almost no effect on either SOD2 acetylation or enzyme activity (Fig 3B). Furthermore, although co-expression of the wild-type SIRT3 consistently led to a twofold increase in SOD2 specific activity, co-expression with catalytic mutant SIRT3H248Y has no effect on SOD2-specific activity (Fig 3C), confirming that SIRT3 activates SOD2 through its deacetylase activity, as opposed to through an indirect effect of their physical association with SOD2. Together, these results indicate that SIRT3 is the main deacetylase that activates SOD2 in vivo.
Figure 3.
SIRT3 deacetylates K68 and activates SOD2. (A) Knockdown of SIRT3 increases SOD2 acetylation and decreases SOD2 activity. Flag-tagged SOD2 was expressed in HEK293T cells, followed by transfection of siRNA oligo-targeting SIRT3. (B) SIRT3 activates SOD2. SOD2 was co-expressed in HEK293T cells with individual HA-tagged SIRTs, purified by Flag beads, followed by enzyme assay and western blotting. (C) Wild-type, but not catalytic mutant SIRT3 activates SOD2. SOD2 was expressed in HEK293T cells together with HA-tagged individual SIRTs as indicated. SOD2 proteins were purified by Flag beads. (D) SIRT3 does not activate K68 mutants of SOD2. Wild-type and mutant SOD2 were co-expressed in HEK293T cells with SIRT3 and purified by Flag beads, followed by enzyme assay and western blotting. (E) SIRT3 decaetylates K68 of SOD2. Acetylation levels and catalytic activities of SOD2 expressed and purified from HEK293T cells with or without SIRT3 knockdown were determined. (F) Knocking down SIRT3 increases endogenous SOD2 acetylation. Acetylation levels of endogenous SOD2 in HEK293T cells with or without SIRT3 knockdown were immunopurified and probed by acetyl lysine antibody. (G) Acetylation levels of K68 of SOD2 expressed in HEK293T cells with either SIRT3 overexpressed or knocked down were determined by iTRAQ. HA, haemagglutinin; HEK, human embryonic kidney; IB, immunoblotting; IP, immunoprecipitation; NAM, nicotinamide; siRNA, short-interfering RNA; SOD2; superoxide dismutase; WB, western blotting; WT, wild type.
To determine whether SIRT3 directly acts on the acetylated K68 of SOD2, we tested the ability of SIRT3 to activate wild-type and K68 mutants of SOD2, and found that co-expression of SIRT3 had no effect on the activity of either K68R or K68Q mutants of SOD2 (Fig 3D). Conversely, knocking down SIRT3 with short-interfering RNA (siRNA) reduced the activity of wild-type SOD2, but had no effect on the activity of either K68R or K68Q mutants (Fig 3E). Moreover, knocking down SIRT3 significantly increased the level of acetylated SOD2 (Fig 3F). In a quantitative iTRAQ mass-spectrometric analysis, we found that overexpression of SIRT3 decreased the percentage of K68-acetylated SOD2 from 54.2% to 35.7%, whereas knockdown of SIRT3 increased the percentage of K68-acetylated SOD2 to 87.7% (Fig 3G). Together, these results demonstrate that SIRT3 deacetylates SOD2 in vivo and that K68 is the primary target of SIRT3.
SIRT3 promotes the activity of SOD2 to reduce ROS levels
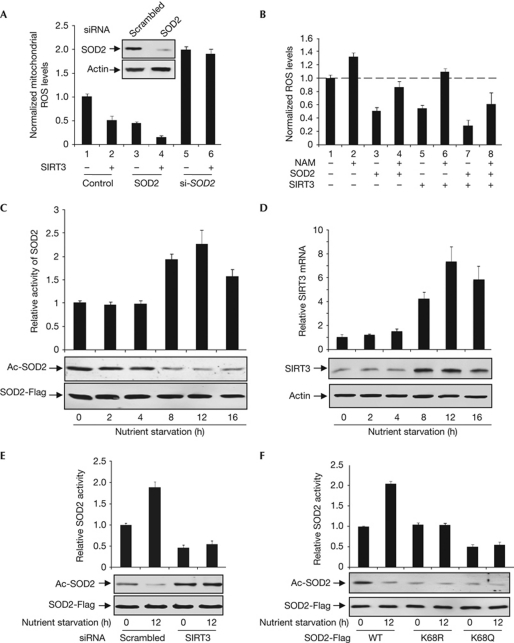
The main biological function of SODs is to remove ROS in cells (Miao & St Clair, 2009). To determine the effect of SOD2 acetylation on its physiological function, we first determined how changes in acetylation levels of mitochondrially localized SOD2 protein affect mitochondrial ROS levels. We measured mitochondrial ROS levels in HEK293T cells with either SOD2 ectopically expressed or knocked down and in combination with SIRT3 co-expression. We found that mitochondrial ROS was reduced by almost 55% by the overexpression of SIRT3 and by 84% by the co-expression of both SIRT3 and SOD2, whereas overexpression of SIRT3 in SOD2-knockdown cells caused no significant mitochondrial ROS reduction (Fig 4A). This supports the idea that SIRT3 enhances the activity of SOD2 to scavenge mitochondrial ROS. We then measured the cellular ROS on gain and loss of function of either SIRT3 or SOD2. We found that overexpression of and knockdown of SOD2 caused a 40% reduction and a 50% increase in cellular ROS levels, respectively (supplementary Fig S3A online). Overexpression and knockdown of SIRT3 caused a 45% decrease and a 28% increase of cellular ROS levels, respectively. Moreover, overexpression of both SIRT3 and SOD2 led to a 65% decrease in ROS levels, showing a synergistic effect of increased SOD2 level and SOD2 deacetylation by SIRT3 in reducing the ROS level. When SOD2 was overexpressed in cells depleted for SIRT3, ROS level, instead of being reduced, was increased by 37%. Similarly, when SIRT3 was overexpressed in cells depleted for SOD2, the cellular ROS level was decreased by only 13%, as opposed to a 43% reduction by SIRT3 overexpression.
Figure 4.
ROS stimulates SIRT3 expression to activate SOD2. (A) SIRT3 reduces mitochondrial ROS levels through SOD2. Plasmids expressing or siRNA targeting SOD2 and SIRT3 were singularly or co-expressed in HEK293T cells, followed by the determination of mitochondrial ROS levels. (B) Inhibition of SIRT activity impairs SOD2 function and SIRT3 to reduce ROS. ROS levels were determined in HEK293T cells with the indicated transfection or treatment. (C) Nutrient starvation decreases SOD2 acetylation level and activates SOD2. Acetylation levels and specific activities of SOD2 expressed in HeLa cells were determined after nutrient starvation by western blotting and enzyme assay, respectively. (D) Nutrient starvation increases SIRT3 mRNA and protein levels. mRNA and protein levels of SIRT3 in HeLa cells after nutrient starvation for different length of times were determined by qRT–PCR and western blotting, respectively. (E) Nutrient starvation regulates SOD2 through SIRT3. Acetylation levels and relative activities of SOD2 expressed in HeLa cells with or without SIRT3 knocked down by siRNA under normal and nutrient starvation conditions were determined. (F) Nutrient starvation regulates SOD2 activity through K68 acetylation. Acetylation levels and relative activity of SOD2, K68R and K68Q mutants expressed under normal or nutrient starvation conditions in HeLa cells were determined. mRNA, messenger RNA; NAM, nicotinamide; ROS, reactive oxygen species; siRNA, short-interfering RNA; SOD2, superoxide dismutase; WT, wild type.
To further corroborate the finding of the role of acetylation in regulating the function of SOD2 in ROS clearance, we ectopically expressed SOD2 and then treated transfected cells with either solvent or NAM. Treatment of cells with NAM alone increased the ROS level by 32% (Fig 4B; comparing lane 1 and lane 2), supporting the function of an NAM-sensitive deacetylase in ROS reduction. Whereas ROS was reduced by 48% in cells ectopically expressing SOD2 (lane 3), the effect of SOD2 expression on ROS was reversed by NAM treatment (lane 4). Similarly, ROS was reduced by 45% in cells overexpressing SIRT3 (lane 5) and was further increased by 10% when the SIRT3-expressing cells were treated with NAM (lane 6), supporting the role of SIRT3 in activating SOD2 to decrease cellular ROS.
Nutrient starvation and ROS stimulate SIRT3 and SOD2
Cell metabolism, especially respiration, is closely associated with mitochondrial and cellular ROS production. We determined the levels of both acetylation and enzyme activity of SOD2 in nutrient-starved cells and found that combined glucose and amino-acid deprivation decreased the levels of SOD2 acetylation (Fig 4C). Associated with the decrease of SOD2 acetylation, SOD2 enzyme activity was increased by 1.9-fold and 2.3-fold after 8 and 12 h of nutrient deprivation. Concurrently, the levels of SIRT3 messenger RNA (mRNA) were stimulated by as much as 3.2- and 7.3- fold after 8- and 12-h nutrient deprivation, respectively. Consistently, SIRT3 protein levels increased (Fig 4D). These results suggest a molecular mechanism—stimulation of SIRT3 gene expression—for the deacetylation and activation of SOD2 by SIRT3 during nutrient deprivation. We next examined the effect of nutrient starvation on SOD2 acetylation and catalytic activity and found that when SIRT3 was knocked down, nutrient deprivation does not affect either SOD2 acetylation level or activity (Fig 4E). We also found that nutrient deprivation caused deacetylation and activation of wild-type SOD2, but had little effect on either acetylation levels or activities of K68Q and K68R mutants (Fig 4F). These two lines of evidence link the nutrient-deprivation effect on SOD2 activity to SIRT3 and K68 acetylation of SOD2.
Treatment of cells with DMNQ, a reagent that is known to increase mitochondria ROS level, decreased SOD2 acetylation and increased SOD2 activity in a time-course-dependent manner, but it had little effect on the steady-state levels of ectopically expressed SOD2 and it increased SIRT3 mRNA and protein levels (supplementary Fig S3B,C online). These results demonstrate that increase of intracellular ROS stimulates SIRT3 gene expression and increases SIRT3 protein levels, leading to decreased SOD2 acetylation and increased SOD2 enzyme activity. Therefore, our data suggest a possible negative feedback mechanism to counterbalance the increase of intracellular ROS by activating SOD2 through SIRT3 induction.
SOD2 is a homotetrameric enzyme and forms a unique intersubunit 4-helix bundle interface that is important not only for the assembly, but also for stabilizing binding to both metal co-factor Mn3+ and substrate superoxide. A ring of 11 positively charged residues, including K68, is formed surrounding the active-site channel (Borgstahl et al, 1992). Acetylation at K68 is expected to decrease its hydrophilicity and thus reduces the affinity with either Mn3+ and/or superoxide, providing a probable structural basis for the negative regulation of SOD2 enzyme activity by acetylation, as well as suggesting that acetylation regulation at K68 might be unique to SOD2.
Deletion of Sirt3 resulted in a marked elevation of acetylation of several mitochondrial proteins (Lombard et al, 2007), suggesting the presence of many substrates of SIRT3 in mitochondria and a broad function of SIRT3 in metabolic regulation. Although Sirt3-null mice are surprisingly normal and lack a tumour phenotype under the normal (that is, without stress) condition, Sirt33−/− mouse embryonic fibroblasts have increased superoxide levels and are susceptible to transformation by a single oncogene (Ras or Myc). Remarkably, overexpression of SOD2 prevents the oncogene-induced immortalization of Sirt3−/− mouse embryonic fibroblasts, suggesting a genetic dependency of SIRT3 on SOD2 in controlling cell proliferation and suppressing cell immortalization (Kim et al, 2010). Several substrates of SIRT3 have been identified, including acetyl-CoA synthetase, succinate dehydrogenase flavoprotein subunit and long-chain acyl CoA dehydrogenase, but none is directly involved in the oxidative stress response. Our demonstration that SOD2 is a direct substrate of SIRT3 and that SIRT3-mediated deacetylation activates SOD2 provides a molecular basis for the function of SIRT3–SOD2 controlling the level of intracellular superoxide and thus cell growth. Recently, two groups reported a similar finding that SIRT3 deacetylates SOD2 to enhance its activity in response to nutrient or ionizing radiation stress (Qiu et al, 2010; Tao et al, 2010). Interestingly, our study and these reports each identified a different site of acetylation in SOD2; we found that K68 is an important site for SIRT3 regulation in human cells, whereas Tao et al identified K122 and Qiu et al identified K53 and K89 as the main sites of Sirt3 deacetylation in mouse cells. It remains to be determined whether the differences are between human and mouse cells, in stress conditions, or whether SIRT3-mediated deacetylation and activation of SOD2 involves several sites. It is also interesting to note that whereas arginine substitution of K53/89 or K122 increased SOD2 activity, K68R mutation did not (Fig 1D,G), suggesting that K68, in addition to acting as a site for acetylation, also contributes to SOD2 enzyme activity that cannot be substituted by an arginine because of different side chains. The molecular basis for this contribution remains to be determined.
Methods
SOD2 assay. The SOD2 activity assay was carried out with the SOD Assay Kit using water-soluble tetrazolium salt (WST-1) as a substrate and following the manufacturer's instruction (Dojindo Molecular Technology Inc.).
In vitro deacetylation. His-CobB was purified with nickel beads and stored at −80°C in 10% glycerol. For in vitro deacetylation assay, 5 μg of SOD2-Flag proteins was incubated with 15 μg His-cobB proteins in HEPES buffer (HEPES 40 mM; MgCl2 1 mM; DTT 1 mM; NAD+ 5 mM) at 30°C for 6 h.
Measurement of intracellular ROS level. ROS production was determined by incubating the HEK392T cells in PBS containing 20-μM fluorescent dye 2′,7′-dichlorofluorescein diacetate (Sigma) at 37 °C for 30 min, followed by flow-cytometric analysis.
Measurement of mitochondrial superoxide levels. Superoxide production was determined by measuring MitoSOX (5 μM) (Invitrogen) oxidation in cells, following the manufacturer's instructions. Cells were cultured as described above and incubated for 10 min at 37 °C before being trypsinized, resuspended and measured by flow cytometry.
iTRAQ quantification. SOD2-Flag proteins were immunopurified from HEK293T cells alone or in combination with SIRT3 knockdown and gel-purified. Equal molar ratios of purified SOD2 proteins and internal control peptides were labelled by iTRAQ labelling reagents (ABI), mixed and subjected to LTQ-OrbiTrap analysis under PQD mode. The resultant mass spectrometry spectra (supplementary Fig S2 online) were used to determine the peptide identity and abundance. The relative abundance of a peptide was calculated by comparing the intensities of the corresponding tag (reporter ion m/z at 117 for SOD2, 118 for SOD2+SIRT3, 119 for peptide and 121 for SOD2+shSIRT3).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
This work is supported by the 985 Programme from the Chinese Ministry of Education, State Key Development Programmes of Basic Research of China (2009CB918401, 2009CB918600), Chinese National Science Foundation grants (31030042, 30971485/C0706, 31030042) and National Institutes of Health grants to K.-L.G. (CA132809) and Y.X. (GM067113).
Footnotes
The authors declare that they have no conflict of interest.
References
- Blander G, Guarente L (2004) The Sir2 family of protein deacetylases. Annu Rev Biochem 73: 417–435 [DOI] [PubMed] [Google Scholar]
- Borgstahl GE, Parge HE, Hickey MJ, Beyer WF Jr, Hallewell RA, Tainer JA (1992) The structure of human mitochondrial manganese superoxide dismutase reveals a novel tetrameric interface of two 4-helix bundles. Cell 71: 107–118 [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther T, Olsen JV, Mann M (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834–840 [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ (2000) Oxidants, oxidative stress and the biology of ageing. Nature 408: 239–247 [DOI] [PubMed] [Google Scholar]
- Guan KL, Xiong Y (2010) Regulation of intermediary metabolism by protein acetylation. Trends Biochem Sci 36: 108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC et al. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 23: 607–618 [DOI] [PubMed] [Google Scholar]
- Kim HS et al. (2010) SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 17: 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB et al. (2007) Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol 27: 8807–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L, St Clair DK (2009) Regulation of superoxide dismutase genes: implications in disease. Free Radic Biol Med 47: 344–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Brown K, Hirschey MD, Verdin E, Chen D (2010) Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab 12: 662–667 [DOI] [PubMed] [Google Scholar]
- Smith BC, Hallows WC, Denu JM (2008) Mechanisms and molecular probes of sirtuins. Chem Biol 15: 1002–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R et al. (2010) Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell 40: 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E (2008) Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell 31: 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S et al. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327: 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.