Abstract
Despite the importance of microRNAs (miRNAs) in gene regulation, it is unclear how the miRNA–Argonaute complex—or miRNA-induced silencing complex (miRISC)—can regulate the translation of their targets in such diverse ways. We demonstrate here a direct interaction between the miRISC and the ribosome by showing that a constituent of the eukaryotic 40S subunit, receptor for activated C-kinase (RACK1), is important for miRNA-mediated gene regulation in animals. In vivo studies demonstrate that RACK1 interacts with components of the miRISC in nematodes and mammals. In both systems, the alteration of RACK1 expression alters miRNA function and impairs the association of the miRNA complex with the translating ribosomes. Our data indicate that RACK1 can contribute to the recruitment of miRISC to the site of translation, and support a post-initiation mode of miRNA-mediated gene repression.
Keywords: ALG-1, hAGO2, RACK1, miRISC recruitment, miRNA
Introduction
Initially discovered in Caenorhabditis elegans, microRNAs (miRNAs) have emerged as common cellular components with conserved functions in both animals and plants. In all species, miRNAs associate with Argonaute proteins to form the core–effector complex, known as the miRNA-induced silencing complex (miRISC), which is able to alter protein synthesis and/or induce messenger RNA (mRNA) destabilization. In animals, there are several members of the Argonaute gene family, all of which are essential for small-RNA-mediated silencing pathways (reviewed in Hutvagner & Simard, 2008).
It has been reported that the miRISC can influence translation in distinct ways (reviewed in Filipowicz et al, 2008). Interestingly, in Drosophila, the identity of the Argonaute protein associated with miRNA can dictate the mechanism that will lead to translational inhibition (Iwasaki et al, 2009). This indicates that the Argonaute constituent of the miRISC, as well as its interacting proteins, contribute to regulating protein expression by different mechanisms.
In this study, we identify the receptor for activated C-kinase (RACK1) as an interactor of both C. elegans and human miRISC, and demonstrate the importance of RACK1 for miRNA-mediated gene silencing in both systems. We observe that the loss of RACK1 affects the association of miRNA and Argonaute with translating ribosomes, suggesting that this component of the 40S ribosomal subunit can mediate the recruitment of the miRISC to the active site of translation.
Results and Discussion
RACK1 interacts with the miRISC of C. elegans
To gain a better insight into the miRNA pathway, we carried out a two-hybrid screen to identify the proteins that interact with ALG-1; one of the two Argonaute proteins that are essential for miRNA-mediated gene regulation in C. elegans (Grishok et al, 2001). Among the proteins interacting with ALG-1, we identified K04D7.1, the orthologue of the mammalian protein RACK1 (supplementary Fig S1 online). C. elegans RACK1 (ceRACK1) has also been identified by mass spectrometry as a constituent of the ALG-1 complex (Chan & Slack, 2009). To confirm the relevance of this interaction, we generated tagged recombinant ALG-1 and ceRACK-1 proteins and performed glutathione S-transferase (GST) pulldown assays. When compared with beads coupled to GST protein, we observed that GST-tagged ALG-1 efficiently pulled down His-ceRACK-1 (Fig 1A). This suggests that ceRACK1 is a bona fide interactor of ALG-1 in C. elegans.
Figure 1.
RACK1 interacts with constituents of C. elegans and the human miRISC. (A) Recombinant ceRACK-1 interacts with GST-tagged ALG-1. Western blot analysis of GST pulldowns of His-tagged ceRACK-1 incubated with GST or GST-ALG-1 and probed with His antibody. Input represents the equivalent of 10% of the His-RACK1 used for pulldown. (B) ceRACK-1 associates with the miRISC in C. elegans. Extracts from MJS10 strain were incubated with the let-7-complementary (let-7), lin-4-complementary (lin-4) or nonspecific (ctl) tethered 2′-O-methyl oligonucleotide (2′-O-Me oligo). Beads were washed, and bound GFP∷ALG-1 and HA∷ceRACK-1 were detected by western blotting. (C) Interaction of ceRACK-1, but not RPS-12, with the miRISC is partly resistant to RNase treatment. Strains expressing tagged proteins were incubated with the let-7-complementary or nonspecific (control) tethered 2′-O-methyl oligonucleotide. Beads were washed and treated (+) or not (−) with RNase, followed by the detection of GFP∷ALG-1, HA∷ceRACK-1 and HA∷RPS-12 by western blotting. In B and C, input represents the equivalent of 5% of the total extract incubated with tethered oligonucleotides. Dashed lines indicate that unrelated lanes have been removed between samples. Relative intensities of the signal are shown under each lane in B and C. (D) Knockdown of ceRACK-1 causes heterochronic defects similar to those observed for alg-1(RNAi). Adult alae defects and bursting vulva were scored on young adult animals fed with bacteria expressing either control, rack-1 or alg-1 double-stranded RNA. Developmental (dev.) defects observed include one or more of the following: larval lethality, moulting defects, vulva and gonadal morphogenesis defects. The number (n) of animals scored is indicated. Asterisk: as alg-2(lf); rack-1(RNAi) animals exhibit severe larval lethality and developmental arrest, sufficient numbers of adult-stage animals could not be obtained to reliably scored vulval bursting; however, alae defects were scored for 60 animals that managed to reach the adult stage. (E) Representative adult alae defect (Nomarski optics) observed in rack-1(RNAi) animals. The white bars show the region of adult cuticles lacking alae or with abnormal alae morphology. Magnification is × 630. (F) Extra seam cells (top panel) and defect in seam-cell fusions (arrows; bottom panel) are observed in adult rack-1(RNAi) animals. The magnification of both pictures is × 1,000. GFP, green fluorescent protein; GST, glutathione S-transferase; HA, haemagglutinin; miRISC, miRNA-induced silencing complex; NA, not applicable; ND, not determined; RACK, receptor for activated C-kinase; RNAi, RNA interference; WT, wild type.
To address whether this interaction can occur in vivo, we generated a transgenic worm expressing green fluorescent protein (GFP)∷ALG-1 and haemagglutinin (HA)∷ceRACK-1 to overcome the lack of specific antibodies. Using a whole-worm lysate prepared from the transgenic C. elegans, we immunoprecipitated GFP∷ALG-1 with HA∷ceRACK-1 and conversely detected HA∷ceRACK-1 in the GFP∷ALG-1 pulldown (supplementary Fig S2B online). To determine whether ceRACK-1 can also interact with miRNAs, we used a 2′-O-methylated RNA affinity matrix to trap sequence-specific small RNA complexes that are not bound to target mRNAs (Hutvágner et al, 2004; Yigit et al, 2006). By using this method, we found that both ALG-1 and ceRACK-1 are associated with let-7 and lin-4 miRNAs (Fig 1B). To determine whether the interaction between ceRACK1 and the miRISC is only a general interaction with the ribosomes, we generated transgenic animals expressing a HA-tagged 40S ribosomal protein. We observed that although the RNase treatment almost abolished the interaction between the let-7 miRNA complex and the 40S subunit RPS-12 (Fig 1C; supplementary Fig S2C online), a significant fraction of ceRACK-1 remains associated with the let-7 complex (Fig 1C). Therefore, our findings provide evidence that free miRISC interacts with the 40S ribosomal subunit, and ceRACK-1 contributes to this interaction.
RACK1 is important for miRNA function in C. elegans
In C. elegans, the miRNA pathway has an important role in the control of animal development. The loss of function of genes associated with this pathway results in pleiotropic phenotypes, probably reflecting their roles in the activity of all C. elegans miRNAs (Grishok et al, 2001; Denli et al, 2004; Hammell et al, 2009; Bussing et al, 2010). To examine whether rack-1 is important for the miRNA pathway in C. elegans, we depleted rack-1 in animals using RNA interference (RNAi) feeding delivery (animals carrying loss-of-function alleles of rack-1 gene are embryonic lethal; data not shown). Although the depletion of the 40S ribosomal subunit rps-12 leads mostly to embryonic and larval lethality (data not shown), rack-1(RNAi) shows developmental-timing delay, including heterochronic phenotypes that include defects in adult alae (Fig 1D–F). We also found that a significant proportion of rack-1(RNAi) animals burst from the vulval opening after L4 moult, a phenotype characteristic of let-7-family miRNA mutants (Abbott et al, 2005: Fig 1D). All these phenotypes are similar to those that we observed in alg-1(RNAi) animals and are enhanced in alg-2(lf) single-mutant animals (Fig 1D). The similarity of the phenotypes resulting from the loss of function of rack-1 and the depletion of core components of the miRNA pathway, the synergy observed with alg-2, and the other Argonaute essential for this pathway in C. elegans (Grishok et al, 2001), supports the conclusion that rack-1 functions in the C. elegans miRNA pathway.
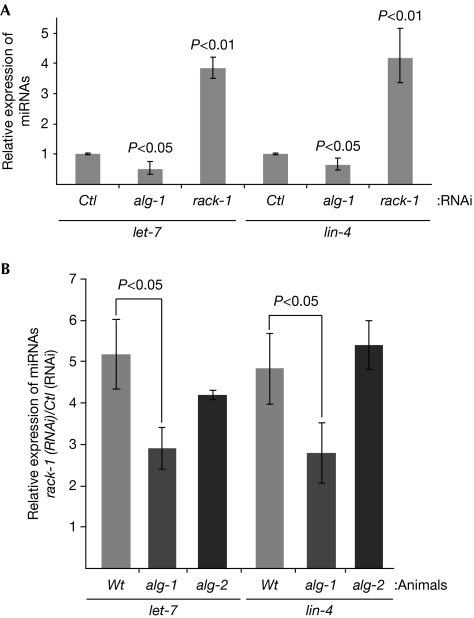
To better understand the function of rack-1 in the miRNA pathway, we decided to monitor the miRNA levels of C. elegans that was subjected to rack-1 RNAi. Although, as reported recently (Kato et al, 2009), we observed a significant decrease in the amount of miRNAs in alg-1(RNAi) animals (Fig 2A), the knockdown of rack-1 in C. elegans led to an increase in let-7 and lin-4 miRNA levels (Fig 2A). Interestingly, this accumulation of miRNAs was attenuated in the absence of alg-1 but was unaffected in alg-2(lf) animals (Fig 2B). This indicates that the accumulation of miRNAs observed in rack-1(RNAi) animals requires the Argonaute ALG-1, but not ALG-2.
Figure 2.
ceRACK-1 affects the level of miRNA in an alg-1-dependent manner. (A) Although alg-1(RNAi) reduces miRNA levels, rack-1(RNAi) leads to an accumulation of miRNAs. (B) ALG-1, but not ALG-2, is important to accumulate miRNAs in rack-1(RNAi) animals. The miRNA levels were measured relative to the small nucleolar RNA (sn2841) using quantitative reverse transcription–PCR (TaqMan Assays) in adult animals fed with bacteria expressing either control (Ctl), alg-1 or rack-1 double-stranded RNA, as indicated. In all RNAi conditions, no significant changes were observed in the level of the control RNA sn2841 (data not shown). The error bars represent the 95% confidence interval from three independent experiments and a Student's two-sided t-test was applied on the normalized Ct values to obtain P-values. miRNA, microRNA; RACK, receptor for activated C-kinase; RNAi, RNA interference; Wt, wild type.
Although rack-1 and alg-1 knockdowns result in similar heterochronic phenotypes (Fig 1D), the opposite effect on miRNA levels suggests that ALG-1 and ceRACK-1 are not functioning at the same point in the miRNA pathway. It is likely that RACK1 is not required for loading and stabilization of miRNAs. The simplest explanation for the accumulation of miRNAs in rack-1(RNAi) animals is that the lack of RACK1, which causes failed regulation, could result in a slower turnover of assembled miRISCs. Interestingly, it has been reported that the interaction between the miRNA and its mRNA target leads to the degradation of the miRNA (Ameres et al, 2010). Thus, the accumulation of miRNAs observed in the absence of ceRACK-1 might indicate that a large amount of ALG-1–miRNA complex cannot reach its mRNA target, leading to miRNA accumulation.
RACK1 interacts with the human miRISC
Because RACK1 is a highly conserved protein in eukaryotes (supplementary Fig S3 online), we next asked whether RACK1 associates with Argonautes in mammals. Although we detected ribosomal RNAs, the ribosomal 40S subunit RPS3, human Ago2 (hAgo2) and miRNAs such as let-7 and miR-21 in the RACK1 immunoprecipitate from HeLa cell lysate, we did not observe an interaction between hAgo1 and RACK1 (Fig 3; supplementary Fig S2D,E online). This is surprising as hAgo1 is a member of the Argonaute gene family that is able to bind to miRNAs (Liu et al, 2004) and is also involved in translational repression. However, the mechanism of its action—which could be different from hAgo2—is not known (Schmitter et al, 2006). The treatment of the samples with RNase A does not abrogate the association between RACK1 and the miRISC, suggesting that part of the interaction is either direct or mediated by other proteins (Fig 3B; supplementary Fig S2D online). Thus, as observed in C. elegans, components of the mammalian miRISC interact in vivo with RACK1.
Figure 3.
RACK1 interacts with human Ago2 and miRNAs. (A) Human RACK1 binds to both Ago2 and miRNAs. RACK1 was immunoprecipitated with monoclonal RACK1 antibody (RACK1) and non-conjugated Protein A beads as a negative control. Input represents the equivalent of 4% of the total extract used for immunoprecipitation. The immunoprecipitate was tested for the presence of RACK1, hAgo2 and hAgo1 by western blotting and let-7 miRNA by northern hybridization. (B) RACK1 interaction with hAgo2 and the miRNA miR-21 is RNA independent. Immunoprecipitations were performed with mouse monoclonal antibodies raised against RACK1 and GFP (used as a negative control) as indicated. Input represents 10% of total lysate used for the immunoprecipitation. The immunoprecipitates were treated with RNase A for 1 h at 4°C. The supernatant (S) and the immunoprecipitate (B) samples, with or without RNase treatment, were tested for the presence of hAgo2, RACK1 and miR-21. GFP, green fluorescent protein; hAgo1/2, human Ago1/2; IP, immunoprecipitation; miRNA, microRNA; RACK, receptor for activated C-kinase; RNAi, RNA interference.
Human miRNA gene silencing requires RACK1
Next, we tested whether RACK1 is required for miRNA-mediated translational repression in mammalian cells. When cells were treated with short-interfering RNA (siRNA)-targeting RACK1, a marked increase in the expression of the endogenous IMP1 and RAS proteins—two characterized let-7 targets in human cells—was observed (Johnson et al, 2005; Selbach et al, 2008; Fig 4A). However, RACK1 knockdown did not alter the steady-state level of IMP1 mRNA, suggesting post-transcriptional regulation (supplementary Fig S4A online). To demonstrate that the effect of RACK1 on translation requires miRNAs, we carried out dual luciferase assays with a miRNA-regulated reporter construct that contains eight tandem let-7 sites (Iwasaki et al, 2009) and a luciferase reporter that contains part of the let-7-targeted HMGA2 3′UTR. The knockdown of RACK1 with three independent siRNAs significantly and specifically altered the expression of both luciferase reporters (Fig 4B; supplementary Fig S5 online). To show that this effect is specific to RACK1, we altered the expression of the human 40S ribosomal subunit RPS3. The knockdown of RPS3 resulted in a general inhibition of translation, without any specific effect on the miRNA-targeted luciferase reporter (supplementary Fig S6 online). Moreover, when we used a reporter that contains three target sites complementary to endogenous let-7a, we observed no change in luciferase expression (supplementary Fig S7 online). Therefore, RACK1 seems to be required to mediate miRNA-dependent translational repression, but not for RNAi.
Figure 4.
RACK1 is required for miRNA silencing in human cells. (A) RACK1 is required for the silencing of the human IMP1 and RAS. Lysates from RNAi-treated cells were immunoblotted with RACK1, IMP1 and RAS antibodies, as indicated. Tubulin immunoblot acted as a loading control. (B) RACK1 affects the translation of a reporter that carries miRNA target sites. Renilla luciferase constructs that contain a 3′UTR with either eight let-7 miRNA target sites or no let-7 miRNA target sites were transfected in RNAi-treated HeLa cells. Firefly luciferase was used as an internal control. The graph represents the quantification of the dual luciferase assay from five repeats and the error bars represent standard propagated errors and significance that were analysed with a Student's t-test. RL:8 let-7 sites: reporter contains eight tandem let-7 miRNA target sites. RL:no let-7 site: reporter without let-7 miRNA site. Right panel, detection of RACK1 shows the efficiency of the knockdown. Tubulin was used as a loading control. Ctl, control; miRNA, microRNA; RACK, receptor for activated C-kinase; RL, renilla luciferase; RNAi, RNA interference; siRNA, short-interfering RNA.
RACK1 RNAi impairs miRISC association with polysomes
As RACK1 has been identified as a core component of the ribosome (reviewed in Nilsson et al, 2004), we decided to test whether RACK1 contributes to the recruitment of the miRISC to the translational machinery. We carried out sucrose gradient fractionation to monitor distribution of the let-7 miRNA in rack-1(RNAi) and control(RNAi) animals. Although neither the polysome distribution in the rack-1(RNAi) population (supplementary Fig S8C online) or the distribution of let-7 mRNA targets such as lin-41 and daf-12 is significantly affected (Reinhart et al, 2000; Slack et al, 2000; Vella et al, 2004; Grosshans et al, 2005; Fig 5A), the amount of let-7 miRNA associated with polysomes is reduced, compared with control(RNAi) animals (Fig 5A; P<0.005).
Figure 5.
RACK1 is important to recruit the miRISC to translating ribosomes. (A) The level of let-7 and let-7 targets, lin-41 and daf-12, in each fractionation collected from the sucrose gradient was monitored by quantitative reverse transcription–PCR. Mean fold change of let-7 and mRNA target RNA molecules associated with polysomes fractions of rack-1(RNAi) animals relative to animals exposed to control (ctl) is shown. Two independent experiments were performed in replicates. Student's two-sided t-test was used to assess the significance of the polysomal distribution of let-7 miRNA between control(RNAi) and rack-1(RNAi) animals. Before the t-test, the Shapiro–Wilk test confirmed that the data were normally distributed. (B) Ribosomal proteins were pelleted with a sucrose cushion from HeLa cells that were treated with control or RACK1 siRNAs, and the pellet-associated hAgo2 was quantified and presented as a ratio of the sum of the free and polysome-associated hAgo2. The graph is derived from five independent experiments carried out with three independent RACK1 siRNAs. Error bars represent the standard deviation, and a Student's t-test was performed to determine the significance of the data. hAgo2, human Ago2; miRNA, microRNA; mRNA, messenger RNA; RACK, receptor for activated C-kinase; RNAi, RNA interference; siRNA, short-interfering RNA.
Human Argonautes have been shown to be associated with polysomes, as their association with heavy fractions is abrogated by puromycin treatment (Nottrott et al, 2006). We also observed that hAgo2 cofractionates with RACK1 on the polysomes, by using the sucrose gradient fractionation approach (supplementary Fig S2F online). We tested whether the presence of hAgo2 with polysomes requires RACK1. To monitor the amount of hAgo2 associated with ribosomes, we pelleted ribosomal complexes by using a sucrose cushion (Halbeisen et al, 2009). Compared with control, the level of hAgo2 copelleted with ribosomes is decreased when RACK1 is depleted by RNAi (Fig 5B; supplementary Fig S8B online), suggesting that RACK1 contributes to the recruitment of hAgo2 to ribosomes.
This data suggests that RACK1 contributes to miRNA-mediated gene regulation at a post-initiation step. Indeed, RACK1 is a stoichiometric component of the 40S ribosome that is positioned at the exit channel to mediate these types of regulations (Sengupta et al, 2004; Coyle et al, 2009). This could explain the mechanism by which miRNA regulates target gene expression at the elongation step (Olsen & Ambros, 1999; Kim et al, 2004; Nelson et al, 2004; Maroney et al, 2006; Nottrott et al, 2006), and how miRNAs could regulate through target sites in coding regions (Hafner et al, 2010). In Saccharomyces cerevisiae, RACK1 is required to recruit Scp160p to specific mRNAs and thus modulate their translations (Baum et al, 2004). These observations and our results suggest that RACK1 functions as an evolutionarily conserved molecular adaptor on ribosomes to recruit a variety of regulators of mRNA translation—such as miRISC—and facilitates their interactions with the translational machinery at diverse steps of translation. The RACK1-dependent miRNA-mediated gene regulation is probably different from canonical GW182-dependent miRNA-mediated gene regulation, and it might also be a fail-safe mechanism that can capture miRNA-targeted mRNAs that escape regulation at the initiation step.
Methods
Nematode methods. C. elegans strains were grown under standard conditions (Brenner, 1974). Transgenic lines MJS10 and MJS17 were produced by microinjection, as described in Mello & Fire (1995). RNAi experiments were conducted with synchronized animal populations, as described previously (Chendrimada et al, 2007).
C. elegans polysome fractionation. C. elegans polysome fractionation was performed as described by Ding & Grosshans (2009). Quantitative detection of mRNAs and miRNAs were performed using TaqMan Gene Expression and microRNA Assay kits (Applied Biosystems), respectively.
Human cells sucrose cushion. HeLa lysates (lysis buffer: 1% NP-40, 10-mM HEPES (pH 7.4), 150-mM KCl, 5-mM MgCl2, 0.25-mM DTT, 50-μM cycloheximide, 0.4-U/μl RNAsin and protease inhibitors) were pretreated with cyclohexamide and spun down (10,000 g, 10 min, 4°C), and equivalent amounts of supernatant were layered onto a 0.5-M sucrose cushion. Samples were then spun at 107,400 g for 45 min at 4°C in an Optima Max Ultracentrifuge.
siRNA transfection in human cells. Cells were plated at 1.25 × 105 per well of a six-well plate. For each well, 200 pmol of siRNA was diluted in 175 μl of Opti-MEM (GIBCO). A volume of 2 μl of oligofectamine (Invitrogen) was diluted in 13-μl Opti-MEM and incubated at 20°C for 5 min. The siRNA mixture and the oligofectamine mixture were mixed and incubated at 20°C for 20 min. The sample was then added to the well and mixed gently. The media were replaced after 4–6 h.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank V. Ambros, R. Utley, A. Sobala and members of our laboratories for comments on the manuscript. We also thank G. Meister for hAgo1 and hAgo2 antibodies and E. Paquet for statistical support. Some nematode strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. This work was funded by the Wellcome Trust and European Framework 6 SIROCCO consortium fund (G.H.) and the Canadian Institutes of Health Research (CIHR; M.J.S.). S.B. is a participant in the Wellcome Trust PhD programme. G.H. is a Wellcome Trust Career Development fellow and M.J.S. is a CIHR New Investigator.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V (2005) The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell 9: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD (2010) Target RNA-directed trimming and tailing of small silencing RNAs. Science 328: 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum S, Bittins M, Frey S, Seedorf M (2004) Asc1p, a WD40-domain containing adaptor protein, is required for the interaction of the RNA-binding protein Scp160p with polysomes. Biochem J 380: 823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussing I, Yang JS, Lai EC, Grosshans H (2010) The nuclear export receptor XPO-1 supports primary miRNA processing in C. elegans and Drosophila. EMBO J 29: 1830–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SP, Slack FJ (2009) Ribosomal protein RPS-14 modulates let-7 microRNA function in Caenorhabditis elegans. Dev Biol 334: 152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R (2007) MicroRNA silencing through RISC recruitment of eIF6. Nature 447: 823–828 [DOI] [PubMed] [Google Scholar]
- Coyle SM, Gilbert WV, Doudna JA (2009) Direct link between RACK1 function and localization at the ribosome in vivo. Mol Cell Biol 29: 1626–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ (2004) Processing of primary microRNAs by the microprocessor complex. Nature 432: 231–235 [DOI] [PubMed] [Google Scholar]
- Ding XC, Grosshans H (2009) Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J 28: 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34 [DOI] [PubMed] [Google Scholar]
- Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ (2005) The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev Cell 8: 321–330 [DOI] [PubMed] [Google Scholar]
- Hafner M et al. (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbeisen RE, Scherrer T, Gerber AP (2009) Affinity purification of ribosomes to access the translatome. Methods 48: 306–310 [DOI] [PubMed] [Google Scholar]
- Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V (2009) nhl-2 modulates microRNA activity in Caenorhabditis elegans. Cell 136: 926–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ (2008) Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 9: 22–32 [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ, Mello CC, Zamore PD (2004) Sequence-specific inhibition of small RNA function. PLoS Biol 2: E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Kawamata T, Tomari Y (2009) Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol Cell 34: 58–67 [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ (2005) RAS is regulated by the let-7 microRNA family. Cell 120: 635–647 [DOI] [PubMed] [Google Scholar]
- Kato M, de Lencastre A, Pincus Z, Slack FJ (2009) Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol 10: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G (2004) Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA 101: 360–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Maroney PA, Yu Y, Fisher J, Nilsen TW (2006) Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol 13: 1102–1107 [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A (1995) DNA transformation. Method Cell Biol 48: 451–482 [PubMed] [Google Scholar]
- Nelson PT, Hatzigeorgiou AG, Mourelatos Z (2004) miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA 10: 387–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Sengupta J, Frank J, Nissen P (2004) Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep 5: 1137–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott S, Simard MJ, Richter JD (2006) Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol 13: 1108–1114 [DOI] [PubMed] [Google Scholar]
- Olsen PH, Ambros V (1999) The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol 216: 671–680 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906 [DOI] [PubMed] [Google Scholar]
- Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, Svoboda P, Filipowicz W (2006) Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res 34: 4801–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Sengupta J, Nilsson J, Gursky R, Spahn CM, Nissen P, Frank J (2004) Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat Struct Mol Biol 11: 957–962 [DOI] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G (2000) The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 5: 659–669 [DOI] [PubMed] [Google Scholar]
- Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ (2004) The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev 18: 132–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC (2006) Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127: 747–757 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.