Abstract
Telomeric repeat-containing RNA (TERRA) has been implicated in the control of heterochromatin and telomerase. We demonstrate that yeast TERRA is regulated by telomere-binding proteins in a chromosome-end-specific manner that is dependent on subtelomeric repetitive DNA elements. At telomeres that contain only X-elements, the Rap1 carboxy-terminal domain recruits the Sir2/3/4 and Rif1/2 complexes to repress transcription in addition to promoting Rat1-nuclease-dependent TERRA degradation. At telomeres that contain Y′ elements, however, Rap1 represses TERRA through recruitment of Rif1 and Rif2. Our work emphasizes the importance of subtelomeric DNA in the control of telomeric protein composition and telomere transcription.
Keywords: non-coding RNA, Rap1, telomere, TERRA
Introduction
Telomeres are essential heterochromatic structures at chromosome ends that ensure genome stability, mediate chromosome positioning and regulate the lifespan of cells that lack telomerase. In Saccharomyces cerevisiae, telomeric DNA is bound by the repressor/activator protein Rap1. Rap1 is essential for telomere-length regulation, chromosome-end protection and silencing of subtelomeric inserted reporter genes (Kyrion et al, 1993; Shore, 1994; Marcand et al, 1997; Pardo & Marcand, 2005). Rap1 elicits many of its functions through the recruitment of Sir proteins (Sir2, 3 and 4) and Rif proteins (Rif1 and 2) by its carboxy-terminal domain (Hardy et al, 1992; Moretti et al, 1994; Cockell et al, 1995; Wotton & Shore, 1997). The histone deacetylase Sir2 functions with Sir3 and Sir4 to promote silencing at telomeres (Rine & Herskowitz, 1987; Aparicio et al, 1991), whereas Rif1 and Rif2 regulate telomere length by mediating the Rap1-counting mechanism for telomerase control (Hardy et al, 1992; Shore, 1994; Wotton & Shore, 1997). In addition to telomeric DNA and its associated proteins, yeast telomeres contain the non-coding telomeric repeat-containing RNA (TERRA), which is transcribed by the RNA polymerase II (RNAPII; Luke et al, 2008).
TERRA is conserved from yeast to humans (Luke & Lingner, 2009) and localized at telomeres in mammalian cells (Azzalin et al, 2007; Schoeftner & Blasco, 2008; Zhang et al, 2009), indicating that it might have an important telomeric function. Precise TERRA regulation promotes genomic stability, as both depletion and accumulation of TERRA have been reported to cause telomeric abnormalities (Azzalin et al, 2007; Deng et al, 2009). Moreover, several findings indicate that TERRA might regulate telomerase (reviewed by Luke & Lingner, 2009). For example, TERRA-mimicking RNA oligonucleotides inhibit telomerase activity in vitro (Schoeftner & Blasco, 2008; Redon et al, 2010). When the function of the yeast nuclear 5′–3′ exonuclease Rat1 is reduced, TERRA accumulates and telomeres shorten because telomerase-mediated elongation is impaired (Luke et al, 2008). Finally, forced telomere transcription (by using the Gal 1,10-promoter) leads to shortening of the transcribed telomere in cis (Sandell et al, 1994).
Although the function of TERRA is unknown, aspects of its regulation have been characterized. In human cells, the nonsense-mediated RNA decay machinery negatively regulates TERRA at chromosome ends (Azzalin et al, 2007; Chawla & Azzalin, 2008), whereas the DNA methyltransferases DNMT3b and DNMT1 methylate TERRA promoters within CpG islands and downregulate its expression (Nergadze et al, 2009). The MLL histone methyltransferase promotes TERRA transcription, probably through a p53-dependent mechanism (Caslini et al, 2009). In yeast, degradation of TERRA is regulated by the 5′–3′ exonuclease Rat1, whereas the poly(A) polymerase Pap1 contributes to its stability (Luke et al, 2008).
Here, we demonstrate that TERRA regulation relies on the presence or absence of Y′ elements, which are repetitive sequences found in approximately 50% of the subtelomeric regions (Chan & Tye, 1983; Walmsley et al, 1984; Louis, 1995). At telomeres containing Y′ elements, Rap1, in conjunction with its interacting partners Rif1 and Rif2, regulates TERRA. Conversely, at telomeres that only contain the so-called X-element, Rap1 promotes both TERRA repression through Rap1 binding to the SIR (2/3/4) complex and Rat1-mediated TERRA degradation. Therefore, subtelomeric repetitive elements determine distinct Rap1-mediated regulatory pathways for TERRA transcription and degradation.
Results And Discussion
Differential regulation of TERRA at Y′ and X-only telomeres
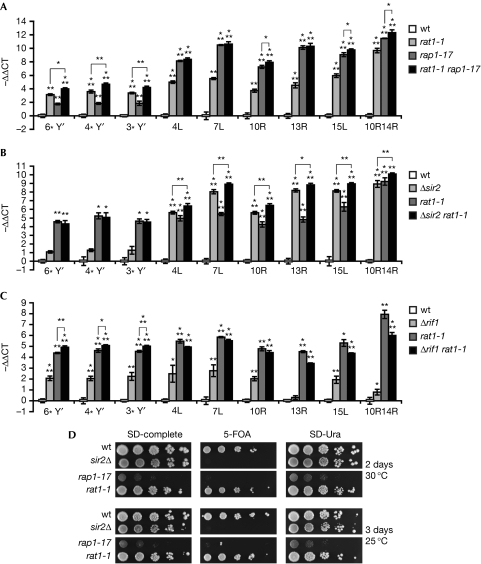
We developed reverse transcription coupled to quantitative real-time PCR protocols to measure TERRA derived from Y′-containing and Y′-lacking chromosome ends. For Y′ telomeres, three sets of primer pairs measure TERRA from different subsets of chromosomes that contain the highly conserved Y′ elements in their subtelomeres (Fig 1A). Conversely, primer pairs with single chromosome-end specificity could be designed to measure TERRA transcribed from several chromosome ends harbouring only an X-element (X-only, from telomere 4L, 7L, 10R, 13R, 15L or 10R14R; Fig 1). As Rap1 binds directly to the double-stranded telomere repeats (Berman et al, 1986; Shore & Nasmyth, 1987) and functions as either an activator or a repressor of transcription (Shore, 1994), we sought to determine whether Rap1 is a regulator of TERRA transcription. TERRA levels were analysed in the C-terminally truncated rap1-17 mutant (Δ663–827 aa) in which both Sir2/3/4 and Rif1/2 can no longer be recruited to chromosome ends (Shore, 1994). We found that TERRA levels were upregulated at all telomeres, with a much greater effect (more than 100-fold) at X-only-containing telomeres (Fig 1B). To determine whether the Sir2/3/4 complex or Rif1 and Rif2 were responsible for TERRA repression, we prepared RNA from sir2/3/4 deletion mutants and from rif1Δ and rif2Δ deletion mutants. We found that the sir2Δ, sir3Δ and sir4Δ mutants strongly derepressed TERRA at X-only-containing telomeres, with weaker effects at Y′ telomeres (Fig 1C). By contrast, the rif1Δ mutant affected TERRA levels at all telomeres (Fig 1D). The rif2Δ mutation derepressed TERRA only slightly; however, when combined with rif1Δ, TERRA derepression was stronger than that in the single mutants at all tested telomeres (Fig 1D). Overall, the analysis suggests that TERRA repression at X-only telomeres is mostly mediated by Sir2, Sir3 and Sir4, whereas Rif1 and Rif2, individually, only contribute slightly to TERRA repression at both X and Y′ telomeres. At Y′ telomeres, Sir proteins have a smaller role and TERRA repression is mostly mediated by Rif1 and, to a lesser extent, by Rif2. Thus, the transcriptional control of Y′ TERRA is similar to that of experimentally inserted subtelomeric reporter genes at Y′ telomeres, which do not rely on Sir2/3/4 (Pryde & Louis, 1999). Furthermore, it has been demonstrated by chromatin immunoprecipitation that the Sir3 protein does not localize to Y′ telomeres (Zhu & Gustafsson, 2009). Rif1 and Rif2 might be regulating Y′ TERRA by transcription or through its rate of turnover.
Figure 1.
Regulation of TERRA by Rap1 complexes. (A) Representation of the Y′ and X-only telomeres indicating the positions of the primers (black bars) used in qRT–PCR analysis. Arrows represent the telomeric tract. Distances to the telomeric tract (bases) are indicated below. (B) Rap1 negatively regulates TERRA. qRT–PCR analysis of Y′ and X-only TERRA in wt and rap1-17 cells. Average values of three independent biological replicates normalized against actin with standard deviation are shown as fold change over wt. The wt value is arbitrarily set to 1. Statistical analyses were calculated using Student's t-test (*P<0.05, **P<0.01 and ***P<0.001). (C) Sir2, 3 and 4 are negative regulators of TERRA at X-only telomeres. qRT–PCR analysis of Y′ and X-only TERRA in indicated cells. Average values were normalized and expressed as in (B). (D) Rif1 and Rif2 affect both Y′ and X-only TERRA levels. qRT–PCR analysis of Y′ and X-only TERRA in indicated cells. Average values were normalized and expressed as in (B). qRT–PCR, quantitative real-time PCR; TERRA, Telomeric repeat-containing RNA; wt, wild type.
Rap1 controls many TERRA regulatory pathways
Rap1-mediated regulation of TERRA might involve control of TERRA transcription and/or degradation. The rat1-1 5′–3′ exonuclease mutant was previously found to stabilize TERRA (Luke et al, 2008). To detect a possible collaboration between Rap1 complexes and Rat1, we combined the rat1-1 mutation with rap1-17, sir2Δ or rif1Δ mutants (Fig 2). Although TERRA levels in rap1-17/rat1-1 double mutants were slightly higher than those in the respective single mutants at all Y′ telomeres, at some X-only telomeres there was no additivity. Therefore, rap1-17 and rat1-1 are epistatic, to an extent, indicating that a portion of TERRA repression through the C-terminus of Rap1 might be due to the promotion of TERRA degradation through Rat1, in addition to exerting a Rat1-independent role for the repression of TERRA (Fig 2A). When testing sir2Δ and rif1Δ in combination with rat1-1, we again observed differences between Y′ and X-only telomeres. The combination of rat1-1 with sir2Δ led to a slight increase in TERRA levels at X-only telomeres, compared with the respective single mutants (Fig 2B). A different image was obtained when testing rif1Δ with rat1-1 (Fig 2C). The rat1-1 mutation was epistatic with rif1Δ at X-only telomeres but not at Y′ telomeres, in which the effects were additive. Together, the results support a model in which Sir2 promotes TERRA repression at X-only telomeres in a Rat1-independent manner. Indeed, rat1-1 mutants do not share the silencing defects of sir2Δ mutants (Fig 2D), which supports the notion that these proteins perform independent functions at telomeres. Rif1 also represses TERRA independently of Rat1; however, unlike sir2Δ mutants, the additive effect is only seen at Y′ telomeres. Therefore, Rif1 and Sir2 probably, repress TERRA at Y′ and X-only telomeres, respectively, through transcriptional repression and not through the promotion of RNA degradation. At X-only telomeres, however, Rif1 might also promote TERRA turnover, as Rif1 and Rat1 function in the same genetic pathway in terms of TERRA repression.
Figure 2.
Y′ and X-only telomeres are differentially regulated. (A) qRT–PCR analysis of Y′ and X-only TERRA in wt, rat1-1, rap1-17 and rat1-1/rap1-17 cells. Average values of three independent biological replicates normalized against actin with standard deviation are shown as fold change over wt. The wt value is arbitrarily set to 1. Statistical analyses were calculated using Student's t-test (*P<0.05, **P<0.01 and ***P<0.001). (B) qRT–PCR analysis of Y′ and X-only TERRA in the indicated strains showing that Sir2 and Rat1 negatively regulate TERRA from X-only telomeres through different pathways. Average values were normalized and expressed as in (A). (C) qRT–PCR analysis of Y′ and X-only TERRA in the indicated strains showing that Rif1 and Rat1 negatively regulate Y′ TERRA through different pathways. Average values were normalized and expressed as in (A). (D) A URA3 cassette was integrated at the subtelomeric region of telomere 7L in wt, rap1-17, rat1-1 and sir2Δ strains. The strains were grown overnight at 25 °C in YPD and spotted onto the indicated plates in 10-fold serial dilutions. Plates were incubated at either 30°C or 25°C and were photographed following 2–3 days of incubation. qRT–PCR, quantitative real-time PCR; TERRA, Telomeric repeat-containing RNA; wt, wild type; YPD, yeast extract, peptone and dextrose.
The rap1-17 mutation increases the half-life of TERRA
The above genetic interactions suggested that Rap1 promotes TERRA degradation, at least partly, through Rat1. Consistent with the conclusion that TERRA is a RNAPII transcript (Luke et al, 2008), we observed that a common mutation of both the RNAPI and III subunit rpc40 did not affect TERRA half-lives (supplementary Fig S1 online). Therefore, we tested this hypothesis by combining the rap1-17 mutation with a temperature-sensitive allele of the RNAPII subunit, rpb1-1. In this strain, we were able to turn off transcription by inactivating RNAPII after a shift from permissive (25°C) to non-permissive temperature (39°C). We analysed TERRA stemming from different subsets of Y′ telomeres and found that, following RNAPII inactivation, Y′ TERRA was degraded with a half-life of 13–18 min (Table 1; supplementary Fig S2A online). We also followed the messenger RNA half-life from actin (ACT1) for comparison (Table 1; supplementary Fig S2A online). TERRA half-lives were also determined for transcripts from X-only telomeres, yielding a similar value of 12–28 min (Table 2; supplementary Fig S3 online). When RNAPII was inactivated in a rap1-17 background, the half-life of Y′-derived TERRA increased approximately threefold from 13–18 to 35–49 min, whereas ACT1 RNA degradation rates were not increased in this mutant (Table 1; supplementary Fig S2B online). Deletion of RIF1 did not result in an increase in TERRA half-life at Y′ telomeres, consistent with our epistasis analysis of TERRA levels (Table 1; supplementary Fig S2C online; Fig 2C). We were unable to measure TERRA half-life in sir2Δ rpb1-1 strains, as the sir2 deletion suppressed rpb1-1 for unknown reasons (data not shown). It should be noted that in some of the above experiments an rpc40/rpb1-1 double mutant was used to determine TERRA half-life, as we noticed that either RNAPI or III can transcribe X-only TERRA, to a small extent, after RNAPII was inactivated over prolonged periods; however, this was not the case in the rap1-17 mutants. In summary, the threefold increase of TERRA half-life in the rap1-17 mutant, and the several 100-fold increase of total TERRA (Fig 1B), is consistent with the notion that Rap1 is repressing TERRA through several pathways.
Table 1. Half-life of the indicated RNAs in the indicated strains.
| Strains |
Half-life (min)
‡
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
6*Y′
|
4*Y′
|
3*Y′
|
ACT1
|
|||||
| n1 | n2 | n1 | n2 | n1 | n2 | n1 | n2 | |
| rpb1-1 | 18 | 18 | 15 | 13 | 13 | 14 | 30 | 30 |
| rpb1-1 rap1-17 | 45 | 53 | 44 | 39 | 39 | 32 | 23 | 22 |
| rpb1-1 rif1Δ | 13 | 12 | 10 | 8 | 14 | 14 | 36 | 27 |
| ‡Determined by the equation t1/2=0.693/k, where k is the rate constant for RNA decay. | ||||||||
Table 2. Half-life of non-Y′ TERRAs and ACT1 in rpc40 rpb1-1.
| RNAs |
Half-life (min)
*
|
|
|---|---|---|
| n1 | n2 | |
| 4 L | 14 | 16 |
| 7 L | 21 | 27 |
| 10R | 12 | 11 |
| 13R | 23 | 33 |
| 15 L | 14 | 15 |
| 10R14R | 13 | 18 |
| ACT1 | 27 | 34 |
| *Determined by the equation t1/2=0.693/k, where k is the rate constant for RNA decay. | ||
No effect of telomere length on TERRA levels
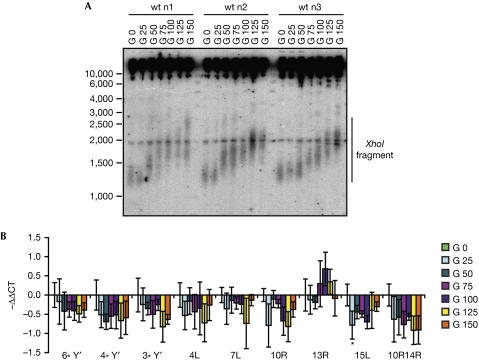
The rap1-17 mutant has several phenotypes, including aberrantly long telomeres as well as loss of silencing in the subtelomeres (Shore, 1994). Similarly, rif1Δ and rif2Δ strains have long telomeres, whereas sirΔ strains have less pronounced changes in telomere length (Palladino et al, 1993; Askree et al, 2004; Gatbonton et al, 2006). To test whether telomere lengthening affects TERRA levels, we over-elongated telomeres by tethering telomerase to telomeres on expression of a Cdc13–Est2 fusion protein (Evans & Lundblad, 1999; Fig 3A). However, telomere over-elongation did not show marked differences in TERRA levels (Fig 3B). If anything, TERRA levels decreased slightly. Consistently, we did not observe changes in RNAPII occupancy at telomeres after telomere lengthening, when assessed by chromatin immunoprecipitation (supplementary Fig S4A online). When we assessed TERRA levels during telomere shortening in tel1Δ cells, we observed a slight (less than twofold) increase in TERRA levels (supplementary Fig S4B online). Overall, TERRA levels do not correlate positively with telomere length in S. cerevisiae.
Figure 3.
Telomere length does not effect TERRA levels. (A) Telomere elongation was obtained by tethering telomerase to telomeres, using the Cdc13–Est2 fusion protein. Southern blot analysis of Y′ telomeres from three strains expressing a Cdc13–Est2 fusion protein on plasmid, propagated for 25, 50, 75, 100, 125 and 150 generations (G) at 30°C, after which the plasmid was shuffled out from the cells. Genomic DNA was digested by XhoI. (B) qRT–PCR analysis of Y′ and X-only TERRA in the same cells as in (A). Average values of three independent biological replicates normalized against actin with standard deviation are shown as fold change over wt. The wt value is arbitrarily set to 1. Statistical analyses were calculated using Student's t-test (*P<0.05). qRT–PCR, quantitative real-time PCR; TERRA, Telomeric repeat-containing RNA; wt, wild type.
In summary, we have shown that TERRA is negatively regulated at the level of transcription and degradation by Rap1, which recruits several factors to different telomeres in a differential manner, depending on whether they have a Y′ element. The telomere-binding protein Rap1 seems to be crucial for all levels of TERRA regulation, with the most pronounced effects observed at X-only telomeres. At Y′ telomeres, Rap1 regulates TERRA both by the promotion of degradation and through a degradation-independent mechanism that is dependent on Rif1 and Rif2. As TERRA half-life is not increased by rif1Δ and as the effects of the rat1-1 mutation on Y′ TERRA were exacerbated in the rat1-1/rif1Δ double mutant, it seems likely that Rif1 represses transcription (Figs 2C, 4) at Y′ telomeres. At X-only telomeres, Rap1 promotes telomeric repression through both Sir2/3/4 and Rif1/2 complexes (Fig 4). However, the genetic interactions place Rap1 in the Rat1-mediated 5′–3′ RNA degradation pathway, suggesting that, in addition to repressing telomere transcription through recruitment of Sir proteins, Rap1 also promotes Rat1-mediated TERRA turnover. As Rap1 is a highly conserved protein throughout evolution (Li et al, 2000; Chen et al, 2011), these findings in S. cerevisiae might provide a model for TERRA regulation throughout the eukaryotic domain.
Figure 4.
A model for Rap1-negative regulation of TERRA at X-only telomeres and Y′ telomeres. Upper panel: Rap1 associated with telomeric repeats (dashed line) affects TERRA levels at X-only telomeres through both Rat1-mediated degradation (probably through Rif1 and Rif2) and transcriptional silencing through the Sir2/3/4 complex. Lower panel: at Y′-containing telomeres, TERRA is regulated by Rap1 through the Rif1 and Rif2 proteins (degradation independent), as well as by the nuclear 5′–3′ exonuclease Rat1. RNAPII, RNA polymerase II; TERRA, Telomeric repeat-containing RNA.
Methods
Strain constructions. All yeast strains used in this study were derived from the BY4741 background and are listed in supplementary Table S1 online.
Spotting assay. Yeast cultures grown to stationary phase were diluted to 1 × 107 cells/ml (or 5 × 106 cells/ml for 5-fluoroorotic acid plates); 10-fold dilutions were spotted on the indicated selective plates and grown at different temperatures for 3 days (see figure legends).
RNA preparation for quantitative real-time PCR. For RNA isolation, cells were grown at 30°C in 15 ml of medium until OD600 (optical density measured at 600 nm wavelength) reached 0.6–0.8. For RNA half-life experiments, cells were grown in YPD (yeast extract, peptone and dextrose) media at 25°C until early log phase, centrifuged and resuspended in 50-ml Falcon tubes, rapidly shifted to 39°C by adding the same volume of YPD preheated to 53°C and incubated at 39°C in a water bath. Samples of 2 ml were taken at indicated times. We found that three consecutive DNase I treatments using the RNase-Free DNase Set (QIAGEN) were required to completely remove telomeric DNA from total RNA. The first DNase I treatment was performed in solution, as recommended by the supplier. DNase I-treated RNA was precipitated, digested a second time on column using the RNeasy kit (QIAGEN) as recommended by the supplier, purified, and the third DNase treatment was again performed on the column. The reverse transcription was performed with 3 μg of RNA, using the SuperScript III Reverse Transcriptase (Invitrogen). TERRA was reverse-transcribed using the CA oligonucleotide (reverse telomeric) as follows: 3 μg RNA, 769 μM of each of the four deoxyribonucleotide triphosphates, 10 μM CA oligonucleotide and 2 μM ACT1 R oligonucleotide in a final reaction volume of 13 μl was incubated for 1 min at 90°C, followed by a fast cooling step during 1 min to 55°C. In all, 4 μl of 5 × First-Strand Reverse RT buffer, 1 μl of 0.1 M dithiothreitol, 1 μl of 40 U/μl RNasin Plus RNase inhibitor (Promega) and 1 μl of SuperScript III RT (200 units/μl) were added when the reaction was at 55°C and further incubated at 55°C for 60 min and 70°C for 15 min. Reverse transcription of mRNA with oligo(dT)15 was performed as described above, except that 500-ng oligo(dT)15 was used; the mixture was first incubated for 5 min at 65°C and then chilled on ice. The RT reaction was performed at 50°C. For qPCR, the cDNA was diluted 2.5 times in H2O. A volume of 2 μl was quantified in a final volume of 20 μl by real-time PCR with the Power SYBR Green PCR Master mix (Applied Biosystems) using an Applied Biosystems 7900 HT Fast Real-Time PCR System. Primers were used at a final concentration of 0.2–0.6 μM. The reactions were incubated for 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. All primer sequences and final concentrations used, as well as the TERRA that they amplified, are listed in supplementary Table S2 online. The TERRA identity in amplified products was verified using TOPO cloning (Invitrogen), followed by sequencing of at least 6–10 sequences per strain. Statistical significance was determined using a two-tailed Student's t-test.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank C. Boone, V. Lundblad, S. Marcand, R. Young, E. Louis, V. Zakian, J. Berman and D. Shore for reagents. We also thank A. Porro for critical reading of the manuscript. V.P. is supported by an EMBO postdoctoral fellowship. Work in J.L.'s laboratory is supported by the Swiss National Science Foundation, the European Community's Seventh Framework Programme FP7/2007-2011 (grant agreement number 200950) and a European Research Council advanced investigator grant (grant agreement number 232812). Work in B.L.'s lab is supported by the Netzwerk Alterns-Forschung, which is funded from the Ministerium für Wissenschaft, Forschung und Kunst Baden-Württemberg.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aparicio OM, Billington BL, Gottschling DE (1991) Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66: 1279–1287 [DOI] [PubMed] [Google Scholar]
- Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ (2004) A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci USA 101: 8658–8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 [DOI] [PubMed] [Google Scholar]
- Berman J, Tachibana CY, Tye BK (1986) Identification of a telomere-binding activity from yeast. Proc Natl Acad Sci USA 83: 3713–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caslini C, Connelly JA, Serna A, Broccoli D, Hess JL (2009) MLL associates with telomeres and regulates telomeric repeat-containing RNA transcription. Mol Cell Biol 16: 4519–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Tye BK (1983) A family of Saccharomyces cerevisiae repetitive autonomously replicating sequences that have very similar genomic environments. J Mol Biol 168: 505–523 [DOI] [PubMed] [Google Scholar]
- Chawla R, Azzalin CM (2008) The telomeric transcriptome and SMG proteins at the crossroads. Cytogenet Genome Res 122: 194–201 [DOI] [PubMed] [Google Scholar]
- Chen Y et al. (2011) A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat Struct Mol Biol 18: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M, Palladino F, Laroche T, Kyrion G, Liu C, Lustig AJ, Gasser SM (1995) The carboxy termini of Sir4 and Rap1 affect Sir3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J Cell Biol 129: 909–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM (2009) TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell 35: 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SK, Lundblad V (1999) Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120 [DOI] [PubMed] [Google Scholar]
- Gatbonton T, Imbesi M, Nelson M, Akey JM, Ruderfer DM, Kruglyak L, Simon JA, Bedalov A (2006) Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet 2: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CF, Sussel L, Shore D (1992) A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev 6: 801–814 [DOI] [PubMed] [Google Scholar]
- Kyrion G, Liu K, Liu C, Lustig AJ (1993) RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev 7: 1146–1159 [DOI] [PubMed] [Google Scholar]
- Li B, Oestreich S, de Lange T (2000) Identification of human Rap1: implications for telomere evolution. Cell 101: 471–483 [DOI] [PubMed] [Google Scholar]
- Louis EJ (1995) The chromosome ends of Saccharomyces cerevisiae. Yeast 11: 1553–1573 [DOI] [PubMed] [Google Scholar]
- Luke B, Lingner J (2009) TERRA: telomeric repeat-containing RNA. EMBO J 28: 2503–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J (2008) The Rat1p 5′–3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell 32: 465–477 [DOI] [PubMed] [Google Scholar]
- Marcand S, Gilson E, Shore D (1997) A protein-counting mechanism for telomere length regulation in yeast. Science 275: 986–990 [DOI] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D (1994) Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev 8: 2257–2269 [DOI] [PubMed] [Google Scholar]
- Nergadze SG, Farnung BO, Wischnewski H, Khoriauli L, Vitelli V, Chawla R, Giulotto E, Azzalin CM (2009) CpG-island promoters drive transcription of human telomeres. RNA 15: 2186–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser SM (1993) SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75: 543–555 [DOI] [PubMed] [Google Scholar]
- Pardo B, Marcand S (2005) Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J 24: 3117–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde FE, Louis EJ (1999) Limitations of silencing at native yeast telomeres. EMBO J 18: 2538–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon S, Reichenbach P, Lingner J (2010) The noncoding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res 38: 5797–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J, Herskowitz I (1987) Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 116: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Gottschling DE, Zakian VA (1994) Transcription of a yeast telomere alleviates telomere position effect without affecting chromosome stability. Proc Natl Acad Sci USA 91: 12061–12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA (2008) Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 10: 228–236 [DOI] [PubMed] [Google Scholar]
- Shore D (1994) RAP1: a protean regulator in yeast. Trends Genet 10: 408–412 [DOI] [PubMed] [Google Scholar]
- Shore D, Nasmyth K (1987) Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell 51: 721–732 [DOI] [PubMed] [Google Scholar]
- Walmsley RW, Chan CS, Tye BK, Petes TD (1984) Unusual DNA sequences associated with the ends of yeast chromosomes. Nature 310: 157–160 [DOI] [PubMed] [Google Scholar]
- Wotton D, Shore D (1997) A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev 11: 748–760 [DOI] [PubMed] [Google Scholar]
- Zhang L, Ogawa Y, Ahn J, Namekawa S, Silva S, Lee JT (2009) Telomeric RNAs mark sex chromosomes in stem cells. Genetics 182: 685–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Gustafsson CM (2009) Distinct differences in chromatin structure at subtelomeric X and Y′ elements in budding yeast. PLoS ONE 4: e6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.