Abstract
Mutations in HepA-related protein (HARP, or SMARCAL1) cause Schimke immunoosseous dysplasia (SIOD). HARP has ATP-dependent annealing helicase activity, which helps to stabilize stalled replication forks and facilitate DNA repair during replication. Here, we show that the conserved tandem HARP (2HP) domain dictates this annealing helicase activity. Furthermore, chimeric proteins generated by fusing the 2HP domain of HARP with the SNF2 domain of BRG1 or HELLS show annealing helicase activity in vitro and, when targeted to replication forks, mimic the functions of HARP in vivo. We propose that the HARP domain endows HARP with this ATP-driven annealing helicase activity.
Keywords: annealing helicase, HARP, replication, RPA, Schimke immunoosseous dysplasia
Introduction
Mutations in HepA-related protein (HARP); also known as DNA-dependent ATPase A and SMARCAL1 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a-like 1)) cause Schimke immunoosseous dysplasia (SIOD), a rare, autosomal recessive disease characterized by spondyloepiphyseal dysplasia, T-cell deficiency and focal segmental glomerulosclerosis (Boerkoel et al, 2002; Deguchi et al, 2008). HARP preferentially binds to forked DNA structures as opposed to single-stranded DNA (ssDNA) or double-stranded DNA (dsDNA), and the ATPase activity of HARP is stimulated by forked DNA structures (Muthuswami et al, 2000; Yusufzai & Kadonaga, 2008). Moreover, HARP possesses ATP-driven annealing helicase activity in vitro and can rewind complementary ssDNA bound by replication protein A (RPA; Yusufzai & Kadonaga, 2008). Recently, several groups including ours have proposed that the annealing helicase activity of HARP might dictate its role in S-phase-specific DNA damage response and that HARP is involved in protecting stalled replication forks by minimizing the accumulation of ssDNA regions and facilitating the repair of collapsed replication forks during DNA replication (Yusufzai & Kadonaga, 2008; Bansbach et al, 2009; Ciccia et al, 2009; Postow et al, 2009; Yuan et al, 2009; Yusufzai et al, 2009).
HARP is a member of the conserved SNF2 family of proteins that are involved in chromatin remodelling and is required for transcriptional regulation, replication, recombination and DNA repair (Coleman et al, 2000). All members of the SNF2 family are characterized by the presence of the SWI/SNF helicase domain (SNF2 domain), but do not always exhibit helicase activity. Studies have shown that some of these proteins use the energy of ATP hydrolysis to translocate along the DNA and thus remodel chromatin by restructuring nucleosomes (Bork & Koonin, 1993; Eisen et al, 1995; Ristic et al, 2001; Saha et al, 2002; Durr et al, 2005; Fan et al, 2005).
HARP is an ATP-driven annealing helicase. The physiological relevance of HARP's annealing helicase activity was revealed by the finding that several mutations observed in SIOD patients result in mutant HARP proteins that have defective ATPase and annealing helicase activity, suggesting that the annealing helicase activity of HARP is important for its function in vivo (Boerkoel et al, 2002; Clewing et al, 2007; Yusufzai & Kadonaga, 2008). This speculation is supported by recent findings indicating that the S-phase functions of HARP also require its ATPase and annealing helicase activity (Bansbach et al, 2009; Ciccia et al, 2009; Postow et al, 2009; Yuan et al, 2009; Yusufzai et al, 2009). As all SNF2 family members have ATPase activity, we are interested in what determines the annealing helicase activity of HARP. In this study, we show that the conserved tandem HARP (2HP) domain of HARP protein dictates its annealing helicase activity. This activity is transferrable because when the SNF2 domain of Brahma-related gene 1 (BRG1); also known as SMARCA4) or helicase, lymphoid specific (HELLS); also known as SMARCA6, two other members of the SNF2 gene family is fused with the 2HP domain of HARP, the chimeric proteins have annealing helicase activity in vitro. Furthermore, the chimeric proteins are fully functional in vivo as they complement the function of HARP in HARP-depleted cells.
Results And Discussion
HARP domain is required for HARP function in vivo
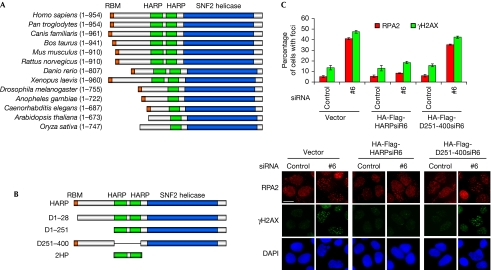
To gain a better understanding of the annealing helicase activity of HARP, we aligned the human HARP sequence with its orthologues from other species. HARPs consist of approximately 700–950 amino acids and have three highly conserved regions: the amino-terminal RPA-binding motif (RBM; not found in Arabidopsis thaliana or Oryza sativa), the central single HARP (HP) or 2HP domain (approximately 60 residues each) and the carboxy-terminal conserved SNF2 domain (approximately 400 residues; Fig 1A).
Figure 1.
The evolutionarily conserved HARP domain is crucial for HARP function. (A) All HARP orthologues contain the HARP domain and the SNF2 domain. (B) Schematic representation of wild-type HARP and the mutants used in this study. (C) The mutant lacking the 2HP domain could not restore HARP function in vivo. U2OS-derivative cells stably expressing siRNA-resistant wild-type HARP (HA-Flag-HARPsiR6) or the mutant lacking the 2HP domain (HA-Flag-D251-400siR6) were transfected with the indicated siRNAs; 72 h later, cells were subjected to immunostaining using RPA2 and γH2AX antibodies as indicated. Foci-positive cells were quantified by counting a total of 200 cells per sample. Data are presented as mean±s.d. from three independent experiments. Scale bar, 10 μm. DAPI, 4′,6-diamidino-2-phenylindole; HA, haemagglutinin; HARP, HepA-related protein; RBM, RPA-binding motif; RPA, replication protein A; siRNA, short interfering RNA.
Many mutations identified in SIOD patients are mapped to the conserved 2HP and SNF2 domains (Boerkoel et al, 2002; Clewing et al, 2007), suggesting that both domains are involved in the pathogenesis of SIOD. On the basis of these conserved domains and the mutations or deletions identified in SIOD patients, we generated a series of internal-deletion and truncation mutants of HARP (HARP D1–28, D1–251, D251–400 and 2HP; Fig 1B) and examined whether these mutants would affect the in vivo and in vitro functions of HARP. Studies have shown that the N-terminal RBM and the SNF2 domain of HARP are involved in stabilizing replication forks and that HARP-depleted cells display longer stretches of ssDNA during DNA replication, leading to collapsed replication forks and accumulation of dsDNA breaks (Bansbach et al, 2009; Ciccia et al, 2009; Postow et al, 2009; Yuan et al, 2009; Yusufzai et al, 2009). The appearance of RPA foci can be used as a surrogate marker for the accumulation of ssDNA regions in the cell (Zou & Elledge, 2003; Jazayeri et al, 2006). The frequency of dsDNA breaks can be monitored by the formation of phosphorylated H2AX (γH2AX) foci. After endogenous HARP was depleted, formation of RPA and γH2AX foci was enhanced (Fig 1C). Interestingly, the expression of siRNA-resistant wild-type HARP, but not of the D251–400 mutant, which lacks the 2HP domain, suppressed the formation of RPA and γH2AX foci. As a control, we show that the expression of D251–400 mutant is similar to that of wild-type HARP (supplementary Fig S1A online). This result and our previous data suggest that all three conserved domains of HARP are required for its functions in vivo.
HARP domain determines the annealing helicase activity
We asked whether the enhanced formation of RPA and γH2AX foci observed in cells expressing the HARP D251–400 mutant (lacking the 2HP domain) was due to compromised DNA binding, DNA-dependent ATPase or the annealing helicase activity of this mutant. Both glutathione-S-transferase (GST)-tagged full-length HARP and its internal-deletion mutant proteins (Fig 2A) displayed DNA-binding activity with fork DNA (Fig 2B). The D1–28 and D1–251 mutants showed a modest reduction in DNA-binding activity, which could be due to the different oligomeric states of wild-type and mutant HARP proteins. However, size-exclusion chromatography of the wild-type and D1–28 mutant of HARP shows that both these proteins exist as monomers in solution (supplementary Fig S1B online). Consistent with earlier studies (Muthuswami et al, 2000; Yusufzai & Kadonaga, 2008), we found that fork DNA stimulates the ATPase activity of full-length HARP and its deletion mutants (Fig 2C). The observed reduction in the ATPase activity of the D1–28 and D1–251 mutants could be attributed to the reduced DNA-binding activity of these mutants (Fig 2B). The 2HP domain alone did not show DNA-binding or ATPase activity (Fig 2B,C). The 2HP domain also does not interact with the RPA:DNA complex or the SNF2 domain of HARP (supplementary Figs S1C–E,S2A online).
Figure 2.
The 2HP domain dictates the annealing helicase activity of HARP. (A) SDS–PAGE profile of purified wild-type and mutant HARP proteins. The indicated purified wild-type HARP and its internal-deletion mutants were analysed by 10% SDS–PAGE. (B) Wild-type and mutant HARP proteins bind to fork DNA. Fork DNA was incubated with increasing concentrations of the indicated wild-type and mutant HARP proteins, and reaction products were analysed by 8% native PAGE. (C) ATPase activity of wild-type and mutant HARP is stimulated by fork DNA. The indicated wild-type and mutant HARP proteins hydrolysed increasing amounts of ATP with increasing concentrations of fork DNA. The graph shows the percentage of ATP hydrolysed against the concentration of fork DNA. Data are plotted as mean±s.d. from three independent experiments. (D) Deleting the 2HP domain abolishes the annealing helicase activity of HARP. The annealing helicase assay was carried out with 100 nM HARP, 2HP, D1–28, D1–251 and D251–400 in the presence (top panel) or absence (bottom panel) of ATP. UTP was used as control in the absence of ATP. All reactions contained DNA, RPA and topoisomerase I. HARP, HepA-related protein; PAGE, polyacrylamide gel electrophoresis; RPA, replication protein A.
Next, we examined the annealing helicase activity of wild-type and mutant HARP using a previously described assay (Yusufzai & Kadonaga, 2008). A partly unwound DNA substrate was generated by treating plasmid DNA with RPA in the presence of topoisomerase I. Addition of wild-type HARP, D1–28 and D1–251 mutants catalysed the annealing of RPA-unwound DNA only in the presence of ATP (Fig 2D). However, the D251-400 mutant of HARP failed to do so (Fig 2D). Furthermore, two other HARP internal deletion mutants (D288–366 and D366–383) identified in SIOD patients (Clewing et al, 2007) that contain a partial or an intact HP domain failed to function as an annealing helicase in vitro (supplementary Fig S2B online), suggesting that the tandem HP domains are required to direct the annealing helicase activity of human HARP. Together with the in vivo study (Fig 1C), these data imply that the conserved 2HP domain is not involved in DNA-binding or ATPase activity, but determines the annealing helicase activity of HARP in vitro and is required for its function in stabilizing replication forks in vivo.
Chimeric proteins display annealing helicase activity
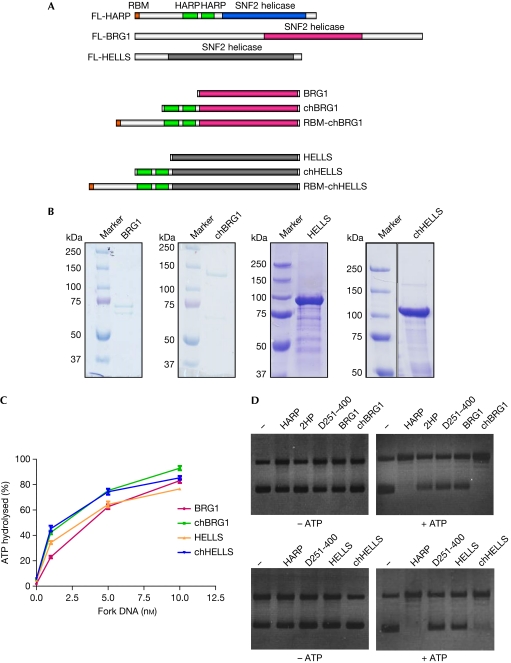
As the 2HP domain alone lacks DNA-binding, ATPase or annealing helicase activity (Fig 2), we suggest that this domain has to function together with the SNF2 domain of HARP to carry out its annealing helicase activity. We asked whether the SNF2 domain of HARP has unique features or whether the 2HP domain of HARP can be transferred to other SNF2 family proteins. We generated chimeric proteins by fusing the 2HP domain with other proteins that belong to the SNF2 family. BRG1 is the central catalytic component of several multi-subunit chromatin-remodelling complexes (Reisman et al, 2009). Similarly to other members of the SNF2 family, BRG1 harbours the conserved SNF2 domain, but lacks the 2HP domain (Fig 3A) and does not have annealing helicase activity (Yusufzai & Kadonaga, 2008). We created a chimera, chBRG1, in which the 2HP domain of HARP was fused with the SNF2 domain of BRG1 (Fig 3A). GST-fused SNF2 domain of BRG1 and GST-chimeric BRG1 (GST-chBRG1; Fig 3B) both displayed DNA-dependent ATPase activity (Fig 3C). Although BRG1 does not have annealing helicase activity, the chimeric protein chBRG1 showed annealing helicase activity comparable to that of HARP (Fig 3D).
Figure 3.
Chimeric proteins have annealing helicase activity similar to that of wild-type HARP. (A) Schematic representation of chimeric constructs used in this study. (B) SDS–PAGE profile of purified chimeric proteins. GST-tagged BRG1, chBRG1, HELLS and chHELLS were purified from Sf9 cells and analysed by 10% SDS–PAGE. (C) BRG1, chBRG1, HELLS and chHELLS all display DNA-dependent ATPase activity. The graph shows the percentage of ATP hydrolysed against the concentration of fork DNA. Data are plotted as mean±s.d. from three independent experiments. (D) chBRG1 and chHELLS have annealing helicase activity. The annealing helicase assay was carried out using the indicated purified proteins in the absence (left panels) or presence (right panels) of ATP. UTP was used as a control in the absence of ATP. All reactions contained DNA, RPA and topoisomerase I. BRG1, Brahma-related gene 1; HARP, HepA-related protein; HELLS, helicase, lymphoid specific; PAGE, polyacrylamide gel electrophoresis; RBM, RPA-binding motif; RPA, replication protein A.
To confirm these results we generated another chimera, chHELLS (Fig 3A), in which the 2HP domain of HARP was fused with the SNF2 domain of HELLS, a protein that has a role in cellular proliferation and leukaemogenesis (Raabe et al, 2001). Again, both HELLS and chHELLS displayed DNA-dependent ATPase activity (Fig 3C). However, chHELLS, but not HELLS, exhibited annealing helicase activity similar to that of full-length HARP (Fig 3D; supplementary Fig S1F online). Furthermore, we detected that HARP and the chimeric proteins exhibited annealing helicase activity on DNA coated with Escherichia coli single-strand DNA-binding protein (SSB), indicating that HARP or these chimeric proteins have general annealing helicase activity that does not depend on or require any specific SSBs (supplementary Fig S2C online). Together, these results suggest that the 2HP domain of HARP protein determines the annealing helicase activity of HARP. It functions together with the SNF2 domain, which is involved in ATP binding and hydrolysis, and promotes the annealing of complementary ssDNA strands.
Chimeric proteins carry out HARP function in vivo
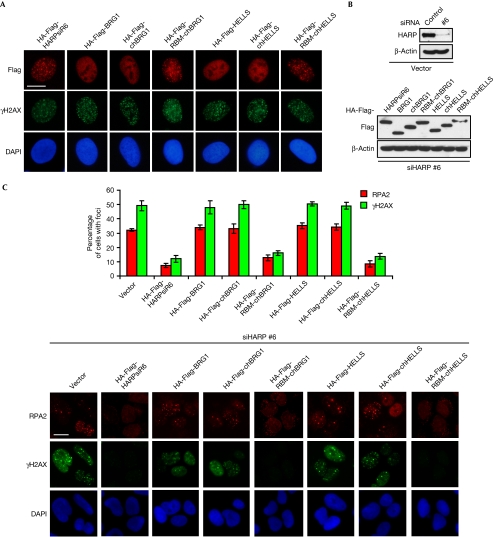
We wondered whether the chimeric proteins could be substituted for HARP in vivo if, similarly to HARP, they had annealing helicase activity in vitro. HARP is recruited to stalled replication forks by means of its direct interaction with RPA, although RPA-binding activity is not required for its annealing helicase activity in vitro (Bansbach et al, 2009; Ciccia et al, 2009; Yuan et al, 2009; Yusufzai et al, 2009). To ensure that the chimeric proteins were correctly localized in the cell, we generated siRNA-resistant fusion constructs (RBM-chBRG1 and RBM-chHELLS) in which RBM was fused to the chimeric proteins (Fig 3A). The mutants with RBM, similarly to wild-type HARP, formed nuclear foci after hydroxyurea treatment (Fig 4A). The expression of these mutants and the knockdown of endogenous HARP in U2OS cells were confirmed by western blot analysis (Fig 4B). Expression of siRNA-resistant RBM-chBRG1 and RBM-chHELLS suppressed the enhanced formation of RPA and γH2AX foci after endogenous HARP was depleted (Fig 4C). However, the SNF2 domain alone (BRG1 and HELLS) or the chimeras lacking RBM (chBRG1 and chHELLS) failed to restore the function of HARP in vivo (Fig 4C). Moreover, we generated two fusion proteins, RBM-BRG1 and RBM-HELLS, without 2HP domain. Although these fusion proteins formed nuclear foci after hydroxyurea treatment, they could not restore HARP function in vivo (supplementary Fig S3 online). In addition, RBM-chBRG1 and RBM-chHELLS, purified from mammalian cells, had annealing helicase activity similar to that of HARP in vitro (supplementary Fig S2D online). Again, these data suggest that the 2HP domain is required and dictates the annealing helicase activity and function of HARP protein.
Figure 4.
Chimeric proteins containing RBM can restore HARP function in vivo. (A) U2OS-derivative cells stably expressing HA-Flag-tagged fusion proteins or the SNF2 domain alone (BRG1 and HELLS), chimeras lacking RBM (chBRG1 and chHELLS), and chimeras with RBM (RBM-chBRG1 and RBM-chHELLS) were generated. The hydroxyurea-induced foci-forming abilities of these fusion proteins were determined by immunostaining using the indicated antibodies. (B) The endogenous and exogenous expression levels of HARP and the chimeric proteins were confirmed by immunoblotting using the indicated antibodies. The extracts were prepared from cells transfected with indicated siRNAs. (C) U2OS-derivative cells were transfected with the indicated siRNAs; 72 h later, cells were subjected to immunostaining using RPA2 and γH2AX antibodies. Foci-positive cells were quantified by counting a total of 200 cells per sample. Data are presented as mean±s.d. from three independent experiments. Scale bars, 10 μm. BRG1, Brahma-related gene 1; DAPI, 4′,6-diamidino-2-phenylindole; HA, haemagglutinin; HARP, HepA-related protein; HELLS, helicase, lymphoid specific; RBM, RPA-binding motif; RPA, replication protein A; siRNA, short interfering RNA.
Stalled replication forks might arise during normal chromosome replication or in the presence of DNA lesions (Walter & Newport, 2000; Tercero & Diffley, 2001; Katou et al, 2003; Pacek et al, 2006). These stalled replication forks, if they are left unrepaired, are unstable because of the presence of long stretches of ssDNA. They can collapse into deleterious structures that prevent resumption of DNA replication and lead to unscheduled recombination, resulting in cell death or genomic instability. Although the mechanisms involved in protecting stalled replication forks are unknown, it has been speculated that the annealing helicase activity of HARP might have a role in either stabilizing stalled forks or mediating the repair of collapsed replication forks, as it can rewind RPA-coated ssDNA regions to form more stable duplex DNA (Yusufzai & Kadonaga, 2008; Bansbach et al, 2009; Ciccia et al, 2009; Postow et al, 2009; Yuan et al, 2009; Yusufzai et al, 2009). In this study, we demonstrate that the evolutionarily conserved HARP domain determines the annealing helicase activity required for the in vivo and in vitro functions of HARP. Moreover, the HARP domain is a distinct functional domain because it can be transferred to other SNF2 family members. Structural analysis is needed to uncover the properties of the HARP domain and the catalytic residues responsible for the annealing helicase activity. Another protein with annealing helicase activity, annealing helicase 2 (AH2), previously termed ZRANB3 (zinc-finger, RAN-binding domain containing 3), has been recently identified (Yusufzai & Kadonaga, 2010), implying that there might be a subset of the SNF2 family of proteins that process annealing helicase activity. Similarly to HARP, AH2 displays DNA-dependent ATPase activity and catalyses ATP-dependent rewinding of RPA-coated ssDNA. AH2 lacks the N-terminal RBM found in HARP, but harbours a conserved SNF2 domain and HNH motif (Yusufzai & Kadonaga, 2010). Alignment of HARP domains with AH2 reveals a putative ‘HARP-like’ domain in the AH2 protein (residues 712–820; supplementary Fig S4 online). This ‘HARP-like’ domain has several residues that are conserved in all of the HARP proteins (data not shown). It would be interesting to test whether this putative ‘HARP-like’ domain is crucial for the function of AH2 as an annealing helicase.
Methods
Plasmid constructs, antibodies, cell culture, transfection and siRNAs, DNA substrates protein purification in insect cells, electrophoretic mobility shift assay and the ATPase assay used in this study are described in the supplementary information online.
Immunostaining. Cells cultured on coverslips were washed with PBS, pre-extracted with 0.5% Triton X-100 solution for 3 min and fixed with 3% paraformaldehyde for 12 min. Coverslips were washed with PBS and then immunostained with primary antibodies in 5% goat serum for 60 min. Coverslips were then washed and incubated with secondary antibodies conjugated to rhodamine or fluorescein isothiocyanate for 60 min. Cells were then stained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize nuclear DNA. The coverslips were mounted onto glass slides with antifade solution and visualized using a Nikon Eclipse E800 fluorescence microscope with a Nikon Plan Fluor × 40 oil objective lens (NA 1.30) at 27 °C. Cells were photographed using a SPOT camera (Diagnostic Instruments) and analysed using Photoshop software (Adobe).
Annealing helicase assay. The annealing helicase assay was carried out as described previously (Yusufzai & Kadonaga, 2008), with the following modifications. Supercoiled pUC19 DNA (0.2 μg) was incubated with 2.5 μg of purified RPA or E. coli SSB (Epicentre Biotechnologies; supplementary Fig S2C online) in the presence of 40 mM Tris–HCl (pH 8.0), 20 mM NaCl, 1 mM EDTA, 5 mM MgCl2 and 5 mM dithiothreitol at 37°C for 45 min. The reaction mixture was then treated with 2 units of E. coli topoisomerase I (New England BioLabs) and incubated at 37°C for another 30 min. The indicated proteins (at 100 nM) were added, and the mixture was incubated for a further 30 min at 37°C. The reaction was terminated with a solution of SDS–EDTA. The products were extracted with an equal volume of chloroform and resolved on 1.2% agarose gel at 40 V/cm for 5 h. The gel was then stained with ethidium bromide.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank all colleagues in the Chen laboratory for discussion and technical assistance. This work was supported in part by grants from the National Institutes of Health to J.C. (CA089239, CA092312 and CA100109). J.C. is also a recipient of an Era of Hope Scholar award from the Department of Defense (W81XWH-05-1-0470) and is a member of the MD Anderson Cancer Center (CA016672). G.G. and J.Y. generated the reagents and designed and carried out the experiments. J.C. advised on the design of the experiments. G.G., J.Y. and J.C. were responsible for the preparation of the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D (2009) The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev 23: 2405–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerkoel CF et al. (2002) Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet 30: 215–220 [DOI] [PubMed] [Google Scholar]
- Bork P, Koonin EV (1993) An expanding family of helicases within the ‘DEAD/H’ superfamily. Nucleic Acids Res 21: 751–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Bredemeyer AL, Sowa ME, Terret ME, Jallepalli PV, Harper JW, Elledge SJ (2009) The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev 23: 2415–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewing JM et al. (2007) Schimke immunoosseous dysplasia: suggestions of genetic diversity. Hum Mutat 28: 273–283 [DOI] [PubMed] [Google Scholar]
- Coleman MA, Eisen JA, Mohrenweiser HW (2000) Cloning and characterization of HARP/SMARCAL1: a prokaryotic HepA-related SNF2 helicase protein from human and mouse. Genomics 65: 274–282 [DOI] [PubMed] [Google Scholar]
- Deguchi K et al. (2008) Neurologic phenotype of Schimke immuno-osseous dysplasia and neurodevelopmental expression of SMARCAL1. J Neuropathol Exp Neurol 67: 565–577 [DOI] [PubMed] [Google Scholar]
- Durr H, Korner C, Muller M, Hickmann V, Hopfner KP (2005) X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell 121: 363–373 [DOI] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res 23: 2715–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HY, Trotter KW, Archer TK, Kingston RE (2005) Swapping function of two chromatin remodeling complexes. Mol Cell 17: 805–815 [DOI] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP (2006) ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8: 37–45 [DOI] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Muthuswami R, Truman PA, Mesner LD, Hockensmith JW (2000) A eukaryotic SWI2/SNF2 domain, an exquisite detector of double-stranded to single-stranded DNA transition elements. J Biol Chem 275: 7648–7655 [DOI] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC (2006) Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21: 581–587 [DOI] [PubMed] [Google Scholar]
- Postow L, Woo EM, Chait BT, Funabiki H (2009) Identification of SMARCAL1 as a component of the DNA damage response. J Biol Chem 284: 35951–35961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raabe EH, Abdurrahman L, Behbehani G, Arceci RJ (2001) An SNF2 factor involved in mammalian development and cellular proliferation. Dev Dyn 221: 92–105 [DOI] [PubMed] [Google Scholar]
- Reisman D, Glaros S, Thompson EA (2009) The SWI/SNF complex and cancer. Oncogene 28: 1653–1668 [DOI] [PubMed] [Google Scholar]
- Ristic D, Wyman C, Paulusma C, Kanaar R (2001) The architecture of the human Rad54-DNA complex provides evidence for protein translocation along DNA. Proc Natl Acad Sci USA 98: 8454–8460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR (2002) Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev 16: 2120–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF (2001) Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557 [DOI] [PubMed] [Google Scholar]
- Walter J, Newport J (2000) Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase α. Mol Cell 5: 617–627 [DOI] [PubMed] [Google Scholar]
- Yuan J, Ghosal G, Chen J (2009) The annealing helicase HARP protects stalled replication forks. Genes Dev 23: 2394–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kadonaga JT (2008) HARP is an ATP-driven annealing helicase. Science 322: 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kadonaga JT (2010) Annealing helicase 2 (AH2), a DNA-rewinding motor with an HNH motif. Proc Natl Acad Sci USA 107: 20970–20973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kong X, Yokomori K, Kadonaga JT (2009) The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev 23: 2400–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.