In his seventeenth-century classic, Novum Organum, Francis Bacon wrote, “we cannot command nature except by obeying her” (Bacon, 2010). Although our knowledge of living systems is much improved since Bacon's time, we are still far from understanding—or commanding—all the complex mechanisms of life. To take full advantage of living organisms for the benefit of mankind, we will need to understand those mechanisms to the furthest possible extent. To do so will require that the concept of information and the theories of information science take a more-prominent role in the understanding of living systems.
Two decades ago, Rolf Landauer (1991) argued that “information is physical” and ought to have a role in the scientific analysis of reality comparable to that of matter, energy, space and time. This would also help to bridge the gap between biology and mathematics and physics. Although it can be argued that we are living in the ‘golden age’ of biology, both because of the great challenges posed by medicine and the environment and the significant advances that have been made—especially in genetics and molecular and cell biology—we feel that information as an essential aspect of life has been neglected, or at least misunderstood.
…we feel that information as an essential aspect of life has been neglected, or at least misunderstood
Here we propose to look at living organisms as information gathering and utilizing systems (IGUSs; Gell-Mann, 1994) that process information about their environment to make decisions about their actions. To do so, we require a definition of ‘information’ that transcends the informal use of the term in the biology literature, in which it is frequently used to refer to stored data, or to some mysterious quality carried by matter or energy. Instead, we define information as a quantity that is transferred—without necessarily being conserved—during a measurement process, as correlations are created between the states of the measured and measuring systems. Such a view of information has increasingly been considered by physicists as an extension of and alternative to the more-traditional concept of entropy (Caves, 1993; Landauer, 1991; Zurek, 1989). In support of Landauer's idea, Toyabe and colleagues recently claimed to have converted information directly into energy (Toyabe et al, 2010). This definition contrasts with, for example, the measure of how many bits can be carried by a memory device such as DNA—also called information capacity—and with the most-compressed description of a specific arrangement of bits, such as a genome, which is also called algorithmic information.
Theoretical biology has not yet embraced such a notion of information, although the field has advanced beyond the idea of open, non-equilibrium systems that sustain structures and functions as matter and energy flow through them. Recent attempts to describe life have used large sets of coupled, non-linear differential equations (Tomita et al, 1999); considered large, interacting complex networks of genes and proteins corresponding to functional modules (Oltvai & Barabási, 2002); or included the concept of a set of digital-like connected modules that represent biochemical functions, within which information—in the informal sense—flows between modules (Nurse, 2008). These concepts, although useful, are overly mechanistic and fall short of capturing life's unbounded spontaneity and adaptability.
We therefore summon Maxwell's Demon (MxD; Leff & Rex, 2002) and its distant relative the ratchet (Serreli et al, 2007), and apply these to biology. The original Demon was the brain-child of Scottish physicist James Clerk Maxwell. In 1867, in a letter to Tait, Maxwell proposed a thought experiment to “show that the Second Law of Thermodynamics has only a statistical certainty” (Leff & Rex, 2002). Maxwell conceived of a device to identify and separate fast and slow particles in order to set up a temperature or pressure gradient, thus producing an asymmetrical gas configuration to yield useful work with an apparent decrease in the entropy of the Universe. Although imaginary, MxD has become a crucial tool for physicists who seek to understand information and its processing.
Our demon is somewhat different: it is an existing biological mechanism that uses information from its surroundings to create a locally ordered environment, while obeying the second law of thermodynamics. Thus, by definition, our MxD is an IGUS. As information transfer implies a change in the state of the MxD device itself, it turns out that in order to operate again, the MxD first has to be reset to its original state; this step carries an entropy cost, usually with an associated energy input, a fact that has an important role, as discussed here.
In a biological context, we postulate the existence of MxDs in every cell, but with a different role than their counterparts from physics: biological Demons use information from within the cell to counteract the inexorable effects of ageing, as well as permitting the creation of young progeny from old cells. This is based on a view of cells as machines that make copies of themselves through organized coupling of the processes of replication—copying the DNA ‘program’—and reproduction: a slightly imperfect duplication of the metabolic machinery and cellular structures.
…biological Demons use information from within the cell to counteract the inexorable effects of ageing, as well as permitting the creation of a young progeny from old cells
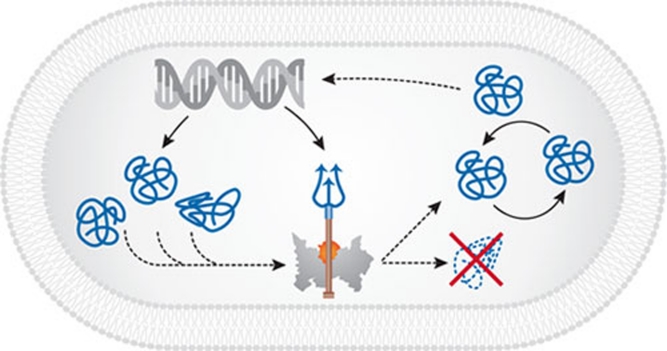
The proteins that implement metabolism and replication degrade with time for a variety of reasons: repeated transitions between metabolic states, hot or cold temperatures, reactive oxygen species, glycation, or other aggressive chemical reactions. Perhaps as importantly, some amino-acid residues undergo spontaneous isomerization that causes functional decay in a way that is conceptually reminiscent of the half-life of radioactive isotopes. In order for life to go on, these aged and dysfunctional metabolites must be identified and either discarded, degraded or repaired during normal cell operation, and even more so during cell reproduction, in which the segregation of new and old molecules guarantees the survival of new daughter cells. Enter Maxwell's Demon: a mechanism that differentiates functional and dysfunctional proteins, perhaps by measuring their shapes and vibration spectra (Schwartz & Schramm, 2009), and segregates normal proteins from defective ones using existing cellular structures, or by generating new ones (Fig 1).
Figure 1.
A schematic of Maxwell's Demon operating within a cell. MxD molecule (with a trident), coded by DNA (helix) along with other proteins, distinguishes functional and damaged proteins. The latter are contained or discarded. MxD, Maxwell's Demon.
How can we identify the entities, proteins or complexes that play the role of MxDs? If they exist, they must be in genomes. They must also be essential for life to perpetuate in a sustainable way. Hence, the corresponding functions should be encoded in all genomes. If it was possible, an easy way to find them would be to search for genes that are common to all genomes. Unfortunately, because life conserves functions but not structures, there are no genes common to the 1,000 or so complete bacterial genomes (Lagesen et al, 2010) that have been sequenced so far, let alone all the other genomes of eukaryotic organisms.
Fortunately, analysis of gene persistence—the tendency of genes to be present in a quorum of genomes—has allowed us to identify the paleome, a common set of genes that code for the essential functions that maintain life (Danchin et al, 2007). Most of these functions are easily accounted for as they are involved in the construction, reproduction and maintenance of cellular structures and the replication of genomes. Yet, some functions, thought to be involved in maintenance or degradation, are found to use energy in a way that is not obviously linked to their role. Hence, the unexpected use of apparently wasted energy can be seen as an evidential ‘smoking gun’ for MxDs of the sort we describe.
…the unexpected use of apparently wasted energy can be seen as an evidential ‘smoking gun’ for Maxwell's Demons of the sort we describe
One example of such energy ‘waste’ might be a kinetic proofreading mechanism during translation (Gueron, 1978; Hopfield, 1974), which involves information transfer, memory and reset. Reading a codon of the genetic message requires interaction with a cognate-adaptor molecule—a transfer RNA loaded with the amino acid corresponding to the codon—on the ribosome, a nanomachine that translates messenger RNA codon after codon, in a ratchet-like manner. There is enough energy in the chemical bond that links the amino acid to the transfer RNA to permit polymerization into a protein. Yet, a further step is required, with an apparently useless consumption of energy. The loaded transfer RNA is bound to a factor (EFTu) that tests the accuracy of the decoding process; there is competition between transfer RNAs at the entry site in the ribosome, and incorrect transfer RNAs are more common than correct ones. When the correct one is bound, free EFTu is released at the expense of a molecule of energy-rich GTP. Energy consumption is only used at this stage to achieve a more-‘ordered’ state of genetic-code use during translation. At the least, this is the work of an IGUS, and at best it could be the earliest MxD (unknowingly) discovered in a cellular process.
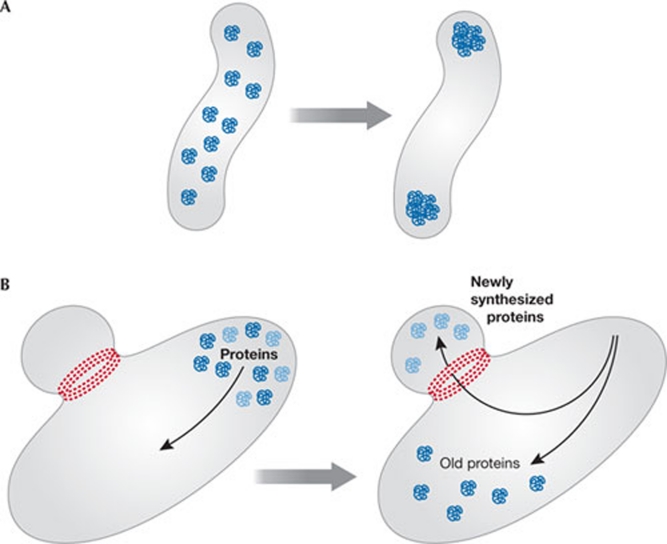
How then might other biological MxDs manifest themselves? All cells, including bacteria, age and eventually deteriorate in unfavourable conditions. As this happens, proteins aggregate into potentially harmful arrangements. Remarkably, these protein aggregates do not distribute randomly within the cell. In some cases, they are transported by an energy-dependent process to the poles of Escherichia coli cells, where they can either be stored and discarded when the cell dies, or be more easily degraded by energy-dependent proteases (Rokney et al, 2009). In other cells—budding yeast, for example—old proteins are not included in newly formed buds, but remain within the mother cell, which acts as a bin. A family of GTP-dependent proteins—septins—seems to be involved in the process; in the absence of septins, old proteins do not remain in the mother cell (Barral, 2010; Hu et al 2010; Kim et al 2010). A similar process has been demonstrated in Caenorhabditis elegans cells, in which a mother cell divides into a cell carrying old protein and a young egg cell (Goudeau & Aguilaniu, 2010). We believe that these are legitimate examples of non-trivial, Demon-like information processing in biology (Fig 2).
Figure 2.
Examples of biological Maxwell's Demons. (A) Protein aggregates are transported to Escherichia coli cell poles for easy disposal. (B) Septin rings prevent old cell products from entering young yeast buds.
Another candidate that deserves further study is the stabilization of neural synapses. It is generally accepted that learning and memory are based on a selective process that retains functioning synapses and discards the others. A variety of processes have been proposed to explain this, but the most obvious one is an energy-dependent system that would tag receptors of functional neurotransmitters, leaving the remainder vulnerable to degradation (Milholland & Gordon, 2007).
The typical modus operandi of an MxD is to quickly use any information acquired—resulting in a correlated change of its own state—to produce a desirable action—for example, to generate work, or to restore the functionality of a degraded protein—and then to be reset to a neutral state, in which it can perform another measurement. In other cases, measurements might accumulate through a ratchet-like mechanism. Instead of taking immediate action, the device keeps modifying its state and becomes a de facto record of a sequence of measurements. This is probably what happens in natural selection. In contrast to the intracellular situation, organisms repeatedly ‘measure’ their environment over generations, leading to a cumulative record of mutations and recombinations in the genome of surviving members of a species. The negative version of this effect is known as Muller's ratchet.
Our proposal is for truly interdisciplinary, long-term collaborations in which biological experiments are inspired by cutting-edge research in theoretical physics
To further test these ideas, one can explore the information-processing behaviour of the paleomic energy-dependent gene products under a variety of conditions. A practical approach could investigate the evolution of synthetic-biology constructs. Bacteria engineered to produce new metabolic pathways would be grown on media with just enough nutrients to maintain colonies, together with a metabolite that is chemically related to some step in the pathway, but which they cannot use directly. As the genome of the cell ages, we expect that continuous turnover of old gene products will occasionally replace some proteins with counterparts that permit the corresponding colonies to resume growth, using the new metabolite. This can be helped by transcription, which considerably increases the local mutation rate of expressed genes (Kim & Jinks-Robertson, 2009). Manipulating MxD genes in this experiment will modulate innovation, leading to either colonies that progressively die out if they fail to produce progeny with adaptive mutations, or that grow faster in the opposite case.
This would be only the first step of an ambitious research programme in which biologists and physicists naturally collaborate, the latter providing knowledge of information theory and related subjects. Our proposal is for truly interdisciplinary, long-term collaborations in which biological experiments are inspired by cutting-edge research in theoretical physics. The potential benefit of such a programme will be an understanding of living systems as active IGUSs, rather than as mere autonomous mechanisms. Not pursuing these ideas might considerably slow the advance of biology, especially if we fail to recognize apparently unrelated observations as manifestations of a single, fundamental principle (Danchin et al, 2007; Rokney et al, 2009; Barral, 2010; Goudeau & Aguilaniu, 2010).
Creators of synthetic constructs will need to harness the function of specific Maxwell's Demons for their own goals
The view presented here has implications for the ‘engineering’ branch of biology: synthetic biology. If, as we surmise, biological systems accumulate information in a myopic way—MxDs are local devices without grand designs—then they will evolve by tinkering in their own way, rather than by fulfilling the design goals of humans. If this is the case, the scaling up of synthetic-biology processes might hit snags. The MxDs we have seen operate within a narrow cellular context; this highlights the need for a theory of semantic information (for example, Floridi, 2004) as part of the proposed programme to achieve greater command of biological entities. In particular, it will be essential to understand which measurements are required to trigger specific biochemical behaviours, especially in cases in which several functions overlap. Creators of synthetic constructs will need to harness the function of specific MxDs for their own goals. This will require that biology also informs those investigating the foundations of information theory, so that they make the best educated guesses to enhance their chances of success. We regard the acceptance and adoption of information as the focus of a fascinating and challenging collaboration between biologists, mathematicians and physicists in the twenty-first century.
Philippe M Binder
Antoine Danchin
Acknowledgments
P.M.B. acknowledges the support of Research Corporation and A.D. aknowledges the support of the Microme FP7 KBBE-2007-3-2-08-222886-2 grant.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bacon F (2010) Novum Organum (reprint and translation of the 1620 Latin original). Charleston, SC, USA: Forgotten books [Google Scholar]
- Barral Y (2010) Septins at the nexus. Science 329: 1289–1290 [DOI] [PubMed] [Google Scholar]
- Caves CM (1993) Information and entropy. Phys Rev E 47: 4010–4017 [DOI] [PubMed] [Google Scholar]
- Danchin A, Fang G, Noria S (2007) The extant core bacterial proteome is an archive of the origin of life. Proteomics 7: 875–889 [DOI] [PubMed] [Google Scholar]
- Floridi L (2004) Outline of a theory of strongly semantic information. Mind Mach 14: 197–222 [Google Scholar]
- Gell-Mann M (1994) The Quark and the Jaguar. San Francisco, CA, USA: WH Freeman [Google Scholar]
- Goudeau J, Aguilaniu H (2010) Carbonylated proteins are eliminated during reproduction in C. elegans. Aging Cell 9: 991–1003 [DOI] [PubMed] [Google Scholar]
- Gueron M (1978) Enhanced selectivity of enzymes by kinetic proofreading. Am Sci 66: 202–208 [PubMed] [Google Scholar]
- Hopfield JJ (1974) Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Nat Acad Sci USA 71: 4135–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Milenkovic L, Jin H, Scott MP, Nachury MV, Spiliotis ET, Nelson WJ (2010) A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science 329: 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S (2009) dUTP incorporation into genomic DNA is linked to transcription in yeast. Nature 459: 1150–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK et al. (2010) Planar cell polarity acts through septins to control collective cell movement and ciliogenesis. Science 329: 1337–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K, Ussery DW, Wassenaar TM (2010) Genome update: the 1000th genome—a cautionary tale. Microbiology 156: 603–608 [DOI] [PubMed] [Google Scholar]
- Landauer R (1991) Information is physical. Phys Today 44: 23–29 [Google Scholar]
- Leff H, Rex AF (2002) Maxwell's Demon 2. London, UK: Taylor and Francis [Google Scholar]
- Milholland RB, Gordon H (2007) A role for acetylcholine receptors in their own aggregation on muscle cells. Devel Neurobiol 67: 999–1008 [DOI] [PubMed] [Google Scholar]
- Nurse P (2008) Life, logic and information. Nature 454: 424–426 [DOI] [PubMed] [Google Scholar]
- Oltvai ZN, Barabási AL (2002) Life's complexity pyramid. Science 298: 763–764 [DOI] [PubMed] [Google Scholar]
- Rokney A, Shagan M, Kessel M, Smith Y, Rosenshine I, Oppenheim AB (2009) E. coli transports aggregated proteins to the poles by a specific energy-dependent process. J Mol Biol 392: 589–601 [DOI] [PubMed] [Google Scholar]
- Schwartz SD, Schramm VL (2009) Enzymatic catalysis and the nature of transition state barrier crossing. Nat Chem Biol 5: 551–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serreli V, Lee C-F, Kay ER, Leigh DA (2007) A molecular information ratchet. Nature 445: 523–527 [DOI] [PubMed] [Google Scholar]
- Tomita M et al. (1999) E-CELL: Software environment for whole-cell simulation. Bioinformatics 15: 72–84 [DOI] [PubMed] [Google Scholar]
- Toyabe S, Sagawa T, Ueda M, Muneyuki E, Sano M (2010) Experimental demonstration of information-to-energy conversion and validation of the generalized Jarzynski equality. Nat Phys 6: 988–992 [Google Scholar]
- Zurek WH (1989) Thermodynamic cost of computation, algorithmic complexity and the information metric. Nature 341: 119–124 [Google Scholar]