Abstract
Phytochromes are plant photoreceptors that regulate plant growth and development with respect to the light environment. Following the initial light-perception event, the phytochromes initiate a signal-transduction process that eventually results in alterations in cellular behavior, including gene expression. Here we describe the molecular cloning and functional characterization of Arabidopsis FHY1. FHY1 encodes a product (FHY1) that specifically transduces signals downstream of the far-red (FR) light-responsive phytochrome A (PHYA) photoreceptor. We show that FHY1 is a novel light-regulated protein that accumulates in dark (D)-grown but not in FR-grown hypocotyl cells. In addition, FHY1 transcript levels are regulated by light, and by the product of FHY3, another gene implicated in FR signaling. These observations indicate that FHY1 function is both FR-signal transducing and FR-signal regulated, suggesting a negative feedback regulation of FHY1 function. Seedlings homozygous for loss-of-function fhy1 alleles are partially blind to FR, whereas seedlings overexpressing FHY1 exhibit increased responses to FR, but not to white (WL) or red (R) light. The increased FR-responses conferred by overexpression of FHY1 are abolished in a PHYA-deficient mutant background, showing that FHY1 requires a signal from PHYA for function, and cannot modulate growth independently of PHYA.
Keywords: Arabidopsis, phytochrome, FHY1, FHY3, far-red light, signal transduction
Light controls the growth of plants through a network of photoreceptors (Kendrick and Kronenberg 1994). These photoreceptors display distinct photosensory and physiological properties, with growth responses to different light wavelengths being attributable to different individual photoreceptors (Kendrick and Kronenberg 1994; Quail et al. 1995). The phytochromes are a small family of red (R)/far-red (FR)-absorbing plant photoreceptors, the best understood of which are phytochrome A (PHYA) and phytochrome B (PHYB). The phytochromes exist in two distinct but interconvertible forms, R-absorbing PR and FR-absorbing PFR, and conversion from PR to PFR initiates phytochrome signaling (Quail et al. 1995).
Recent studies have resulted in several important advances in our understanding of phytochrome function. First, it has become clear that photoconversion of cytoplasmic PR to PFR causes translocation of phytochrome into the nucleus (Sakamoto and Nagatani 1996; Kircher et al. 1999; Yamaguchi et al. 1999). Thus, activation of phytochrome signaling brings phytochrome into the vicinity of the genes that it regulates. Second, PFR interacts with transcriptional regulators such as PIF3 (Ni et al. 1998, 1999; Halliday et al. 1999; Martínez-Garcia et al. 2000; Zhu et al. 2000). Together, these advances have resulted in an emerging general model for phytochrome action, whereby phytochromes perceive light, enter the nucleus, interact with transcriptional regulators, and thus activate gene transcription (Martínez-Garcia et al. 2000). Other studies have also identified important cytoplasmic events in phytochrome signaling, events that possibly influence the nuclear localization of phytochrome (Choi et al. 1999; Fankhauser et al. 1999).
The phytochromes are differentially responsive to light of different wavelengths. For example, in laboratory conditions, Arabidopsis PHYA is uniquely responsible for the response of seedlings to continuous FR (cFR) (Dehesh et al. 1993; Whitelam et al. 1993; Quail et al. 1995). PHYA-deficient mutant seedlings are completely blind to cFR, and exhibit the etiolated phenotypes characteristic of wild-type seedlings grown in continuous dark (cD) (Dehesh et al. 1993; Whitelam et al. 1993). Mutations in FHY1, FHY3, and other loci confer cFR-specific, altered PHYA-signaling phenotypes (Whitelam et al. 1993; Hoecker et al. 1998, 1999; Soh et al. 1998, 2000; Hudson et al. 1999; Bolle et al. 2000; Büche et al. 2000; Fairchild et al. 2000; Fankhauser and Chory 2000; Hsieh et al. 2000). Although many of these loci are cloned, the functional relationships between the proteins that they encode and PHYA are largely unknown.
Here we describe the cloning and characterization of FHY1. We show that FHY1 transcript levels are regulated by light, and by the product of FHY3. This is the first demonstration of a functional relationship between two independent FR-signaling loci. Studies using FHY1–GFP fusion proteins show that FHY1 is found in both nucleus and cytoplasm and that its abundance is influenced by light. We also show that overexpression of FHY1 specifically enhances PHYA signaling. Plants overexpressing FHY1 exhibit enhanced responses to cFR, but not to continuous red (cR) or continuous white (cWL) light. Furthermore, the enhanced FR responses exhibited by FHY1-overexpressing plants are abolished in the absence of functional PHYA, thus showing that FHY1 does not itself have intrinsic growth-modulating activity but is involved in the transduction of the PHYA signal in the regulation of downstream FR responses.
Results
Map-based cloning of FHY1
We cloned FHY1 using a map-based approach (Fig. 1A; Materials and Methods). FHY1 was mapped to the bottom of chromosome 2, between markers ve017 and DET2 (Li et al. 1996; Zachgo et al. 1996). Further recombinant mapping using markers from yeast and bacterial artificial chromosome (YAC and BAC) clones located FHY1 on BAC F16C5 (see Materials and Methods). The FHY1 genomic region was sequenced (GenBank accession no. AC004684) and contains several putative open reading frames (ORFs) (Fig. 1A). A contig of cosmids covering ORFs 9–16 did not complement the fhy1-1 mutant (data not shown). In addition, a single telomeric chromosome breakpoint was detected using an ORF-21-derived RFLP marker, thus excluding ORFs 22–24 from being FHY1. We therefore sequenced ORFs 17–21 from the fhy1-1 allele and from the WT progenitor and identified a single base pair deletion in ORF 18 of fhy1-1 (Fig. 1A), thus permitting the tentative identification of FHY1.
Figure 1.
Molecular cloning of FHY1. (A) Genetic and physical map of part of chromosome 2 containing FHY1. FHY1 was mapped using 714 (rec. 1) and 1708 (rec. 2) chromosomes that were recombinant between FHY1 and the markers cop1-6 and nga168, respectively. We positioned FHY1 on a contig of YACs and BACs (see Materials and Methods) and identified a mutation in the sequence of ORF 18 in the fhy1-1 mutant. The structure of FHY1 is shown (thin line, introns; thick line, ORF; filled thick line, noncoding 5′ and 3′ transcribed sequence) with the position of the fhy1-1 mutation. FHY1 corresponds roughly to, but differs substantially from, the last part of the ORF 18 originally identified by the genefinder program. (B) Genomic DNA gel-blot analysis showing the rearrangement in fhy1-2. DNA was digested with BglII and probed with a radiolabeled FHY1 probe. (C) An FHY1 probe reveals FHY1 transcripts in FHY1, fhy1-1 but not fhy1-2 plants. (Upper panel) RNA gel-blot hybridization of RNA from wild type (WT), fhy1-1, and fhy1-2. (Lower panel) UV fluorescence of RNA blotted and hybridized in panel above. (D) A genomic DNA fragment containing FHY1 complements the fhy1-2 phenotype. Seedlings grown 5 d in cFR are shown; 343 is an fhy1-2 homozygote that is also homozygous for an FHY1-containing transgene.
Further screens for mutants exhibiting a FR-specific elongated hypocotyl phenotype had resulted in the identification of the fast-neutron-generated fhy1-2 allele (Materials and Methods). Molecular analysis using an FHY1 probe detected an altered restriction fragment pattern in fhy1-2 genomic DNA (Fig. 1B). Further analysis of fhy1-2 revealed that this mutation is caused by an inversion of an ∼550-kb DNA fragment from chromosome 2. This inversion interrupts FHY1 and disrupts the FHY1 ORF (data not shown). In addition, FHY1 transcripts were detected in FHY1, at reduced level in fhy1-1 plants, but not in fhy1-2 plants (Fig. 1C). These observations suggest that fhy1-2 is a loss-of-function allele of FHY1. Furthermore, because the FR-response phenotypes conferred by fhy1-1 and fhy1-2 are indistinguishable (data not shown), it is likely that fhy1-1 (which potentially encodes a prematurely truncated protein; see below) is also a loss-of-function allele of FHY1. Final proof that the gene tentatively identified as FHY1 is indeed FHY1 was obtained via genetic complementation. When grown in cFR, the fhy1-2 mutant displays an elongated hypocotyl and unexpanded cotyledons (Fig. 1D). A 2.1-kb genomic fragment, spanning 795 bp upstream to 304 bp downstream of the FHY1 coding sequence, fully complemented fhy1-2, conferring a phenotype resembling that of the wild-type control (short hypocotyl and expanded cotyledons in cFR; Fig. 1D).
FHY1 encodes a predicted polypeptide (FHY1) of 23 kD (202 amino acids) that is rich in serine and glutamic acid residues (Fig. 2A). FHY1 has no obvious overall similarity to any protein of known function, and contains putative nuclear localization sequence (NLS) and nuclear export sequence (NES) domains (Fig. 2A; Nigg 1997). fhy1-1 lacks a single adenine residue 671 bp downstream of the FHY1 start codon, resulting in a premature STOP codon (Fig. 2A). Database searches revealed the existence of a second Arabidopsis ORF that encodes a predicted protein that is related in sequence to FHY1, together with FHY1-related ESTs from soybean and tomato (data not shown). Interestingly, FHY1 contains a region of shared homology with part of the PHYA PAS-A domain (Fig. 2B; Quail et al. 1995). The biological significance of this shared homology is unknown, because it does not overlap with the region of the PAS domain that is conserved in PAS-domain proteins involved in a variety of roles from photoperception to circadian clock function (Heintzen et al. 2001).
Figure 2.
FHY1 sequence. (A) Predicted amino acid sequence of FHY1. Arrowheads indicate intron position; the putative NLS is single-underlined; the putative NES is underlined with a dotted line (for consensus NLS and NES sequences, see Nigg 1997). A region of homology with the phytochrome PAS-A domain (Quail et al. 1995) is double-underlined (see B). (B) Sequence alignment of a region conserved between FHY1, PHYA, and phytochrome C (PHYC). Numbers refer to the N-terminal residue position for each respective sequence. Identical residues are shown on a black background, similar residues are shown on a gray background. The SwissProt accession nos. are P14712: Arabidopsis thaliana PHYA [PhyA (A. t.)]; P06593: Avena sativa PHYA [PhyA (A. s.)], and P14714: Arabidopsis thaliana PHYC [PhyC (A. t.)].
FHY1 expression is regulated by light and by FHY3
It seemed likely that seedling FHY1 levels are related to morphogenesis in cFR, for the fhy1-1 loss-of-function allele confers a cFR-specific elongated hypocotyl phenotype (Whitelam et al. 1993). We addressed this possibility in two ways. First, we investigated the environmental and genetic factors that regulate FHY1 gene expression. Second, we studied the effect of increased expression of FHY1 on morphogenesis in cFR (see below).
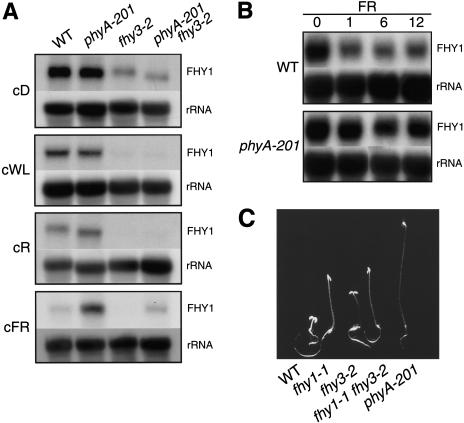
We examined FHY1 mRNA levels in seedlings grown in cD and in various light conditions: cWL, cR, and cFR (Fig. 3A). In wild-type seedlings, FHY1 mRNA levels accumulated to higher levels in cD than in any light condition, suggesting that light negatively regulates FHY1 gene expression. In cFR, but not in cWL or cR, FHY1 transcript levels were higher in PHYA-deficient phyA-201 seedlings than in wild-type seedlings (Fig. 3A). Therefore, PHYA reduces FHY1 mRNA levels in cFR, and other photoreceptors do the same in cWL and cR.
Figure 3.
FHY1 mRNA level is regulated by light and by FHY3. (A) RNA gel-blot analysis of FHY1 transcripts in seedlings grown for 5 d in cD, cWL, cR, or cFR. (B) RNA gel-blot analysis of FHY1 transcripts in wild-type (WT) seedlings grown for 3 d in cD and then exposed to cFR for 0–12 h. In A and B, loading control hybridization with rDNA probe is shown. (C) Phenotype of seedlings grown for 5 d in cFR.
When etiolated wild-type seedlings were moved into cFR, FHY1 transcript levels decreased within an hour of exposure, and then remained unchanged (Fig. 3B). This response was not observed in phyA-201 seedlings (Fig. 3B), and is therefore PHYA-dependent. This rapid PHYA-dependent down-regulation of FHY1 mRNA levels indicates the existence of a negative feedback loop, whereby PHYA influences the abundance of FHY1 transcripts, and possibly of FHY1 itself (see below).
Like the fhy1-1 and fhy3-2 single mutants, the fhy1-1 fhy3-2 double mutant displayed a reduced cFR response (Fig. 3C). However, the cFR-grown fhy1-1 fhy3-2 hypocotyl was only slightly longer than that of fhy1-1 (in both standard and nonsaturating cFR conditions; Fig. 3C; data not shown), and a strongly synergistic, cFR-blind phenotype was not observed. This observation suggests that FHY1 and FHY3 may act in the same pathway, and that, in addition to the FHY1–FHY3 pathway, additional pathways may mediate the cFR response. Alternatively, FHY1 and FHY3 may act in separate pathways. We investigated FHY1 mRNA levels in fhy3-2 seedlings (Fig. 3A). In all conditions, fhy3-2 seedlings accumulated less FHY1 transcript than did wild-type seedlings, showing that FHY3 positively regulates FHY1 mRNA levels, and suggesting that FHY1 acts downstream of FHY3. Furthermore, fhy3-2 seedlings still exhibited light-dependent reductions in FHY1 mRNA levels (Fig. 3A), suggesting that these reductions are not FHY3-dependent.
Because cFR and FHY3 have opposite effects on FHY1 transcript levels, FHY1 expression in a phyA-201 fhy3-2 double mutant was investigated. In cFR, FHY1 mRNA levels in phyA-201 fhy3-2 were intermediate between those of phyA-201 and fhy3-2, and were similar to those of wild type (Fig. 3A). This suggests that PHYA and FHY3 regulate FHY1 transcript levels independently of each other.
Increased FHY1 expression specifically enhances the cFR response
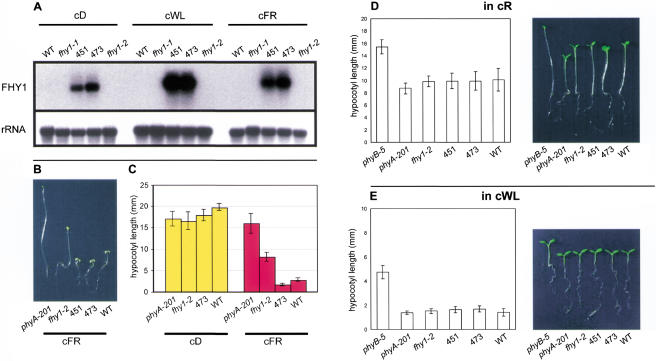
The above observations on FHY1 expression present a paradox: reductions in FHY1 function (in fhy1 loss-of-function mutants) reduce the cFR response, but apparent reductions in FHY1 function (via reduced FHY1 transcript levels) are also part of the cFR response. To resolve this paradox we investigated the consequences of increasing FHY1 expression (in a way that was not subject to down-regulation by light). fhy1-2 plants that expressed FHY1 under the control of the CaMV 35S promoter had elevated FHY1 transcript levels in cD, cWL, and cFR (Fig. 4A). These plants exhibited an enhanced cFR response, being more deetiolated in cFR than wild-type plants (Fig. 4B,C). Therefore, overexpression of FHY1 enhances the FR response above that of wild-type plants. To determine if this effect was FR-specific, plants overexpressing FHY1 were also grown in cD, cR, and cWL (Fig. 4C,D,E, respectively). In all cases, FHY1-overexpressing plants were indistinguishable from wild-type controls, thereby showing that the effect of FHY1 overexpression is light-dependent and FR-specific. Furthermore, these observations suggest that the paradox outlined above can be resolved if FHY1 function is (in normal plants) subject to negative feedback regulation via PHYA at the transcriptional level (as shown above).
Figure 4.
Seedlings overexpressing FHY1 exhibit enhanced cFR responses. (A) Plants containing the 35S::FHY1 construct have elevated levels of FHY1 transcript. (Upper panel) FHY1 transcripts are detectable in 451 and 473 (fhy1-2 lines containing a 35S::FHY1 transgene) but not detectable in wild type (WT), fhy1-1, or fhy1-2 in cD, cWL, or cFR. This was a shorter exposure than in Figure 3A, thus explaining why FHY1 transcripts are visible in cD-grown WT in Figure 3A but not in this figure. (Lower panel) rRNA loading control for hybridization in upper panel. (B) 35S::FHY1;fhy1-2 hypocotyls exhibit enhanced responses to cFR. Seedlings from lines 451 and 473 (35S::FHY1;fhy1-2) have shorter hypocotyls in cFR than WT, fhy1-2, or phyA-201. (C) Histogram shows mean and standard error of hypocotyl lengths of seedlings grown in cD and cFR (n = 25–38). Seedlings grown in cD are indistinguishable from one another. 473 (35S:FHY1;fhy1-2) hypocotyls are significantly shorter than WT hypocotyls in cFR. (D) 35S::FHY1;fhy1-2 hypocotyls do not exhibit enhanced responses to cR. Photograph and histogram of seedlings of various genotypes. Histogram shows mean and standard error of hypocotyl lengths (n = 24–28). For reference, the phyB-5 mutant, which exhibits a long hypocotyl in cR, is shown. The 451 and 473 lines are indistinguishable from WT. (E) 35S::FHY1;fhy1-2 hypocotyls do not exhibit enhanced responses to cWL. Photograph and histogram of seedlings of various genotypes. Histogram shows mean and standard error of hypocotyl lengths (n = 18–23). The 451 and 473 lines are indistinguishable from WT.
FHY1 requires a signal from PHYA to effect a cFR response
We investigated the effect of FHY1 overexpression in the PHYA-deficient phyA-201 mutant. As shown in Figure 5A, phyA-201 plants containing the 35S::FHY1 construct are blind to cFR, and indistinguishable from control phyA-201 plants. To confirm that these plants were expressing the 35S::FHY1 construct, RNA gel-blot analysis showed that the phyA-201 35S::FHY1 line accumulated FHY1 transcripts to levels comparable with that seen in the control 35S::FHY1 line (Fig. 5B). Therefore, overexpression of FHY1 causes enhanced cFR responses in the presence (Fig. 4B,C), but not in the absence (Fig. 5A), of PHYA. FHY1 cannot influence growth in the absence of a signal from PHYA, suggesting that the enhanced FR responses of plants overexpressing FHY1 (in the presence of PHYA) are attributable to a FHY1 overexpression-dependent enhancement of the PHYA signal.
Figure 5.
The enhanced FR responses of seedlings overexpressing FHY1 are PHYA-dependent. (A) Photograph and histogram of seedlings of various genotypes grown in cFR. Histogram shows mean and standard error of hypocotyl lengths (n = 29). phyA-201 and phyA-201 35S::FHY1 seedlings have indistinguishable phenotypes in FR. (B) RNA gel-blot hybridization showing that phyA-201 35S::FHY1 seedlings have similar levels of FHY1 transcripts to 35S::FHY1 seedlings. (Upper panel) FHY1 transcripts; (lower panel) ribosomal RNA (loading control). (C) Hypocotyl lengths of various genotypes grown in high-intensity (HiFR) and low-intensity (LoFR) cFR. Histograms show mean and standard error of hypocotyl lengths (n = 23–32). For comparison, seedlings from lines 451 and 473 (35S::FHY1 fhy1-2 lines) are also shown.
In further experiments, we investigated the consequences of FHY1 overexpression in mutant plants containing a partially defective PHYA (Fig. 5C). The phyA-3 and phyA-4 alleles encode mutant PHYA polypeptides whose function is reduced, but not completely abolished. For example, phyA-3 confers a hypocotyl that is taller than wild type, but shorter than that conferred by the phyA-201 null allele in high-intensity cFR (Hi-cFR), indicating that PHYA-3 function is reduced compared with that of PHYA (Fig. 5C). In low-intensity cFR (Lo-cFR), the phyA-3 and phyA-201 hypocotyls are of a more similar length (Fig. 5C). When the 35S:FHY1 construct was introduced into phyA-3, it partially suppressed the cFR phenotype of phyA-3. In Hi-cFR conditions, the phyA-3 35S::FHY1 line (043) had shorter hypocotyls than the phyA-3 controls, an effect that was less apparent in Lo-cFR (Fig. 5C). These effects are cFR-dependent, because, when grown in cD, these lines were fully etiolated (data not shown). Furthermore, the 35S::FHY1 construct also suppressed the phenotype of phyA-4 (data not shown), indicating that FHY1 overexpression suppresses the general effects of a reduced PHYA signal, rather than any specific effects of the individual mutant PHYA alleles. These observations suggest that overexpression of FHY1 enhances responses to a reduced PHYA signal.
FHY1–GFP fusion proteins can be detected in the hypocotyl cells of etiolated plants
The intracellular localization of FHY1 was investigated using plants containing a transgene construct that expressed a FHY1–GFP fusion protein under the control of the FHY1 promoter. Preliminary experiments showed that this construct complemented the fhy1-2 phenotype, thus showing that the FHY1–GFP fusion protein has FHY1 function (data not shown).
Fluorescence microscopy was used to determine the intracellular location of the FHY1–GFP fusion protein. As shown in Figure 6, strong fluorescence was observed in the nuclei of the hypocotyl cells of cD-grown FHY1–GFP-expressing plants, but not in hypocotyl nuclei of cFR-grown FHY1–GFP-expressing plants, or of cD- or cFR-grown control (nontransgenic) plants. In addition, as shown in Figure 7A, weak fluorescence was also observed in the cytoplasm of the hypocotyl cells of cD-grown FHY1–GFP-expressing plants. This fluorescence was clearly brighter than the background fluorescence seen in nontransgenic controls (Fig. 7A). Therefore, FHY1–GFP is distributed (in a manner similar to that of GFP itself; Fig. 7A) between the nucleus and the cytoplasm of cD-grown hypocotyls, although the nuclear fluorescence is stronger than that of the cytoplasm. In further experiments, we looked to see if the nuclear localization of FHY1–GFP would change rapidly following transfer of seedlings from cD to cFR (Fig. 7B). These experiments showed that the nuclear fluorescence attributed to FHY1–GFP was still detectable 2 h after transfer to cFR. In addition, there appeared to be a slight increase in cytoplasmic fluorescence following 2 h of cFR. However, the fact that clear nuclear fluorescence was retained following the 2-h cFR treatment suggests that nuclear FHY1–GFP is not rapidly relocalized or destroyed following transfer of hypocotyls from cD to cFR. Additional experiments showed that fluorescence due to FHY1–GFP in seedlings first grown in cD is reduced by subsequent light treatments of longer duration. We observed that 24 h of cFR was needed to reduce the fluorescence to almost undetectable levels (data not shown). Interestingly, treatments with cB, cR, or cWL resulted in a more rapid loss, with fluorescence essentially disappearing between 4 and 12 h of the onset of exposure (data not shown).
Figure 6.
FHY1–GFP is detected in cD-grown hypocotyls. Line 133 expresses FHY1–GFP in an fhy1-2 genetic background. GFP is detected in nuclei and cytoplasm of cD-grown 133 hypocotyls (cf. same hypocotyl in A [GFP fluorescence] and B [DAPI-stained nuclei]), but not in WT hypocotyls (that lack the FHY1–GFP expressing transgene, cf. C and D). GFP fluorescence is not detected in cells of cFR-grown 133 or WT hypocotyls (cf. E with F, G with H). All GFP fluorescence images were obtained using the same camera set-up parameters. Therefore, FHY1–GFP accumulates in the cells of cD-grown hypocotyls but not in the cells of cFR-grown hypocotyls. Bar, 100 μm. Seedlings were 3 d old.
Figure 7.
Nuclear FHY1–GFP is not rapidly relocalized or destroyed following transfer of hypocotyls from cD to light. (A) GFP fluorescence is clearly detectable in hypocotyl nuclei and cytoplasm of cD-grown 3-day-old 133 and 35S::GFP seedlings, but not in WT seedlings. (B) Compared with 3-day-old cD-grown 133 seedlings (cD + 2 h cD), GFP fluorescence is still detectable in the hypocotyl nuclei of cD-grown 133 seedlings transferred into cFR for 2 h (cD + 2 h cFR). Bar, 100 μm.
Discussion
Phytochrome action involves light-dependent nuclear targeting, light-dependent phosphorylation of one or more signaling component substrates, and the regulation of nuclear gene expression via transcription factors (Quail et al. 1995; Ni et al. 1998, 1999; Yeh and Lagarias 1998; Choi et al. 1999; Fankhauser et al. 1999; Halliday et al. 1999; Kircher et al. 1999; Yamaguchi et al. 1999; Martínez-Garcia et al. 2000). In addition, there is evidence suggesting the involvement of GTP-binding proteins, cGMP, and Ca2+/calmodulin (CaM) in phytochrome signaling (Neuhaus et al. 1993; Bowler et al. 1994). Although PHYA likely operates via these general mechanisms, there is clear evidence for the existence of additional components, some of them nuclear, that appear to be more specific for PHYA signaling (Hoecker et al. 1999; Hudson et al. 1999; Bolle et al. 2000; Fairchild et al. 2000; Hsieh et al. 2000; Soh et al. 2000). In this paper we describe the isolation and characterization of FHY1, a gene encoding the PHYA-specific signaling component FHY1. Previous experiments indicated that FHY1 links PHYA to gene activation via a cGMP-dependent pathway (Barnes et al. 1996), or that FHY1 functions within a PHYA signaling pathway that involves a Gα subunit (Okamoto et al. 2001). Here we have shown that FHY1 transcript levels are regulated by light and by the product of FHY3, another gene involved in FR-signaling (Whitelam et al. 1993). We have also shown that overexpression of FHY1 confers an exaggerated response to cFR, and unaltered responses to cD, cR, and cWL. This, together with the observation that loss-of-function fhy1 alleles confer a FR-specific phenotype (Whitelam et al. 1993), shows that FHY1 function is specifically related to FR-signaling. Furthermore, we have shown that overexpression of FHY1 confers increased responses to cFR only in the presence of functional PHYA, thus confirming that FHY1 acts in PHYA-signaling and has no intrinsic effect in promoting FR responses.
FHY1 presumably operates within the context of the action of other FR-specific signaling components, such as those encoded by the FHY3, SPA1, PAT1, FIN2, FAR1, FIN219, HFR1, and REP1 loci (Whitelam et al. 1993; Soh et al. 1998; Hoecker et al. 1998, 1999; Hudson et al. 1999; Bolle et al. 2000; Fairchild et al. 2000; Hsieh et al. 2000; Soh et al. 2000). Some of these components have been identified and partially characterized, and it is instructive to attempt to draw together these various observations to provide an overall view of how PHYA-signaling works. As outlined above, PHYA exists in the PR form in the cytoplasm and, upon conversion to PFR, becomes localized in the nucleus. Two of the above loci, PAT1 and FIN219, encode products that appear to be cytoplasmically localized (Bolle et al. 2000; Hsieh et al. 2000). Perhaps PAT1 and FIN219 are involved with cytoplasmic stages of PHYA-signaling, or with the targeting of the PFR form of PHYA to the nucleus. All of the other FR-signaling loci cloned to date appear to encode nuclear factors: SPA1 is a nuclear WD-repeat protein that acts as a negative regulator of FR-signaling (Hoecker et al. 1999), FAR1 is a novel nuclear protein of unknown function that appears to be a positive regulator of FR-signaling (Hudson et al. 1999), and HFR1 (also known as REP1) is a positive regulator of FR-signaling and a member of the basic helix–loop–helix (bHLH) family of DNA-binding proteins (Fairchild et al. 2000; Soh et al. 2000). FHY1, as shown here, is a novel protein that can exist in both nucleus and cytoplasm and that acts as a positive regulator of FR-signaling. Presumably the various nuclear factors act together with PHYA within complexes that regulate the transcription of PHYA-regulated genes. Because some mutants, such as the hfr1 and fhy1 mutants (Whitelam et al. 1993; Johnson et al. 1994; Barnes et al. 1996; Fairchild et al. 2000), affect subsets of the full FR response, it seems possible that individual components may target the regulation of defined subsets of PHYA-regulated genes. In addition, it might be that FHY1 function partially overlaps with that of the FHY1-related gene also found in the Arabidopsis genome, and that loss-of-function alleles at both loci are required to completely abolish FR responses.
We have shown that FHY1–GFP accumulates predominantly in the nucleus (but is also detectable in the cytoplasm) in cD-grown seedlings, is not found in light-grown seedlings, and disappears with relatively slow kinetics from seedlings moved from cD into the light. At present we do not know how these different nucleocytoplasmic distributions relate to FHY1 function. However, FHY1 behavior follows a pattern observed with several light-signaling proteins including phytochrome A, COP1, and HY5 (von Arnim and Deng 1994; Kircher et al. 1999; Osterlund et al. 2000), whereby light-dependent differential stability and/or light-dependent differential subcellular localization are important components of function.
Although several different factors have been identified as FR-signaling components, there is little information concerning how these factors interact with each other, with PHYA, or within the context of information from photoreceptors other than PHYA itself. Here, we have described the results of initial experiments that investigate the effects of some of these factors on the expression of FHY1. First, we have shown that FHY1 expression (at the level of transcript accumulation) is down-regulated by light. PHYA is clearly involved in this process, suggesting negative-feedback regulation of FR-signaling by cFR via PHYA. However, additional photoreceptors are also involved. Regulation of FHY1 mRNA levels by photoreceptors other than PHYA may reflect a point of interaction between the signal-transduction pathways associated with these different photoreceptors and the PHYA signal-transduction pathway. Intriguingly, the FHY1 promoter contains a G-box consensus sequence similar to that bound by PIF3 (Martínez-Garcia et al. 2000; data not shown), perhaps identifying a site of regulation by PHYB, PHYA, or other phytochromes.
The effect of light on the expression of other FR-signaling genes has also been investigated. For example, SPA1 mRNA levels were increased when dark-grown (D) seedlings were transferred to cFR or cR. The effect of cFR on SPA1 transcript levels was PHYA-mediated, whereas the effect of cR was mediated by PHYA, PHYB, and additional phytochromes (Hoecker et al. 1999). Therefore, there are clear parallels between the regulation of FHY1 and SPA1 expression. Both genes display PHYA-mediated cFR regulation of their respective transcript levels, and cR regulates the transcript levels of both genes via phytochromes additional to PHYA. Furthermore, it is interesting to note that cFR promotes the accumulation of SPA1 transcripts (which encode a negative regulator of FR-signaling) and reduces the accumulation of FHY1 transcripts (which encode a positive regulator of FR-signaling). However, this pattern does not hold for all FR-signaling genes, because the levels of FAR1 transcripts (which encode a positive regulator of FR-signaling) are unaffected by light (Hudson et al. 1999) and levels of HFR1 transcripts (which also encode a positive regulator of FR-signaling) are actually increased by cFR (Fairchild et al. 2000).
FHY1 expression is also regulated, in a light-independent fashion, by the FHY3 gene product. This result is significant because it shows, for the first time, a clear relationship between the function of different genes involved in regulating FR-signaling. Further work is needed to determine the precise relationships among PHYA, FHY1, and FHY3. It is possible that FHY1 and FHY3 act in the same pathway. However, the FR phenotype conferred by fhy3 mutants is different from that conferred by fhy1 mutants (Fig. 3C). Therefore, it is unlikely that regulation of FHY1 transcript levels is the sole FR-related function of FHY3. Alternatively, FHY3 could regulate FHY1 transcript levels independently of PHYA-signaling, but additionally be part of a PHYA-signaling pathway that involves PHYA, FHY1, and FHY3. In addition, recent evidence suggests that FHY1 and FHY3 may operate within separate signaling pathways (Okamoto et al. 2001).
We have shown that FHY1 is functional only if PHYA is also functional. Plants overexpressing FHY1 in the absence of functional PHYA do not exhibit the enhanced FR responses exhibited by FHY1-overexpressing plants possessing normal PHYA function. Therefore, the function of FHY1 is clearly associated with the transduction of the PHYA signal, and in the absence of a PHYA signal, FHY1 has no obvious function. In this respect, FHY1 function is similar to that of SPA1, because the phenotype of spa1 mutants is dependent on the presence of functional PHYA (Hoecker et al. 1999). Furthermore, it has been suggested that the spa1 mutations specifically amplify PHYA-signaling and that the function of SPA1 is therefore to diminish PHYA-signaling (Hoecker et al. 1999). Here we have shown that overexpression of FHY1 in partially defective PHYA mutants results in enhancement of the response of plants to the reduced PHYA signals generated by the defective PHYA proteins. Perhaps FHY1 and SPA1 act as counterbalancing amplifying and dampening controls on the transmission of the PHYA signal in the modulation of FR responses. Intriguingly, spa1 mutant seedlings are hyperresponsive to cR, in a PHYA-dependent fashion (Hoecker et al. 1999), but 35S::FHY1 seedlings are not (this study; Fig. 4D). These observations indicate that SPA1 does not inhibit PHYA-signaling via FHY1. Alternatively, overexpression of FHY1 alone is not sufficient to enhance cR-responsiveness.
In conclusion, our observations suggest that FHY1 accumulates in the cells of cD-grown seedlings in preparation for light-activated nuclear import of PHYA, and then acts as part of a specific PHYA signal-transduction mechanism. FHY1 may achieve this by imposing specificity/selectivity on the general transcription factor-mediated phytochrome gene-activation mechanism, and appears to act as a specific amplifier of PHYA signaling.
Materials and methods
Mutant lines
Throughout this paper, the wild type is Arabidopsis thaliana ecotype Landsberg erecta. phyA-201, fhy1-1, and fhy3-1 are as described (Whitelam et al. 1993; Quail et al. 1994). fhy1-2 and fhy3-2 were identified in a fast-neutron mutagenized wild-type population (Lehle Seeds, Round Rock, TX). phyA-3 and phyA-4 (named according to Quail et al. 1994) were identified in an ethyl methane sulfonate-mutagenized wild-type population, and contain missense alleles that confer a substantial reduction in, but not complete loss of, PHYA function (data not shown). All putative double-mutant homozygotes were test-crossed to both parents to confirm their genotypes.
Molecular cloning of FHY1
Mapping populations were generated by crossing fhy1-1 (Landsberg erecta background) with wild type or cop1-6 (Columbia background). DNA from fhy1-1 homozygote F2 or F3 plants was analyzed with cleaved amplified polymorphic sequence (CAPS; Konieczny and Ausubel 1993) and simple sequence length polymorphism (SSLP; Bell and Ecker 1994) markers. FHY1 (FHY1 sequence has been deposited in GenBank, accession no. AF432142).was mapped to the bottom of chromosome 2, with 714 and 1708 recombinant chromosomes identified between FHY1 and the markers cop1-6 and nga168, respectively. We designed a CAPS marker with the gene DET2 (Li et al. 1996; GenBank accession no. U53860) (DdeI cleavage of a PCR product amplified with primers 5′-AAATCCAATACCGGCCCAA GACCA-3′ and 5′-AAATCCAATACCGGCCCAAGACCA-3′). Analysis of the recombinant chromosomes enabled us to place FHY1 between the markers ve017 and DET2. With these two markers and published data (Li et al. 1996; Zachgo et al. 1996), we constructed a new yeast artificial chromosome (YAC) map of the region. Total yeast DNA from the selected CIC YACs (Creusot et al. 1995) and yUP (Ecker 1990) clones was extracted according to Gibson and Somerville (1992). The YAC-ends CIC10E10-R, CIC5G4-L, CIC5G4-R, and yUP2C12-L were isolated by inverse PCR for the right end or by plasmid rescue for the left end (Gibson and Somerville 1992) and converted into RFLP markers. Nylon filters spotted with the IGF (Mozo et al. 1998a,b) and TAMU (Choi et al. 1996) bacterial artificial chromosome (BAC) libraries of Arabidopsis Col0 genomic DNA were screened with the 32P-labeled yUP2C12-L marker. The relative positions of the BACs were determined by using BAC-end derived markers isolated as described (http://www.tigr.org/tdb/at/atgenome/atgenome.html), a BamHI subclone of the BAC F16C5 (B4-1), a PCR product of the ACT1 gene (which corresponds to ORF 12 in Fig. 1; amplified with primers 5′-TGTAGCGCTTTTGTGTCCTTATGG-3′ and 5′-CGGCTT GAGAAATGGTCGGA-3′), and the yUP6B10-R fragment. Analysis of the cop1-6/fhy1-1 recombinants showed that B4-1, an RFLP marker derived from BAC F16C5, mapped centromeric to FHY1, thus showing that F16C5 covers FHY1.
The CD4-14 λZapII cDNA library (Kieber et al. 1993) was screened with a fragment of the second exon of FHY1 (PCR-amplified with primers 18-9R, 5′-TCACATGATCATAAG TAGTAGTAAA-3′; and 18-5F, 5′-CCAGAGGACAGAACAAA CTTAGCA-3′). Fourteen positive clones were isolated from ∼8.5 × 105 pfu, of which 11 were sequenced. These cDNAs represented a single gene (FHY1). Total RNA was extracted from cD-grown wild-type seedlings, and 5′-RACE (Frohman et al. 1988) was performed using the Marathon kit (Stratagene) and primer 18-5F. PCR products were cloned into the pGEM-T easy vector (Promega), and two clones were sequenced. The FHY1 transcript contains an in-frame STOP codon upstream of the first ATG, suggesting that the FHY1 ORF (Fig. 1) represents the complete FHY1 protein.
For molecular complementation with constructs containing FHY1, plant transformations were as described (Clough and Bent 1998).
DNA sequencing was performed using the Big Dye terminator cycle sequencing kit (Perkin Elmer).
Transcript analysis
Total RNA was separated on denaturing gels and transferred onto Hybond-NX (Amersham). An 18-5F/18-9R PCR fragment was cloned into the pGEM-T easy vector (Promega), and used to make an [α-32P]UTP-labeled RNA probe (Riboprobe systems, Promega). The ribosomal control was as in Deng et al. (1991).
Plant growth and light conditions
In vitro seedling growth medium was as described in Estelle and Somerville (1987), but without sugar. For FR, light sources were FL20S.FR74 bulbs (Toshiba), output filtered through black plexiglass (A.S.H Filters Ltd, UK); for R, FL20SS.BRN/18 bulbs (Toshiba, Japan), output filtered through Fire-red filter (Lee Filters, UK), intensity 3 W/m2; for WL, as described in Whitelam et al. (1993). Light intensities were measured with a Li-Cor model LI-185-B radiometer with a Li-Cor pyranometer probe.
Transgenic lines overexpressing FHY1
A PCR product, amplified from a subclone of BAC F16C5 using primers 18-19R (5′-AAGATCTATGCCTGAAGTGGAAGTG-3′; containing a BglII site) and 18-8F (5′-CAGGGATACTCTT GAACA-3′), was cloned in the pGEM-T easy vector (Promega), isolated as a BglII/SacI fragment, and inserted behind the CaMV 35S promoter in a BamHI/SacI opened pBIN121 derivative vector. The construct was introduced into fhy1-2, phyA-3, and phyA-4 mutants. Transformants were kanamycin-selected, and the progeny were tested in cFR. Transformants grown in cD were indistinguishable from untransformed controls. A phyA-4 transformant was crossed with phyA-201; F2 and F3 progeny of this cross were screened for plants homozygous for both the phyA-201 mutation and the transgene.
Transgenic lines expressing FHY1–GFP
The FHY1 gene and 795 bp of upstream sequence were amplified by PCR from a genomic DNA clone, inserted upstream of, and in frame with, the GFP4 coding sequence in a pBIN121 derivative vector, and then transformed into fhy1-2. Seedlings (progeny of primary transformants) were examined under UV light with a Leica DMRXA microscope equipped with filter A (Leica) and filter XF115 (Omega Optical) for GFP and DAPI (4‘,6-diamidino-2-phenylindole) fluorescence, respectively. For DAPI staining of the nuclei, seedlings were dipped in isopropanol for a few seconds, rinsed in water, stained in DAPI (1 μg/mL) for 5 min, washed 2 min in water, and mounted on a microscope slide. Images were acquired using a SPOT camera (Diagnostic Instruments). For the photographs in Figure 7, A and B, the transgenic and control seedlings were mounted side by side on the same slide, and both photographs were taken with the same microscope and camera settings. The 35S::GFP line is in the WS background.
Acknowledgments
We thank Minami Matsui for helping us to obtain appropriate light sources; C. Lister and L. Bent for YACs, BACs, and cosmid libraries (John Innes Centre, Norwich, UK); N. Hartley for DNA sequencing; J. Peng, D. Richards, T. Ait-ali, and others in the Molecular Genetics Department for helpful discussions; L. Nussaume (LMC) for helping with microscopy and for endless support; K. King for helping with the figures; M. Smith for plant care; J. Azimzadeh (INRA Versailles) for the 35S::GFP line; and the Arabidopsis Biological Resource Center, Ohio, for phyA-201, cop1-6, and the CD14-4 cDNA library. This work was supported by BBSRC (Core Strategic Grant to the John Innes Centre; Link grant to G.C.W. and N.P.H.) and EU Framework IV (BIO4-CT-972124). P.P. was supported in part by an EU Human Capital and Mobility Fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL nicholas.harberd@bbsrc.ac.uk; FAX 44-1603-450025.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.205401.
References
- Barnes SA, Quaggio RB, Whitelam GC, Chua N-H. fhy1 defines a branch point in phytochrome A signal transduction pathways for gene expression. Plant J. 1996;10:1155–1161. doi: 10.1046/j.1365-313x.1996.10061155.x. [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bolle C, Koncz C, Chua N-H. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes & Dev. 2000;14:1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Neuhaus G, Yamagata H, Chua N-H. Cyclic GMP and calcium mediate phytochrome phototransduction. Cell. 1994;77:73–81. doi: 10.1016/0092-8674(94)90236-4. [DOI] [PubMed] [Google Scholar]
- Büche C, Poppe C, Schäfer E, Kretsch T. Eid1: A new Arabidopsis mutant hypersensitive in phytochrome A-dependent high-irradiance responses. Plant Cell. 2000;12:547–558. [PMC free article] [PubMed] [Google Scholar]
- Choi S, Creelman RA, Mullet JA, Wing RA. Construction and characterization of a bacterial artificial chromosome library of Arabidopsis thaliana. Weeds World. 1996;2:17–20. [Google Scholar]
- Choi G, Yi H, Lee J, Kwon YK, Soh MS, Shin B, Luka Z, Hahn TR, Song PS. Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature. 1999;401:610–613. doi: 10.1038/44176. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Creusot F, Fouilloux E, Dron M, Lafleuriel J, Picard G, Billault A, Le Paslier D, Cohen D, Chaboue ME, Durr A, et al. The CIC library: A large insert YAC library for genome mapping in Arabidopsis thaliana. Plant J. 1995;8:763–770. doi: 10.1046/j.1365-313x.1995.08050763.x. [DOI] [PubMed] [Google Scholar]
- Dehesh K, Franci C, Parks BM, Seeley KA, Short TW, Tepperman JM, Quail PH. Arabidopsis HY8 locus encodes phytochrome A. Plant Cell. 1993;5:1081–1088. doi: 10.1105/tpc.5.9.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X-W, Caspar T, Quail PH. cop1, a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes & Dev. 1991;5:1172–1182. doi: 10.1101/gad.5.7.1172. [DOI] [PubMed] [Google Scholar]
- Ecker JR. PFGE and YAC analysis of the Arabidopsis genome. Methods. 1990;1:186–194. [Google Scholar]
- Estelle MA, Somerville C. Auxin-resistance mutants of Arabidopsis thaliana with altered morphology. Mol Gen Genet. 1987;206:200–206. [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes & Dev. 2000;14:2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. RSF1, an Arabidopsis locus implicated in phytochrome A signalling. Plant Physiol. 2000;124:39–45. doi: 10.1104/pp.124.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Yeh KC, Lagarias JC, Zhang H, Elich TD, Chory J. PKS1, a substrate phosphorylated by phytochrome that modulates light signalling in Arabidopsis. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson S, Somerville C. Chromosome walking in Arabidopsis thaliana using yeast artificial chromosomes. In: Koncz C, et al., editors. Methods in Arabidopsis research. London, UK: World Scientific Publishing; 1992. pp. 119–143. [Google Scholar]
- Halliday KJ, Hudson M, Ni M, Qin M, Quail PH. poc1: An Arabidopsis mutant perturbed in phytochrome signalling because of a T-DNA insertion in the promoter of PIF3, a gene encoding a phytochrome-interacting bHLH protein. Proc Natl Acad Sci. 1999;96:5832–5837. doi: 10.1073/pnas.96.10.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzen C, Loros J, Dunlap JC. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell. 2001;104:453–464. doi: 10.1016/s0092-8674(01)00232-x. [DOI] [PubMed] [Google Scholar]
- Hoecker U, Xu Y, Quail PH. SPA1: A new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell. 1998;10:19–33. doi: 10.1105/tpc.10.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH. SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science. 1999;284:496–499. doi: 10.1126/science.284.5413.496. [DOI] [PubMed] [Google Scholar]
- Hsieh H-L, Okamoto H, Wang M, Ang L-H, Matsui M, Goodman H, Deng X-W. FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes & Dev. 2000;14:1958–1970. [PMC free article] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signalling. Genes & Dev. 1999;13:2017–2027. doi: 10.1101/gad.13.15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E, Bradley M, Harberd NP, Whitelam GC. Photoresponses of light-grown phyA mutants of Arabidopsis: Phytochrome A is required for the perception of daylength extensions. Plant Physiol. 1994;105:141–149. doi: 10.1104/pp.105.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick RE, Kronenberg GHM. Photomorphogenesis in plants. 2nd ed. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schäfer E, Nagy F. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell. 1999;11:1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Martínez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Mozo T, Fischer S, Meier-Ewert S, Lehrach H, Altmann T. Use of the IGF BAC library for physical mapping of the Arabidopsis thaliana genome. Plant J. 1998a;16:377–384. doi: 10.1046/j.1365-313x.1998.00299.x. [DOI] [PubMed] [Google Scholar]
- Mozo T, Fischer S, Shizuya H, Altmann T. Construction and characterization of the IGF Arabidopsis BAC library. Mol Gen Genet. 1998b;258:562–570. doi: 10.1007/s004380050769. [DOI] [PubMed] [Google Scholar]
- Neuhaus G, Bowler C, Kern R, Chua N-H. Calcium/calmodulin-dependent and -independent phytochrome signal transduction pathways. Cell. 1993;73:937–952. doi: 10.1016/0092-8674(93)90272-r. [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- ————— Binding of phytochrome B to its nuclear signaling partner PIF3 is reversibly induced by light. Nature. 1999;400:781–784. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: Signals, mechanism and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Okamoto H, Matsui M, Deng X-W. Overexpression of the heterotrimeric G-protein α-subunit enhances phytochrome-mediated inhibition of hypocotyl elongation in Arabidopsis. Plant Cell. 2001;13:1639–1652. doi: 10.1105/TPC.010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardthe CS, Wei N, Deng X-W. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Quail PH, Briggs WR, Chory J, Hangarter RP, Harberd NP, Kendrick RE, Koornneef M, Parks B, Sharrock RA, Schäfer E, et al. Spotlight on phytochrome nomenclature. Plant Cell. 1994;6:468–471. doi: 10.1105/tpc.6.4.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D. Phytochromes: Photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Nagatani A. Nuclear localization activity of phytochrome B. Plant J. 1996;10:859–868. doi: 10.1046/j.1365-313x.1996.10050859.x. [DOI] [PubMed] [Google Scholar]
- Soh MS, Hong SH, Hanzawa H, Furuya M, Nam HG. Genetic identification of FIN2, a far-red light-specific signalling component of Arabidopsis thaliana. Plant J. 1998;16:411–419. doi: 10.1046/j.1365-313x.1998.00307.x. [DOI] [PubMed] [Google Scholar]
- Soh M-S, Kim Y-M, Han S-J, Song P-S. REP1, a basic helix-loop-helix protein is required for a branch pathway of phytochrome A signalling in Arabidopsis. Plant Cell. 2000;12:2061–2074. doi: 10.1105/tpc.12.11.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Deng X-W. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. Light-dependent translocation of a phytochrome B–GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol. 1999;145:437–445. doi: 10.1083/jcb.145.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K-C, Lagarias JC. Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachgo EA, Wang ML, Dewdney J, Bouchez D, Camilleri C, Belmonte S, Huang L, Dolan M, Goodman HM. A physical map of chromosome 2 of Arabidopsis thaliana. Genome Res. 1996;6:19–25. doi: 10.1101/gr.6.1.19. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Tepperman JM, Fairchild CD, Quail PH. Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc Natl Acad Sci. 2000;97:13419–13424. doi: 10.1073/pnas.230433797. [DOI] [PMC free article] [PubMed] [Google Scholar]