Abstract
Embryonic stem (ES) cells give rise to all cell types of an organism. Since mutations at this embryonic stage would affect all cells and be detrimental to the overall health of an organism, robust mechanisms must exist to ensure that genomic integrity is maintained. To test this proposition, we compared the capacity of murine ES cells to repair DNA double-strand breaks with that of differentiated cells. Of the 2 major pathways that repair double-strand breaks, error-prone nonhomologous end joining (NHEJ) predominated in mouse embryonic fibroblasts, whereas the high fidelity homologous recombinational repair (HRR) predominated in ES cells. Microhomology-mediated end joining, an emerging repair pathway, persisted at low levels in all cell types examined. The levels of proteins involved in HRR and microhomology-mediated end joining were highly elevated in ES cells compared with mouse embryonic fibroblasts, whereas those for NHEJ were quite variable, with DNA Ligase IV expression low in ES cells. The half-life of DNA Ligase IV protein was also low in ES cells. Attempts to increase the abundance of DNA Ligase IV protein by overexpression or inhibition of its degradation, and thereby elevate NHEJ in ES cells, were unsuccessful. When ES cells were induced to differentiate, however, the level of DNA Ligase IV protein increased, as did the capacity to repair by NHEJ. The data suggest that preferential use of HRR rather than NHEJ may lend ES cells an additional layer of genomic protection and that the limited levels of DNA Ligase IV may account for the low level of NHEJ activity.
Introduction
Embryonic stem (ES) cells are self-renewing cells derived from the blastocyst inner cell mass. They give rise to all cell types of the embryo proper after successive rounds of growth and division before differentiation. Mutations incurred at this early embryonic stage may be detrimental to the overall development of an organism, resulting in possible genetic instability, birth defects, sterility, or death [1]. Thus, these pluripotent cells are likely to have robust mechanisms that minimize accumulation of mutations and maintain genetic integrity.
Consistent with this proposition, murine ES cells have a mutation frequency about 100-fold lower than that of isogenic mouse embryonic fibroblasts (MEFs) [2]. Mouse ES cells also lack a G1 checkpoint [3–6] and are hypersensitive to DNA damage [7–11]. The absence of a G1 checkpoint may allow cells with damaged DNA to enter S-phase where the damage would be exacerbated and promote cell death. Eliminating damaged cells by this mechanism would maintain a pristine stem cell population. In addition to suppression of mutation and enhanced apoptosis, mouse ES cells have a more effective stress defense pathway and are more efficient than their differentiated counterparts in removing endogenous free radicals generated by their rapid proliferation [12]. Finally, ES cells are very efficient in repair of damaged DNA. When repair efficiencies between human ES cells and other cell types in response to different DNA damaging treatments were compared, ES cells had significantly faster repair capacities than somatic cells based on alkaline comet assay [13]. Similar data have been reported for murine ES cells and NIH 3T3 cells after exposure to increasing doses of ionizing radiation [12].
DNA double-strand breaks (DSBs) are highly cytotoxic and have been used as targets for cancer therapeutics [14–18]. Cells have evolved several mechanisms to effectively repair DSBs. Repair by homologous recombination (HRR) uses a template such as that presented by the sister chromatid or homologous chromosome to achieve high fidelity repair and is most active during the S and G2 phases of the cell cycle [19]. Repair by nonhomologous end joining (NHEJ) is a rapid, but error-prone, process that rejoins DSBs and is functional throughout all phases of the cell cycle [19]. A third, but less understood pathway, is microhomology-mediated end joining (MMEJ), which is a mutagenic DSB repair pathway that processes broken DNA ends until repeat sequences are encountered on either side of the break and religated, resulting in large deletions (reviewed in ref. [20]).
The present study compares the capacity of ES cells and differentiated cells to repair DSBs by each of these repair pathways in the context of the hypothesis that ES cells preferentially utilize high fidelity HRR since repair by error-prone NHEJ and MMEJ would accumulate an unacceptable level of mutations. We now show that HRR is the predominant DSB repair pathway in ES cells with minimal contributions by NHEJ. The reverse was true of somatic cells. When ES cells were induced to differentiate, NHEJ became predominant and HRR was significantly reduced. When MMEJ was examined, it was found to be present at similarly low levels in MEFs and ES cells.
Experimental Procedures
Cell culture and microscopy
Primary MEFs were isolated from 13.5 days post coitum (dpc) embryos derived from either 129/Sv or C57Bl/6 mice and were cultured until passages 2–4 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 μg Penicillin, 100 U/mL Streptomycin (Fisher), 1× Nonessential amino acids (Fisher), and 2 mM SG-200 (Fisher). NIH 3T3 cells were grown under the same conditions as MEFs. Murine 129/Sv (J11) and C57Bl/6 (WD44; described in ref. [21]) were cultured as described [5]. All cell lines were maintained in 10% CO2 at 37°C. ES cells were maintained on MEF feeder layers treated with mitomycin-C unless otherwise noted and were separated from feeder layers when harvested for experiments. To differentiate ES cells, cells were seeded on gelatinized plates in the absence of feeders and leukemia inhibitory factor (LIF) and were treated for 5 days with 10 μM all-trans retinoic acid (Sigma). For protein stability studies, ES cells or MEFs were treated with 25 μg/mL cycloheximide (RPI Corp.) and harvested at the indicated time points. For proteasome inhibition studies, ES cells or MEFs were treated with 30 μM MG-132 (RPI Corp.) for 4 h before harvesting. For immunofluorescent microscopy, transfected ES cells were grown on coverslips and fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) before staining with Draq5 (1/3000; Axxora). Cells were observed using a Zeiss Axioplan 2 LSM510 confocal microscope.
Plasmids and transfections
To create pEGFP-tagged Ligase IV, Ligase IV cDNA was amplified by polymerase chain reaction (PCR) using primers 5′-atg gct tcc tca caa act tca caa act gtt-3′ and 5′-cta aag caa ata ctg gtt ttc ctc ctg cag-3′ and inserted into pCR topo 2.1 vector (Invitrogen). The KpnI-XbaI fragment was subsequently cloned in frame into pEGFP-C1 (Clontech) to generate pLig4-EGFP. Site-directed mutagenesis of pLig4-EGFP was used to make the DNA Ligase IV R278H hypomorphic mutant, with primers 5′-gct tga tgg tga gcA cat gca gat gca caa ag-3′ and 5′-ctt tgt gca tct gca tgT gct cac cat caa gc-3′. All sequences were verified by sequencing. Plasmids pCaggs, pBASCE, and pDR-GFP were provided by Maria Jasin (Memorial Sloan-Kettering), and pEGFP-PEM1-AD2 was a gift from Vera Gorbunova (U. Rochester). pCOH-CD4 and pINV-CD4 were gifts from Pascale Bertrand (Commissariat à l'Energie Atomique). The plasmid pCMV/I-SCE1/GFP was provided by Kevin Hiom (MRC Laboratory). Cells were transfected with the plasmids electroporation using the Gene Pulser Xcell (Biorad). Briefly, 5 × 106 cells were transfected in PBS with 40 μg of construct DNA and cotransfected with 5 μg pDsRed Express N1 (Clontech) using settings of 400 V and 250 μF.
Protein isolation and Western blotting
MEFs and ES cells were grown to 60% confluency and harvested by tryptic digestion. Cells were washed with PBS and lysed in cold RIPA buffer (20 mM Tris·HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 1% sodium dodecyl sulfate [SDS], and 0.5% deoxycholic acid) containing a protease inhibitor cocktail (RPI Corp.). Cell extracts were subjected to SDS–polyacrylamide gel electrophoresis using 30–50 μg of total protein, followed by protein transfer to polyvinyl difluoride (PVDF) membranes (Millipore). Blots were probed with antibodies to Ku70 (H-308; 1:1000; Santa Cruz), Ku80 (M-20; 1:1000; Santa Cruz), DNA Ligase IV (H-300; 1:250; Santa Cruz), XRCC4 (C-20; 1:500; Santa Cruz), α-actinin (C-20; 1:500; Santa Cruz), Oct3/4 (H-134; 1/1000; Santa Cruz), Rad51 (H-92; 1:1000; Santa Cruz), Rad52 (C-17; 1:1000; Santa Cruz), Rad54 (N-15; 1:500; Santa Cruz), β-Actin (AC-15; 1/10000; Sigma), DNA Ligase III (7; 1/10000; BD Biosciences), XRCC1 (H-300; 1:1000; Santa Cruz), or Parp-1 (1/1000; Cell Signaling) before incubation with the appropriate IgG-HRP-conjugated secondary antibodies (Santa Cruz) and exposure to X-ray film.
RNA isolation and quantitative PCR
Total RNA was isolated from 5 × 105 MEFs and ES cells using Tri-Reagent (MRC Corp.) and mRNA was isolated using the RNeasy kit (Qiagen). Reverse transcription of mRNA (2 μg) was carried out using the TaqMan Reverse Transcription Reagent kit (Applied Biosystems). An aliquot of cDNA was amplified for 40 cycles on an ABI Prism 7900HT Sequence Detection System with gene-specific primers designed using Primer Express software (Applied Biosystems). Sybr Green dye was used for signal detection. All analyses were carried out in triplicate, and nontemplate controls and dissociation curves were used to ensure specific template amplification. Target gene expression level was calculated as a ratio of the target mRNA relative to the mRNA level for glyceraldehyde-3-phosphate dehydrogenase. Data are presented as the average ± standard error of the mean, normalized to MEFs untreated where appropriate. Primers used were DNA Ligase IV (5′-GTC AAG CCC GAG TGG CTT TTA and 5′-CGG CAC GCA TGT TTT TGT CT-3′) and glyceraldehyde-3-phosphate dehydrogenase (5′-CTC CAC TCA CGG CAA ATT CAA-3′ and 5′-GAT GAC AAG CTT CCC ATT CTC G-3′).
Functional assays and flow cytometry
MEFs or ES cells were transiently transfected by electroporation, which generally introduces DNA as a single copy [22,23]. For transient HRR assays, cells were electroporated with pCaggs (empty vector), circular pDR-GFP, or I-SCE1-linearized pDR-GFP. The transient NHEJ assay utilized circular, or HindIII- or I-SCE1-linearized pEGFP-Pem1-Ad2 transfected into cells. For transient MMEJ, cells were transfected with circular or I-SCE1-linearized pCMV/I-SCE1/GFP [24]. Linearized plasmids were extracted with phenol-chloroform and precipitated with isopropanol before resuspension in TE buffer. Plasmid pDsRed Express N1 (Clontech) was coelectroporated as a transfection efficiency marker for the above transfections. Seventy-two hours posttransfection, cells were trypsinized and resuspended in PBS before live cell sorting.
For assays with integrated reporters, pDR-GFP, pCOH-CD4, or pINV-CD4 were stably transfected into ES cells or NIH 3T3 cells and selected with 10 μg/mL puromycin or 3 μg/mL blasticidin, respectively. To confirm genomic integration of pDR-GFP, DNA was isolated from nontransfected or stably transfected cells with buffer containing 100 mM Tris, pH8.5, 5 mM ethylenediaminetetraacetic acid, 200 mM NaCl, 0.2% SDS, and 100 μg/mL proteinase K. Genomic DNA (1 μg) was used for PCR using primers within the plasmid sequence (5′-GAA CGG CAT CAA GGT GAA CTT CAA-3′ and 5′-TGA AAT TTC TAG ACC AGC CCA CG). β-actin served as a loading control (5′-TGA GAC CTT CAA CAC CCC AG-3′ AND 5′-GAG CCA GAG CAG TAA TCT CC-3′). To confirm the stable integration of pCOH/INV, stable NIH 3T3 cells or ES cells were harvested in tri-reagent (MRC Corp.) and cDNA was reverse transcribed using the superscript III cDNA synthesis kit (Invitrogen), according to the manufacturer's instructions. Approximately 400 ng of cDNA was used to perform PCR using primers for H2Kd (5′-ATG GTA CCG TGC ACG CTG CTC CTG-3′ and 5′-TCA CGC TAG AGA ATG AGG GTC ATG A-3′) and PCNA (5′-AAG AAG GTG CTG GAG GCT CTC AAA−3′ and 5′-TCT GGG ATT CCA AGT TGC TCC ACA-3′).
Stably transfected cells were further transfected with the I-SCE1 expression vector pBasce or were mock transfected with pDsRed, serving as a transfection efficiency marker. Three days posttransfection, cells were harvested by trypsin digestion (pDR-GFP) or with 20 mM ethylenediaminetetraacetic acid in PBS (pCOH/pINV), transferred to PBS (pDR-GFP), or fixed with paraformaldehyde (4% w/v) for 20 min before resuspension in PBS (pCOH/pINV). Cells transfected with pCOH/INV were blocked with 2% BSA in PBS and stained using Alexa Fluor 488 rat anti-mouse CD4 (RM4-5; BD Biosciences) antibody at a dilution of 1/250. Cells were then subjected to analysis by flow cytometry.
For flow cytometry, samples were analyzed using a BD LSRII (BD Biosciences). The instrument was aligned with AlignFlow 2.5 μm beads (Molecular Probes) for the 488 nm laser. Data were collected and analyzed with FACSDiVa software (BD Biosciences). Cells were excited with the 488 nm line of an argon-ion laser using GFP (or Alexa Fluor 488) and DsRed fluorochromes. Log fluorescence was collected for GFP (or Alexa Fluor 488) using a 530/30 band pass filter and for DsRed using a 585/42 band pass filter. In all experiments, 20,000 events were collected, and the ratio of GFP (or Alexa Fluor 488) to DsRed cells was used for data processing. Data presented are the average of at least 3 independent experiments. Error bars represent standard error of the mean.
Results
Proteins involved in HRR but not NHEJ are consistently elevated in ES cells
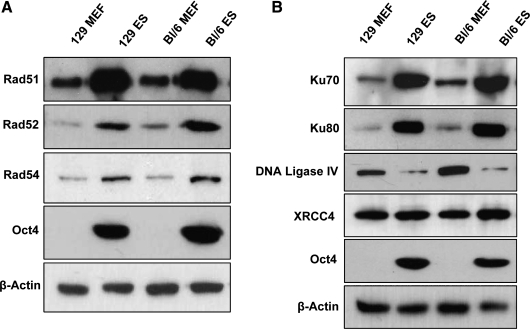
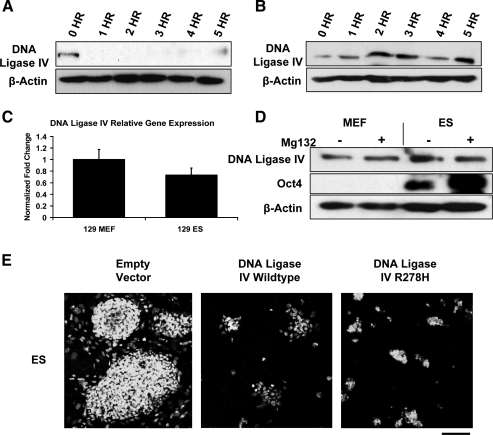
Western blots were performed on whole-cell lysates of ES cells and MEFs to determine whether differences in the levels of proteins involved in DSB repair existed between these cell types and whether such differences might be indicative of function. Proteins involved in HRR, such as Rad51, Rad52, and Rad54, were highly elevated in Oct4-positive undifferentiated ES cell lines from 2 mouse strains, but were low in isogenic MEFs (Fig. 1A). There was no clear pattern of elevation or suppression of proteins involved in NHEJ as a group when ES cells and MEFs were compared (Fig. 1B). The Ku70 and Ku80 proteins were more abundant in ES cells as previously reported [25], and DNA Ligase IV expression was significantly reduced in ES cells, regardless of mouse strain from which they were derived. The level of XRCC4 protein was similar in both cell types.
FIG. 1.
Basal protein expression of predominant DSB repair pathways. Western blots of whole cell lysates from either 129/SV or C57Bl/6 cells representing basal expression of several proteins involved in HRR (A) or NHEJ (B). Oct4 served as a marker of undifferentiated ES cells and β-actin was used as a loading control. DSB, double-strand break; HRR, homologous recombinational repair; NHEJ, nonhomologous end joining; ES cells, embryonic stem cells; MEF, mouse embryonic fibroblast.
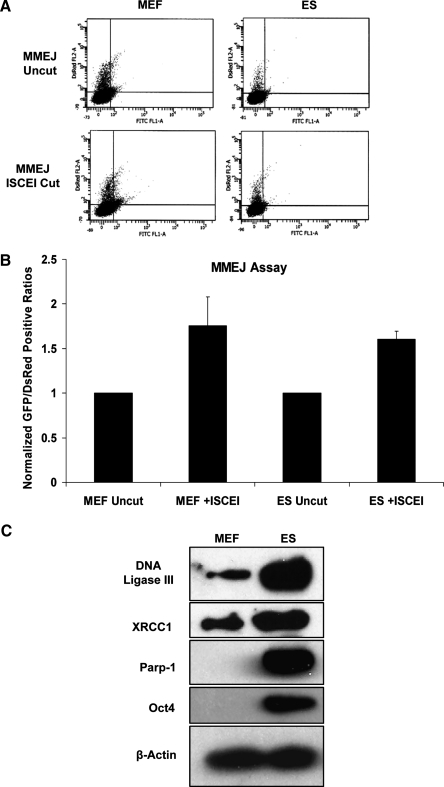
MMEJ provides an alternative pathway for repairing DSBs, but is less understood than either HRR or NHEJ. Although many of the participating proteins in this pathway are still being elucidated, we assessed the relative abundance of some of these proteins in MEFs and ES cells (Fig. 4C). It appears that DNA Ligase III rather than DNA Ligase IV is involved in MMEJ [26]. DNA Ligase III is unstable unless as a heterodimer with XRCC1 [27]. The expression level of the latter, therefore, was also monitored, as was that of Parp-1, which has also been implicated in this pathway [28]. Proteins that participate in MMEJ, including DNA Ligase III, XRCC1, and Parp-1, were all significantly elevated in ES cells compared with MEFs.
FIG. 4.
MMEJ repair capacity. (A) Whole cell lysates from 129/SV MEFs and ES cells were subjected to Western blotting using antibodies for DNA Ligase III, XRCC1, Parp1, Oct4, or β-actin. (B) Scatter diagrams for 129/Sv MEFs and ES cells transiently transfected with pDsRed and uncut or I-SCE1-linearized pCMV/I-SCE1/GFP vector and analyzed by flow cytometry 72 h posttransfection. Repair of this vector should only occur by MMEJ. This vector has a cassette inserted into the GFP gene, which renders it nonfunctional. Digestion with I-SCE1 and subsequent end processing to a repeat sequence at the ends of the cassette will render the GFP gene functional if ends are ligated together. (C) Quantitation of flow cytometric data. Data were accumulated from 10,000 cells per transfection as the total number of GFP+/DsRed+ cells and were normalized to the uncut vector for the respective cell line. MMEJ, microhomology-mediated end joining.
Homologous recombination repair predominates in ES cells
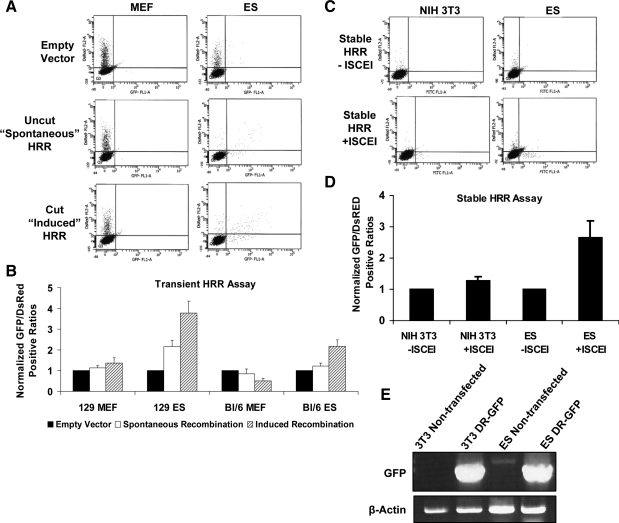
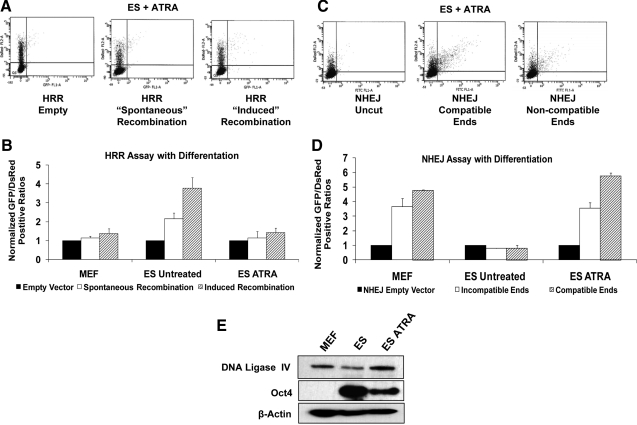
Although protein levels associated with each of the repair pathways may be an indicator of potential functional activity, they are not by themselves sufficient to define the relative activities of each pathway. To assess this, we have taken advantage of several validated reporter plasmids that distinguish between the various DNA repair pathways. These are described separately. The pDR-GFP plasmid [29] was transfected into cells as both a circular plasmid and as linearized with I-SCE1 restriction enzyme to assess relative levels of HRR repair. The plasmid has 2 tandem but inactive GFP repeats, one of which contains the unique I-SCE1 site. Fluorescence is restored when a functional GFP is reconstituted by homologous recombination between the 2 nonfunctional repeats. When ES cells from 129/Sv or C57Bl/6 strains were transfected transiently with the DR-GFP plasmid, either circular or linearized, there was robust HRR activity based on the level of reconstituted fluorescence. In contrast, MEFs transfected with these plasmids showed little activity above that of the control empty vector (Fig. 2B).
FIG. 2.
Quantitation of HRR repair capacity. (A) Representative flow cytometry scatter plots for MEFs or ES cells derived from 129/Sv mice transiently transfected with pCAGGS (empty vector), circular pDR-GFP (spontaneous recombination), or I-SCE1-linearized pDR-GFP (induced recombination) and cotransfected with pDsRed to account for differences in transfection efficiency. Seventy-two hours posttransfection, cells were harvested and live sorted. A minimum of 10,000 cells were sorted per trial; only 5,000 are displayed. Axes represent mean fluorescent intensity on a 5-log scale. The X-axis represents GFP-positive cells; Y-axis is DsRed+ cells. pDR-GFP contains 2 nonfunctional copies of GFP separated by approximately 3 kb. Recombination between the 2 copies can yield functional GFP by gene conversion or a nonfunctional deletion product, which is not measured. Recombination between the 2 copies in the circular plasmid is noted as spontaneous recombination. When the I-SCE1 site located in one of the GFP copies is cut, a DSB in the plasmid occurs, which, if repaired, has induced recombination. (B) Quantitation of HRR activity in MEFs and ES cells from 129/Sv or C57Bl/6 stains. Data were generated based on the ratios of GFP+/DsRed+ cells and normalized to background caused from the empty vector from each cell type. A minimum of 10,000 cells per trial were used; 5,000 events are displayed. (C) Scatter plots of stably integrated DR-GFP in NIH 3T3 cells or 129/Sv ES cells. NIH 3T3 cells were used as a surrogate for primary MEFs, which cannot be selected for with their finite passage number. I-SCE1 expression vector pBaSCE or empty vector pCAGGS was introduced to induce site-specific DSBs and pDsRed was cotransfected for transfection efficiency. Seventy-two hours posttransfection, cells were harvested and sorted live. (D) Quantitation of HRR activity in stable cell lines represented in (C) before or after DSB induction. Data were collected as the total of GFP/DsRed+ cells and were normalized to respective cell lines transfected with empty vector. (E) PCR for GFP or β-actin on genomic DNA from nontransfected and stably transfected and pooled NIH 3T3 and ES cells to demonstrate pDR-GFP genomic integration. PCR, polymerase chain reaction.
To eliminate the possibility that the high HRR in ES cells might be attributed to the extrachromosomal substrate [30], ES cell lines were selected that stably harbored the pDR-GFP plasmid. Since stable transfection of MEFs by electroporation is not possible due to cell death and eventual senescence during selection, as well as to keep transfection conditions the same, we used stably transfected NIH 3T3 cells, which were derived from MEFs, as a surrogate for comparison with ES cells (Fig. 2E). Uncut DR-GFP reported little HR activity in either cell type. After cells were transfected with the pBasce plasmid that encodes the I-SCE1 enzyme, there was a modest increase in HR activity above background in NIH 3T3 cells and a robust increase in the ES cells (Fig. 2D), consistent with results from transient transfections.
NHEJ activity is low in ES cells
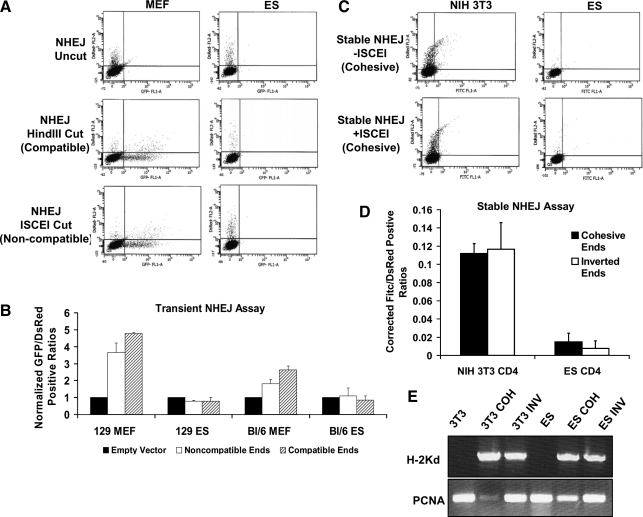
The capacity of ES cells and MEFs to carry out NHEJ was assessed by transfection of cells with the reporter plasmid pEGFP-Pem1-Ad2 [31]. This reporter measures repair of cohesive and noncohesive ends depending upon the restriction enzymes used to digest the vector before transient transfection. The plasmid contains a nonfunctional GFP that is interrupted by a Pem1 intron into which has been inserted an adenovirus AD2 exon sequence flanked by dominant splice acceptor and donor sequences as well as HindIII and I-SCE1 cleavage sites external to the splice sites. When the plasmid is digested with HindIII (complementary ends) or I-SCE1 (noncomplementary ends), the AD2 sequence and its splice sites are removed, so that repair by NHEJ allows juxtaposition of the 2 GFP fragments and reconstitution of GFP fluorescence. MEFs from both strains display robust NHEJ activity regardless of the types of ends available at the site of the break. In contrast, ES cells appear to lack appreciable NHEJ activity (Fig. 3B).
FIG. 3.
NHEJ repair capacity in ES cells and differentiated cells. (A) Scatter plots for MEFs or ES cells from 129/Sv mouse strains transiently transfected with undigested (uncut/empty), HindIII-digested (compatible ends), or I-SCE1-digested (noncompatible ends) pEGFP-PEM1-AD2 vector and cotransfected with pDsRED. This vector contains a single GFP gene interrupted by a pem1 intron. Within the intron there is a killer ad2 exon, which prevents expression of functional GFP. Digestion with either HindIII or I-SCE1, located at the termini of the killer exon, allows for its removal. The pem1 intron can then be repaired by NHEJ and spliced out to render the GFP gene functional. Seventy-two hours posttransfection cells were harvested and analyzed by flow cytometry. At least 10,000 cells were examined per trial; 5,000 events are displayed. Axes are the same as described in Fig. 2. (B) Quantitation of NHEJ data from transiently transfected 129/Sv or C57Bl/6 MEFs and ES cells. Data were collected as total number of GFP+/DsRed+ cells and normalized to empty vector. (C) Representative scatter diagrams of NIH3T3 and 129/Sv ES cells stably transfected with pCOH-CD4 after transient transfection with pDsRed and pCAGGS or pBaSCE and staining with CD4-Fitc-conjugated antibody. (D) Cells were stably transfected with pCOH-CD4 (cohesive ends) or pINV-CD4 (inverted ends). Ends refer to the orientation of 2 I-SCE1 restriction sites within each constructs. Stable transfection leads to expression of H2Kd. When pBaSCE is introduced into stable cells, H2kd is cut out. Repair by NHEJ subsequently promotes expression of a downstream CD4 gene. Background CD4-positive cells from the pCAGGS empty vector were subtracted from total numbers after I-SCE1 introduction. (E) RNA was isolated from nontransfected or blasticidin-resistant pooled colonies of pCOH/pINV-transfected NIH 3T3 cells or 129/Sv ES cells. cDNA was synthesized and semiquantiative PCR was completed to demonstrate the stable integration of the constructs. PCR products were run on 0.8% agarose gels containing 3 μg/mL ethidium bromide. These cells normally do not express the cell surface marker H2Kd, but after transfection of pCOH or pINV, H2Kd expression was robust. PCNA demonstrated similar loading of pcr product per lane.
A third set of reporter plasmids, pCOH-cd4 or pINV-cd4 [32], was used to confirm these results. When these plasmids are digested with I-SCE1, a fragment containing an expressed H2Kd surface antigen is removed and the free DNA ends have either cohesive (pCOH-cd4) or inverted (pINV-cd4) overhanging sequences. If the DSB is repaired by NHEJ, a CD4 marker is transcribed from the CMV promoter and expressed at the cell surface. Expression of this CD4 marker was monitored and quantified by flow cytometry. ES cells and NIH 3T3 were electroporated with the above plasmids, and clones that stably express H2Kd-positive cells were obtained (Fig. 3E). Transfection of stable cells with plasmid pBasce, which expresses I-SCE1 enzyme and thus cleaves the integrated reporters, confirmed that ES cells had barely detectable NHEJ activity, while activity in NIH 3T3 cells was significantly higher (Fig. 3D).
ES Cells and MEFs have MMEJ activity
In addition to HRR and NHEJ, the 2 major pathways for repairing DSBs, there is a third pathway, MMEJ, which is less well characterized. MMEJ requires sequences of microhomology extending 5–25 bases to effectively repair a DSB. Using a reporter plasmid, pCMV/I-SCE1/GFP [24], that has 7 bp repeat sequences flanking an I-SCE1 site, we tested the capacity of MEFs and ES cells to repair an extrachromosomal template digested with I-SCE1 in a transient transfection assay. The MMEJ activity was similar in ES cells and MEFs, despite the elevated abundance of MMEJ proteins in ES cell extracts and activity was low in both cell types (Fig. 4A,B).
Sensitivity of ES cells to changes in DNA Ligase IV levels
DNA Ligase IV is rate-limiting for NHEJ [33–35]. Given its low abundance in ES cells it is reasonable to expect that its low level of expression might, in part, account for the low level of NHEJ activity in these cells. Western blots for DNA Ligase IV in ES cells after cycloheximide treatment showed that the protein had a half-life of less than 1 h (Fig. 5A). The XRCC4 protein that heterodimerizes with DNA Ligase IV in equimolar amounts to promote rejoining of DNA breaks is present at similar levels in ES cells and MEFs. This observation indicates that the low abundance of DNA Ligase IV in ES cells is not the result of protein destabilization due to absence of its binding partner [36], since XRCC4 appears to be in excess. The half-life of DNA Ligase IV protein in MEFs was much longer, remaining stable throughout the cycloheximide treatment time course (Fig. 5B). Since MEFs and ES cells have similar levels of DNA Ligase IV mRNA (Fig. 5C), the regulation of DNA Ligase IV in ES cells appears to be exclusively at the protein level.
FIG. 5.
DNA Ligase IV regulation in mouse ES cells. 129/Sv ES cells (A) or MEFs (B) were treated with 25 μg/mL of cycloheximide to block new protein synthesis or left untreated and harvested at the specified time points for Western blotting with DNA Ligase IV antibody. β-actin served as a loading control. (C) RNA from untreated 129/Sv MEFs and ES cells was isolated and subjected to semiquantitative PCR. Data are graphed as a ratio of DNA Ligase IV/Gapdh. (D) 129/Sv MEFs and ES cells were treated with 30 μM of the proteasome inhibitor MG-132 or left untreated and harvested after 4 h. Western blotting was performed using antibodies against DNA Ligase IV or Oct4 to demonstrate undifferentiated ES cells. β-actin served as a loading control. (E) 129/Sv ES cells were transfected with GFP, GFP-DNA Ligase IV wild type, and GFP-DNA Ligase IV hypomorphic mutant (R278H), and allowed to grow for 72 h. Nuclei were stained with Draq5. While large ES cell colonies are visible in the vector-only-transfected cells, the GFP DNA Ligase IV wild-type and mutant-transfected cells were much fewer in number. Scale bar = 100 μm.
To determine whether ES cells can be stimulated to more actively utilize NHEJ, we attempted to increase the abundance of DNA Ligase IV by treating the cells with the proteasome inhibitor MG-132 to inhibit its proteasome-mediated degradation. While treatment with MG-132 effectively increased the Oct4 protein level in ES cells, it had no effect on the level of DNA Ligase IV in either ES cells or MEFs (Fig. 5D). Attempts to overexpress DNA Ligase IV by transfection with a GFP-tagged Ligase IV cDNA resulted in rapid cell death (Fig. 5E). To assess whether cell death is the result of increased DNA Ligase IV activity, we generated a GFP-tagged hypomorphic R278H Ligase IV variant found in some patients with Ligase IV syndrome. Expression of this variant is normal, but the protein has only 5%–10% of wild-type enzymatic activity [37]. Like the wild-type cDNA, transfection of ES cells with the R278H variant induces cell death, indicating that either a very modest increase in DNA Ligase IV activity can cause cell death, or that a mechanism unrelated to its enzymatic activity is responsible for lethality.
ES cells switch preferred DSB repair pathways when they differentiate
Mouse ES cells predominantly utilize HRR to repair DNA DSBs, whereas somatic cells utilize NHEJ. To ask whether mouse ES cells switch from HRR to NHEJ to repair DSBs when they differentiate into somatic cells, ES cells were induced to differentiate by removal of LIF and culturing with all-trans retinoic acid, which downregulates the LIF receptor [38]. When the pDR-GFP plasmid was introduced into ES cells that had been allowed to differentiate, the HRR activity was significantly reduced to a level comparable to that seen in MEFs (Fig. 6B). Conversely, when pEGFP-PEM1-AD2 was linearized with HindIII or I-SCE1 and transfected into ES cells that had been allowed to differentiate, NHEJ activity was increased to a level that was equal to or greater than that in MEFs (Fig. 6D). Concomitant with the increase in NHEJ activity, Ligase IV protein level in differentiated ES cells was elevated to a level comparable to that in MEFs (Fig. 6E), and presumably to a level not tolerated by undifferentiated ES cells.
FIG. 6.
Differentiated ES cell DSB repair patterns. ES cells were differentiated by treatment with ATRA. (A) Scatter diagrams of transiently transfected differentiated ES cells using vectors pCAGGS and pDR-GFP (A) or pEGFP-PEM1-AD2 (C). (B) Flow cytometry quantitation of differentiated ES cell capacity to repair DSBs by HRR (B) or NHEJ (D) as compared with MEFs or undifferentiated ES cells. Data accumulation and methodology is identical to that of Figs. 3B and 4B. (E) Whole-cell lysates from MEFs, undifferentiated ES cells, or ATRA-treated ES cells were subjected to Western blotting using antibodies to DNA Ligase IV, Oct4, and β-actin. DNA Ligase IV is increased in differentiated cells. Oct4 is still present in these cells, suggesting that complete differentiation has not yet occurred. ATRA, all-trans retinoic acid.
Discussion
ES cells, like germ cells, are capable of differentiating into all cell types of the body. It is critical, therefore, that they have the capacity to repair DNA DSBs rapidly and with minimal residual mutations. When DSBs are introduced, our data demonstrate that ES cells predominantly utilize high-fidelity HRR to repair the lesions rather than more error-prone mechanisms such as NHEJ or MMEJ.
That HRR is the predominant form of DSB repair in mouse ES cells is, perhaps, because a greater proportion of cells occupy S-phase than they do in somatic cells. This interpretation, however, does not account for the fact that NHEJ is equally active in the S-phase of somatic cells [19,39,40] yet is suppressed in ES cells. ES cells have rapid cell cycles with abbreviated G1 and G2 phases [41]. Many proteins involved in HRR are regulated by E2F and therefore express during S-phase [42]. The absence of Rad51 and concomitant HRR activity in Rad51 knockout mice produces very early lethality, around the time of blastocyst development and formation of the inner cell mass, the progenitors of ES cells [43,44]. In contrast, knockouts for key components of the NHEJ machinery are lethal at much later embryonic stages, after the inner cell mass cells would have differentiated [45–49]. In aggregate, transcriptional regulation of genes involved with HRR and the relative timing of fetal lethality in mouse embryos lacking components of HRR and NHEJ is consistent with the importance of HRR in the ES cell population. Further investigation into the regulation of HRR proteins in ES cells, however, revealed that the elevated levels observed are not regulated by E2F-induced transcription alone (Supplementary Fig. S1, available online at www.liebertonline.com/scd), but rather by some other mechanism.
There is indirect evidence to support the contention that repair of DSBs by HRR in ES cells is important for maintenance of their genomic stability, and that NHEJ plays a lesser role in these cells. On the one hand, mouse ES cells lacking XRCC2, an integral component of HRR, display much higher frequencies of chromosome fragmentation and rearrangements than their wild-type counterparts. On the other hand, absence of Ku70, a member of the NHEJ pathway, shows no obvious increase in chromosome fragmentation in ES cells [50]. Further support derives from the observation that ES cells are unable to efficiently repair an integrated reporter plasmid into which 2 double-strand cuts had been introduced about 8.8 kb apart [51]. The reporter had no regions of homology around the break sites requiring utilization of NHEJ for repair rather than HRR. When a reporter with RAG recognition sites was cleaved by RAG recombinase in ES cells, 90% of the repair was reported to occur by NHEJ [52]. When I-SCE1 target sites and I-SCE1 enzyme were used to introduce double-strand breaks in the same cells, NHEJ and HRR were reported to occur at similar frequencies, suggesting that the enzyme and the target sites used to introduce DSBs can affect the choice of repair pathway. Others have suggested that RAG recombinase may direct DSB repair into the NHEJ pathway by a mechanism that is not understood, and that affects overall NHEJ capacity in ES cells [53]. Despite the confounding evidence presented by preferential utilization of NHEJ in ES after introduction of RAG recombinase, the preponderance of evidence argues that under most conditions HHR is the preferred pathway for DSB repair in ES cells.
NHEJ is the predominant pathway by which somatic cells repair DSBs [54,55], and appears to be the predominant form of repair regardless of cell cycle phase [19,39,40]. Our data demonstrate that NHEJ activity is very low in ES cells, and that the abundance of DNA Ligase IV is also low in these cells. When induced to differentiate, its abundance increases, as does NHEJ repair activity, suggesting that the amount of available DNA Ligase IV may be a limiting factor in NHEJ activity. Although low levels of DNA Ligase IV are sufficient for fully functional NHEJ in Hela cells [56], its relative abundance has not been directly compared with that found in ES cells. Maintaining a low level of DNA Ligase IV is unlikely to be the sole mechanism for regulating NHEJ, since this enzyme is also regulated by phosphorylation by DNA-PKCS [57], which also is expressed at low levels in murine ES cells [25]. It appears that an alternative function other than actual ligase activity, or perhaps the DNA Ligase IV protein structure itself, plays a role in promoting ES cell death by an unknown mechanism. This possibility is supported by the finding that ectopic expression of a hypomorphic mutant also leads to cell death and is not inconsistent with other DNA repair pathway proteins playing roles in apoptosis [58–60].
While DNA Ligase IV expression remains low in undifferentiated ES cells, the Ku proteins are elevated when compared with MEFs (Fig. 2B). At least 1 report has confirmed this observation and also described a reduction in the Ku protein levels when ES cells were induced to differentiate [25]. The rationale for the elevated Ku protein in ES cells, which exhibit low NHEJ capacity, likely stems from roles for Ku proteins other than NHEJ. Ku proteins localize to telomeres and have been reported to prevent end-to-end chromosome fusions by suppressing homologous recombination in the t-loop, which is similar in structure to the d-loop intermediate formed during homologous recombination [61–63]. Thus, the elevated level of Ku proteins found in ES cells may reflect a mechanism to minimize mutation by suppressing inappropriate homologous recombination at specific sites of the chromosome, rather than functioning in NHEJ. Further, Ku proteins are also involved in DNA replication [64–68], and their elevation in ES cells is consistent with the notion that ES cells spend a large proportion of their cell cycles in S-phase.
A third but minor pathway for repairing DSBs is MMEJ, which provides an alternative to NHEJ (reviewed in ref. [20]) and remains poorly understood. For example, Ku70 has been implicated in blocking MMEJ in yeast [69], whereas Ku80 appears to promote MMEJ in chinese hampster ovary (CHO) cells [70]. Although these disparate observations are from very different systems, they are, nevertheless, perplexing since Ku70 and Ku80 normally heterodimerize when promoting NHEJ [71]. Since the levels of proteins involved in MMEJ that were assessed are elevated in ES cells compared with MEFs, we anticipated that MMEJ activity would also be elevated. Although ES cells can utilize MMEJ to repair DSBs, the level of activity is not commensurate with the elevated protein levels. It should be noted that many of the proteins involved in MMEJ also participate in base excision repair (BER), which is very active in ES cells and which may account for the high level of these proteins [13].
The finding that ES cells utilize high fidelity HRR to repair DSBs and that somatic cells predominantly utilize error-prone NHEJ is consistent with the observation that somatic cells accumulate an increased mutational burden as a function of age [72–74]. The switch from HRR to NHEJ when ES cells are induced to differentiate further supports the proposition that mouse ES cells have multiple mechanisms to preserve the integrity of their genomes that are lost as they become somatic cells. The consequent accumulation of mutations raises concerns regarding the potential therapeutic use of induced pluripotent stem cells derived from adult somatic cells with acquired mutations. Use of somatic cells from early postnatal individuals or from cord blood might therefore be more advantageous. It will also be informative to see if induced pluripotent stem cells revert to HRR as the predominant DSB pathway or if they retain NHEJ as the preferred mechanism.
Supplementary Material
Acknowledgments
This work was supported in part by grants R01 ES012695 and R01 ES12695-4S1 to P.J.S. from the National Institutes of Health. E.D.T. was supported by training grant T32 ES 007250-21.
Author Disclosure Statement
No competing financial interests exist for any authors involved in this study.
References
- 1.Friedberg EC. Meira LB. Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage Version 7. DNA Repair (Amst) 2006;5:189–209. doi: 10.1016/j.dnarep.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Cervantes RB. Stringer JR. Shao C. Tischfield JA. Stambrook PJ. Embryonic stem cells and somatic cells differ in mutation frequency and type. Proc Natl Acad Sci USA. 2002;99:3586–3590. doi: 10.1073/pnas.062527199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savatier P. Huang S. Szekely L. Wiman KG. Samarut J. Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene. 1994;9:809–818. [PubMed] [Google Scholar]
- 4.Savatier P. Lapillonne H. Jirmanova L. Vitelli L. Samarut J. Analysis of the cell cycle in mouse embryonic stem cells. Methods Mol Biol. 2002;185:27–33. doi: 10.1385/1-59259-241-4:27. [DOI] [PubMed] [Google Scholar]
- 5.Hong Y. Stambrook PJ. Restoration of an absent G1 arrest and protection from apoptosis in embryonic stem cells after ionizing radiation. Proc Natl Acad Sci USA. 2004;101:14443–14448. doi: 10.1073/pnas.0401346101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker KA. Stein JL. Lian JB. van Wijnen AJ. Stein GS. Establishment of histone gene regulation and cell cycle checkpoint control in human embryonic stem cells. J Cell Physiol. 2007;210:517–526. doi: 10.1002/jcp.20903. [DOI] [PubMed] [Google Scholar]
- 7.Corbet SW. Clarke AR. Gledhill S. Wyllie AH. P53-dependent and -independent links between DNA-damage, apoptosis and mutation frequency in ES cells. Oncogene. 1999;18:1537–1544. doi: 10.1038/sj.onc.1202436. [DOI] [PubMed] [Google Scholar]
- 8.Aladjem MI. Spike BT. Rodewald LW. Hope TJ. Klemm M. Jaenisch R. Wahl GM. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr Biol. 1998;8:145–155. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- 9.Van Sloun PP. Jansen JG. Weeda G. Mullenders LH. van Zeeland AA. Lohman PH. Vrieling H. The role of nucleotide excision repair in protecting embryonic stem cells from genotoxic effects of UV-induced DNA damage. Nucleic Acids Res. 1999;27:3276–3282. doi: 10.1093/nar/27.16.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roos WP. Christmann M. Fraser ST. Kaina B. Mouse embryonic stem cells are hypersensitive to apoptosis triggered by the DNA damage O(6)-methylguanine due to high E2F1 regulated mismatch repair. Cell Death Differ. 2007;14:1422–1432. doi: 10.1038/sj.cdd.4402136. [DOI] [PubMed] [Google Scholar]
- 11.Filion TM. Qiao M. Ghule PN. Mandeville M. van Wijnen AJ. Stein JL. Lian JB. Altieri DC. Stein GS. Survival responses of human embryonic stem cells to DNA damage. J Cell Physiol. 2009;220:586–592. doi: 10.1002/jcp.21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saretzki G. Armstrong L. Leake A. Lako M. von Zglinicki T. Stress defense in murine embryonic stem cells is superior to that of various differentiated murine cells. Stem Cells. 2004;22:962–971. doi: 10.1634/stemcells.22-6-962. [DOI] [PubMed] [Google Scholar]
- 13.Maynard S. Swistowska AM. Lee JW. Liu Y. Liu ST. Da Cruz AB. Rao M. de Souza-Pinto NC. Zeng X. Bohr VA. Human embryonic stem cells have enhanced repair of multiple forms of DNA damage. Stem Cells. 2008;26:2266–2274. doi: 10.1634/stemcells.2007-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallick I. Waldron JN. Radiation therapy for head and neck cancers. Semin Oncol Nurs. 2009;25:193–202. doi: 10.1016/j.soncn.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell LR. Albert JM. Lu B. Chemoradiotherapy in locally advanced, unresectable non-small cell lung cancer. Rev Recent Clin Trials. 2009;4:110–121. doi: 10.2174/157488709788185969. [DOI] [PubMed] [Google Scholar]
- 16.Polat B. Fassnacht M. Pfreundner L. Guckenberger M. Bratengeier K. Johanssen S. Kenn W. Hahner S. Allolio B. Flentje M. Radiotherapy in adrenocortical carcinoma. Cancer. 2009;115:2816–2823. doi: 10.1002/cncr.24331. [DOI] [PubMed] [Google Scholar]
- 17.Diavolitsis V. Boyle J. Singh DK. Small W., Jr The role of adjuvant radiation in endometrial cancer. Oncology (Williston Park) 2009;23:342–349. [PubMed] [Google Scholar]
- 18.Mendenhall WM. Indelicato DJ. Scarborough MT. Zlotecki RA. Gibbs CP. Mendenhall NP. Mendenhall CM. Enneking WF. The management of adult soft tissue sarcomas. Am J Clin Oncol. 2009;32:436–442. doi: 10.1097/COC.0b013e318173a54f. [DOI] [PubMed] [Google Scholar]
- 19.Mao Z. Bozzella M. Seluanov A. Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008;7:2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McVey M. Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ware CB. Siverts LA. Nelson AM. Morton JF. Ladiges WC. Utility of a C57BL/6 ES line versus 129 ES lines for targeted mutations in mice. Transgenic Res. 2003;12:743–746. doi: 10.1023/b:trag.0000005246.35812.c8. [DOI] [PubMed] [Google Scholar]
- 22.Boggs SS. Gregg RG. Borenstein N. Smithies O. Efficient transformation and frequent single-site, single-copy insertion of DNA can be obtained in mouse erythroleukemia cells transformed by electroporation. Exp Hematol. 1986;14:988–994. [PubMed] [Google Scholar]
- 23.Baer A. Schubeler D. Bode J. Transcriptional properties of genomic transgene integration sites marked by electroporation or retroviral infection. Biochemistry. 2000;39:7041–7049. doi: 10.1021/bi992957o. [DOI] [PubMed] [Google Scholar]
- 24.Yun MH. Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banuelos CA. Banath JP. MacPhail SH. Zhao J. Eaves CA. O'Connor MD. Lansdorp PM. Olive PL. Mouse but not human embryonic stem cells are deficient in rejoining of ionizing radiation-induced DNA double-strand breaks. DNA Repair (Amst) 2008;7:1471–1483. doi: 10.1016/j.dnarep.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 26.Liang L. Deng L. Nguyen SC. Zhao X. Maulion CD. Shao C. Tischfield JA. Human DNA ligases I and III, but not ligase IV, are required for microhomology-mediated end joining of DNA double-strand breaks. Nucleic Acids Res. 2008;36:3297–3310. doi: 10.1093/nar/gkn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tebbs RS. Thompson LH. Cleaver JE. Rescue of Xrcc1 knockout mouse embryo lethality by transgene-complementation. DNA Repair (Amst) 2003;2:1405–1417. doi: 10.1016/j.dnarep.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Audebert M. Salles B. Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 29.Pierce AJ. Johnson RD. Thompson LH. Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jasin M. Liang F. Mouse embryonic stem cells exhibit high levels of extrachromosomal homologous recombination in a chloramphenicol acetyltransferase assay system. Nucleic Acids Res. 1991;19:7171–7175. doi: 10.1093/nar/19.25.7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seluanov A. Mittelman D. Pereira-Smith OM. Wilson JH. Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc Natl Acad Sci USA. 2004;101:7624–7629. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guirouilh-Barbat J. Rass E. Plo I. Bertrand P. Lopez BS. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc Natl Acad Sci USA. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y. Lamarche BJ. Tsai MD. Human DNA ligase IV and the ligase IV/XRCC4 complex: analysis of nick ligation fidelity. Biochemistry. 2007;46:4962–4976. doi: 10.1021/bi0621516. [DOI] [PubMed] [Google Scholar]
- 34.Lee KJ. Huang J. Takeda Y. Dynan WS. DNA ligase IV and XRCC4 form a stable mixed tetramer that functions synergistically with other repair factors in a cell-free end-joining system. J Biol Chem. 2000;275:34787–34796. doi: 10.1074/jbc.M004011200. [DOI] [PubMed] [Google Scholar]
- 35.Riballo E. Woodbine L. Stiff T. Walker SA. Goodarzi AA. Jeggo PA. XLF-Cernunnos promotes DNA ligase IV-XRCC4 re-adenylation following ligation. Nucleic Acids Res. 2009;37:482–492. doi: 10.1093/nar/gkn957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foster RE. Nnakwe C. Woo L. Frank KM. Monoubiquitination of the nonhomologous end joining protein XRCC4. Biochem Biophys Res Commun. 2006;341:175–183. doi: 10.1016/j.bbrc.2005.12.166. [DOI] [PubMed] [Google Scholar]
- 37.Chistiakov DA. Voronova NV. Chistiakov AP. Ligase IV syndrome. Eur J Med Genet. 2009;52:373–378. doi: 10.1016/j.ejmg.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Tighe AP. Gudas LJ. Retinoic acid inhibits leukemia inhibitory factor signaling pathways in mouse embryonic stem cells. J Cell Physiol. 2004;198:223–229. doi: 10.1002/jcp.10424. [DOI] [PubMed] [Google Scholar]
- 39.Mao Z. Bozzella M. Seluanov A. Gorbunova V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst) 2008;7:1765–1771. doi: 10.1016/j.dnarep.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takashima Y. Sakuraba M. Koizumi T. Sakamoto H. Hayashi M. Honma M. Dependence of DNA double strand break repair pathways on cell cycle phase in human lymphoblastoid cells. Environ Mol Mutagen. 2009;50:815–822. doi: 10.1002/em.20481. [DOI] [PubMed] [Google Scholar]
- 41.White J. Dalton S. Cell cycle control of embryonic stem cells. Stem Cell Rev. 2005;1:131–138. doi: 10.1385/SCR:1:2:131. [DOI] [PubMed] [Google Scholar]
- 42.Bracken AP. Ciro M. Cocito A. Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29:409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Tsuzuki T. Fujii Y. Sakumi K. Tominaga Y. Nakao K. Sekiguchi M. Matsushiro A. Yoshimura Y. Morita T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim DS. Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barnes DE. Stamp G. Rosewell I. Denzel A. Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 46.Frank KM. Sharpless NE. Gao Y. Sekiguchi JM. Ferguson DO. Zhu C. Manis JP. Horner J. DePinho RA. Alt FW. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 47.Karanjawala ZE. Adachi N. Irvine RA. Oh EK. Shibata D. Schwarz K. Hsieh CL. Lieber MR. The embryonic lethality in DNA ligase IV-deficient mice is rescued by deletion of Ku: implications for unifying the heterogeneous phenotypes of NHEJ mutants. DNA Repair (Amst) 2002;1:1017–1026. doi: 10.1016/s1568-7864(02)00151-9. [DOI] [PubMed] [Google Scholar]
- 48.Gu Y. Jin S. Gao Y. Weaver DT. Alt FW. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao Y. Sun Y. Frank KM. Dikkes P. Fujiwara Y. Seidl KJ. Sekiguchi JM. Rathbun GA. Swat W. Wang J. Bronson RT. Malynn BA. Bryans M. Zhu C. Chaudhuri J. Davidson L. Ferrini R. Stamato T. Orkin SH. Greenberg ME. Alt FW. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 50.Griffin C. Waard H. Deans B. Thacker J. The involvement of key DNA repair pathways in the formation of chromosome rearrangements in embryonic stem cells. DNA Repair (Amst) 2005;4:1019–1027. doi: 10.1016/j.dnarep.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 51.Boubakour-Azzouz I. Ricchetti M. Low joining efficiency and non-conservative repair of two distant double-strand breaks in mouse embryonic stem cells. DNA Repair (Amst) 2008;7:149–161. doi: 10.1016/j.dnarep.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 52.Weinstock DM. Jasin M. Alternative pathways for the repair of RAG-induced DNA breaks. Mol Cell Biol. 2006;26:131–139. doi: 10.1128/MCB.26.1.131-139.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cui X. Meek K. Linking double-stranded DNA breaks to the recombination activating gene complex directs repair to the nonhomologous end-joining pathway. Proc Natl Acad Sci USA. 2007;104:17046–17051. doi: 10.1073/pnas.0610928104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karanjawala ZE. Grawunder U. Hsieh CL. Lieber MR. The nonhomologous DNA end joining pathway is important for chromosome stability in primary fibroblasts. Curr Biol. 1999;9:1501–1504. doi: 10.1016/s0960-9822(00)80123-2. [DOI] [PubMed] [Google Scholar]
- 55.Ferguson DO. Sekiguchi JM. Chang S. Frank KM. Gao Y. DePinho RA. Alt FW. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc Natl Acad Sci USA. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Windhofer F. Wu W. Iliakis G. Low levels of DNA ligases III and IV sufficient for effective NHEJ. J Cell Physiol. 2007;213:475–483. doi: 10.1002/jcp.21120. [DOI] [PubMed] [Google Scholar]
- 57.Wang YG. Nnakwe C. Lane WS. Modesti M. Frank KM. Phosphorylation and regulation of DNA ligase IV stability by DNA-dependent protein kinase. J Biol Chem. 2004;279:37282–37290. doi: 10.1074/jbc.M401217200. [DOI] [PubMed] [Google Scholar]
- 58.Lin DP. Wang Y. Scherer SJ. Clark AB. Yang K. Avdievich E. Jin B. Werling U. Parris T. Kurihara N. Umar A. Kucherlapati R. Lipkin M. Kunkel TA. Edelmann W. An Msh2 point mutation uncouples DNA mismatch repair and apoptosis. Cancer Res. 2004;64:517–522. doi: 10.1158/0008-5472.can-03-2957. [DOI] [PubMed] [Google Scholar]
- 59.Toft NJ. Winton DJ. Kelly J. Howard LA. Dekker M. te Riele H. Arends MJ. Wyllie AH. Margison GP. Clarke AR. Msh2 status modulates both apoptosis and mutation frequency in the murine small intestine. Proc Natl Acad Sci USA. 1999;96:3911–3915. doi: 10.1073/pnas.96.7.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bernstein C. Bernstein H. Payne CM. Garewal H. DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: fail-safe protection against carcinogenesis. Mutat Res. 2002;511:145–178. doi: 10.1016/s1383-5742(02)00009-1. [DOI] [PubMed] [Google Scholar]
- 61.Samper E. Goytisolo FA. Slijepcevic P. van Buul PP. Blasco MA. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 2000;1:244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Celli GB. Denchi EL. de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- 63.Hsu HL. Gilley D. Galande SA. Hande MP. Allen B. Kim SH. Li GC. Campisi J. Kohwi-Shigematsu T. Chen DJ. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 2000;14:2807–2812. doi: 10.1101/gad.844000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matheos D. Ruiz MT. Price GB. Zannis-Hadjopoulos M. Ku antigen, an origin-specific binding protein that associates with replication proteins, is required for mammalian DNA replication. Biochim Biophys Acta. 2002;1578:59–72. doi: 10.1016/s0167-4781(02)00497-9. [DOI] [PubMed] [Google Scholar]
- 65.Novac O. Matheos D. Araujo FD. Price GB. Zannis-Hadjopoulos M. In vivo association of Ku with mammalian origins of DNA replication. Mol Biol Cell. 2001;12:3386–3401. doi: 10.1091/mbc.12.11.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rampakakis E. Di Paola D. Zannis-Hadjopoulos M. Ku is involved in cell growth, DNA replication and G1-S transition. J Cell Sci. 2008;121:590–600. doi: 10.1242/jcs.021352. [DOI] [PubMed] [Google Scholar]
- 67.Schild-Poulter C. Matheos D. Novac O. Cui B. Giffin W. Ruiz MT. Price GB. Zannis-Hadjopoulos M. Hache RJ. Differential DNA binding of Ku antigen determines its involvement in DNA replication. DNA Cell Biol. 2003;22:65–78. doi: 10.1089/104454903321515887. [DOI] [PubMed] [Google Scholar]
- 68.Cosgrove AJ. Nieduszynski CA. Donaldson AD. Ku complex controls the replication time of DNA in telomere regions. Genes Dev. 2002;16:2485–2490. doi: 10.1101/gad.231602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Decottignies A. Microhomology-mediated end joining in fission yeast is repressed by pku70 and relies on genes involved in homologous recombination. Genetics. 2007;176:1403–1415. doi: 10.1534/genetics.107.071621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Katsura Y. Sasaki S. Sato M. Yamaoka K. Suzukawa K. Nagasawa T. Yokota J. Kohno T. Involvement of Ku80 in microhomology-mediated end joining for DNA double-strand breaks in vivo. DNA Repair (Amst) 2007;6:639–648. doi: 10.1016/j.dnarep.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 71.Jin S. Weaver DT. Double-strand break repair by Ku70 requires heterodimerization with Ku80 and DNA binding functions. EMBO J. 1997;16:6874–6885. doi: 10.1093/emboj/16.22.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morley AA. The somatic mutation theory of ageing. Mutat Res. 1995;338:19–23. doi: 10.1016/0921-8734(95)00007-s. [DOI] [PubMed] [Google Scholar]
- 73.Turker MS. Somatic cell mutations: can they provide a link between aging and cancer? Mech Ageing Dev. 2000;117:1–19. doi: 10.1016/s0047-6374(00)00133-0. [DOI] [PubMed] [Google Scholar]
- 74.Woodruff RC. Thompson JN., Jr The role of somatic and germline mutations in aging and a mutation interaction model of aging. J Anti-Aging Med. 2003;6:29–39. doi: 10.1089/109454503765361560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.