Abstract
The CI repressor of bacteriophage λ is a model for the role of cooperativity in the efficient functioning of genetic switches. Pairs of CI dimers interact to cooperatively occupy adjacent operator sites at OR and at OL. These CI tetramers repress the lytic promoters and activate transcription of the cI gene from PRM. CI is also able to octamerize, forming a large DNA loop between OR and OL, but the physiological role of this is unclear. Another puzzle is that, although a dimer of CI is able to repress PRM by binding to the third operator at OR, OR3, this binding seems too weak to affect CI production in the lysogenic state. Here we show that repression of PRM at lysogenic CI concentrations is absolutely dependent on OL, in this case 3.8 kb away. A mutant defective in this CI negative autoregulation forms a lysogen with elevated CI levels that cannot efficiently switch from lysogeny to lytic development. Our results invalidate previous evidence that Cro binding to OR3 is important in prophage induction. We propose the octameric CI:OR–OL complex increases the affinity of CI for OR3 by allowing a CI tetramer to link OR3 and the third operator at OL, OL3.
Keywords: CI repressor, octamer, DNA looping, genetic switch, negative autoregulation, bacteriophage λ
Bacteriophage lambda occupies a special place in molecular biology, having been an immensely productive model system in our understanding of many fundamental biological processes (for recent review, see Friedman and Court 2001). Of particular value has been the unraveling of the gene regulatory mechanisms and strategies that comprise lambda's genetic switch, a bistable switch that underlies the phage's ability to efficiently use its two modes of development (Ptashne 1992). In the emerging field of gene network analysis, the lambda regulatory network has often been used as a testing ground for modeling methods (for review, see Hasty et al. 2001). However, despite the extensive characterization of the lambda switch, a few of its features remain unexplained. In the present study we show that some of these features combine in a regulatory mechanism necessary for efficient switching from lysogeny to lytic development.
The key regulator in the λ genetic switch is the CI protein, which binds to the OR and OL regions of the λ genome to repress the lytic promoters during lysogeny. Cooperative binding of a pair of CI dimers at two of the three CI operators at OR (OR1 and OR2) represses PR (Fig. 1; Johnson et al. 1981; Ptashne 1992), and similar binding at two of the three operators at OL (OL1 and OL2) represses the PL promoter (Johnson 1980; Brenowitz et al. 1986). Binding of CI to OR2 also stimulates the intrinsically weak promoter for the cI gene, PRM (Meyer and Ptashne 1980), so that in the lysogenic state CI positively regulates its own synthesis and the presence of CI is maintained. Switching from lysogeny to lytic development, called lysogenic induction or prophage induction, occurs on activation of the host SOS system in response to DNA damage, for example, after treatment with ultraviolet light (UV). RecA bound to ssDNA stimulates the self-cleavage of CI monomers (Roberts and Devoret 1983; Little 1984), removing CI's ability to bind DNA cooperatively and reducing the activity of CI to a point where the lytic promoters become derepressed.
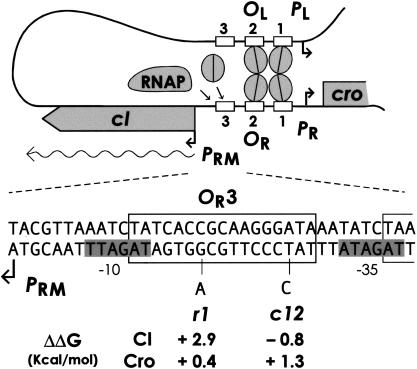
Figure 1.
Regulation of cI transcription in the lysogenic state. The upper cartoon shows a CI octamer occupying the OL1, OL2, OR1, and OR2 operators. Transcription of the cI gene from the PRM promoter by RNA polymerase (RNAP) is stimulated by CI bound at OR2 but repressed by CI bound at OR3. The lower portion shows the sequence of PRM and OR3, and the mutations used in this study with the predicted changes in free energy of binding of CI and Cro.
CI is also capable of negative autoregulation when at high concentrations, repressing the transcription of its own gene from PRM by occupying the third operator at OR, OR3 (Fig. 1; Maurer et al. 1980). However, despite the existence of this regulatory mechanism, the studies of Maurer et al. (1980) revealed very little negative autoregulation of CI—at the lysogenic CI concentration, the activity of PRM was reduced only ∼5%–20% from maximal, and repression of PRM to half its maximal activity required a CI concentration 15 times the normal lysogenic level. It has therefore become accepted that CI negative autoregulation has little or no physiological role in the lysogenic state (Johnson et al. 1981; Ptashne 1992; Darling et al. 2000). Thus, it may be that OR3 functions primarily as a site for repression of PRM by Cro protein (Johnson et al. 1981; Ptashne 1992), with CI binding to OR3 being a by-product of CI and Cro's ability to bind to similar DNA sequences.
The function of the third operator at OL, OL3, is also unclear. The contribution of OL3 to CI repression of PL is probably small (Brenowitz et al. 1986), and Cro binding to OL3 is unlikely to affect PL, so that two CI operators should suffice at OL. It has been suggested that having three CI operators at OL and OR provides a redundancy in CI repression of the lytic promoters that makes it more difficult for virulent strains of λ to arise by mutation (Johnson et al. 1981).
Recently, Révet et al. (1999) demonstrated that CI is able to form a long DNA loop between OL and OR. They showed by electron microscopy that a DNA loop can form in vitro between two pairs of CI dimers spaced 2.5 or 2.9 kb apart (the natural OL and OR spacing is 2.4 kb). Using reporter studies, they showed that the presence of OL1 and OL2 at a distance of 3.6 kb from PR enhanced CI repression of PR in vivo fourfold. The requirement for four CI operators led Révet et al. (1999) to propose that the DNA looping in their experiments was mediated by a CI octamer, as shown in Figure 1. This is consistent with the association properties of CI in solution (Senear et al. 1993) and with the crystal structure of the CI C-terminal domain (Bell et al. 2000). However, the physiological relevance of CI octamerization is not clear (Senear et al. 1993; Bell et al. 2000; Koudelka 2000) because PR is 98.5% repressed by lysogenic CI concentrations even in the absence of OL (Meyer et al. 1980).
Here we show by reporter studies that CI at its lysogenic concentration does repress PRM, but only in the presence of OL, placed, in this case, 3.8 kb away. A phage with a mutation in OR3 that eliminates the ability of CI to repress PRM forms a lysogen that produces elevated CI levels and is defective in prophage induction by UV. We propose that the formation of the OL–OR loop through CI octamerization juxtaposes OR3 and OL3 such that they can be occupied cooperatively by a CI tetramer, allowing effective repression of PRM. This mechanism acts to limit the CI concentration in the lysogenic state and permit efficient switching into lytic development.
Results
Mutations in OR3 affect the efficiency of prophage induction
We reasoned that if CI negative autoregulation is not significant in lysogeny, as widely believed, then mutations in OR3 that alter the ability of CI to repress PRM should have little or no effect on the properties of the lysogenic state. Mutations designed to increase or decrease CI repression of PRM were therefore introduced into OR3 (Fig. 1). The analyses by Takeda and coworkers of the effects of base pair changes in OR1 on the strengths of CI and Cro binding (Sarai and Takeda 1989; Takeda et al. 1989) were used as a guide, with the reservation that the effects may be different in the context of OR3. We chose r1 as a mutation that should markedly decrease CI binding with little effect on Cro binding. The r1 mutation has been previously shown to strongly reduce repression of PRM by CI (Maurer et al. 1980). The c12 change should improve CI binding slightly while decreasing Cro binding. The mutations were created in a plasmid vector and then the wild-type and mutant operator regions were recombined in vivo into λ+ to give λwt, λr1, and λc12.
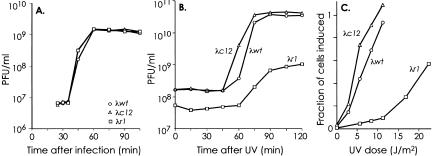
The three phages formed indistinguishable turbid plaques on indicator bacteria. Phage production kinetics after infection of the bacterial strain C600 were unaffected by the mutations (Fig. 2A). Monolysogens of C600 and NK7049 were obtained without difficulty and the UV-inducibility of the wild-type and mutant prophages in these lysogens was compared. Phage production after treatment of the C600 lysogens with 7.6 J/m2 UV was much less efficient in the λr1 lysogen, whereas the λc12 mutant was induced slightly better (Fig. 2B). Compared with the λwt lysogens, fewer λr1 lysogens were induced (29% of wild type: 95% confidence limits 14%–52%, n = 4), the average number of phage produced per induced lysogen was much less (9% of wild type: 95% confidence limits 2%–24%), and the time required to release phage particles was longer (∼60 min compared with 45 min). The fraction of NK7049 λr1 lysogens induced after UV doses of 5.6–11.2 J/m2 was ∼10% of λwt, whereas the λc12 mutant was induced better than λwt at all doses (Fig. 2C), the maximal difference being 1.7-fold at 5.6 J/m2 (P < 0.05, 2-tailed t-test, n = 4). Thus the r1 and c12 mutations at OR3 have significant and opposite effects on the ability of λ to exit the lysogenic state in response to UV light.
Figure 2.
Defective ultraviolet (UV) induction of OR3 mutant prophages. (A) Infection of C600. Plaque-forming units (PFU) per milliliter after infection of C600 with λwt, λr1, or λc12 (averages of two experiments). (B) UV-induced phage production from C600 lysogens. PFU per milliliter produced after UV irradiation (7.6 J/m2) of C600 lysogenic for λwt, λr1, or λc12 (averages of four experiments). (C) UV-induced phage production from NK7049 lysogens. Fraction of lysogens producing phage versus UV dose (averages of four experiments).
Mutations in OR3 affect the CI concentration in the lysogenic state
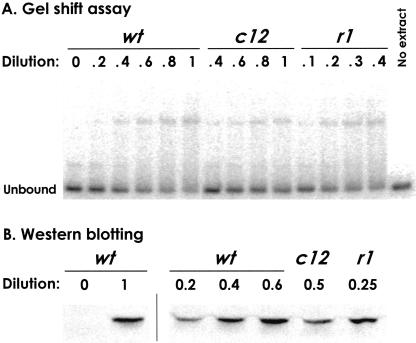
The effects of the OR3 mutations on prophage induction could be explained if the concentration of CI in the lysogenic state was increased by the r1 mutation and decreased by the c12 mutation, because this would hinder or assist, respectively, clearance of CI activity from the cell by activated RecA. To test this, we directly measured CI levels in the λwt, λr1, and λc12 NK7049 lysogens by two methods: a gel mobility shift assay for DNA binding activity and Western blotting (Fig. 3). In both methods, extracts from λwt lysogenic cells and nonlysogenic cells were mixed to produce a set of standards, and extracts from the λr1 and λc12 lysogens were diluted in nonlysogenic extract and compared with these standards. This allowed determination of CI levels or activities in wild-type lysogenic units (WLU), that is, relative to the wild-type lysogen. The two methods gave good agreement. The gel shift assays gave CI levels in the λr1 and λc12 lysogens as 2.8 and 0.6 CI WLU, respectively (95% confidence limits: r1 2.43–3.14 and c12 0.51–0.67, n = 4). Immunoblotting gave CI levels in the λr1 and λc12 lysogens as 2.3 and 0.6 CI WLU, respectively (95% confidence limits: r1 2.10–2.53, n = 5, and c12 0.45–0.74, n = 6). Thus, the OR3 mutations have strong effects on the lysogenic CI concentration.
Figure 3.
Effect of OR3 mutations on lysogenic CI levels. (A) Gel shift quantitation of CI concentrations in λ lysogens. Extracts from NK7049 λwt, λr1, and λc12 lysogens (with plasmids pUHA1 and pZC320) were diluted in nonlysogenic extract; the dilution factor indicates the proportion of lysogenic extract in each reaction. The fraction of the 154-bp OL-containing DNA fragment bound in each lane was measured, allowing determination of the CI levels in the r1 and c12 extracts relative to wt, that is, in CI wild-type lysogenic units (CI WLU, see Materials and Methods). (B) Western blotting to quantitate CI concentrations in λ lysogens. Extracts were as in (A) except that pZC320 was sometimes replaced by pZE15. The two leftmost tracks are from a separate blot and only a portion of the blots is shown.
CI transcriptional autoregulation in the absence of OL
The effects of the r1 and c12 mutations on the concentration of CI in the lysogenic state are consistent with their expected effects on CI repression of PRM. The c12 mutation should increase CI repression of PRM, leading to a lower CI expression in the lysogen; the r1 mutation should decrease CI repression of PRM and increase CI levels. However, the magnitude of the r1 effect, an ∼250% increase in CI levels, was inconsistent with the finding of Maurer et al. (1980) that no more than 20% repression of PRM occurred in the lysogenic state. To try to resolve this discrepancy, we repeated the experiment of Maurer et al. (1980) by examining the activity of the wild-type and mutant PRM promoters in vivo in response to a range of CI concentrations.
To report on the activity of PRM, we cloned DNA fragments from the −123 to the +62 positions of the promoter (+42 to –143 of PR) upstream of lacZ and placed them in single copy in the bacterial chromosome of NK7049 as part of a λ prophage (Fig. 4A), using a modification of the system of Simons et al. (1987). Note that the reporter integrates at the λatt site but, since it carries the immunity region of φ21 (imm21), it does not make λ CI and does not contain λ OR or OL. To test the effect of a range of CI concentrations on PRM, we supplied CI to these reporters from either of two plasmids carrying the cI gene under the control of the wild-type Plac promoter. Plasmid pZC320cI is a single copy, mini-F based plasmid (Shi and Biek 1995), and pZE15cI has a copy number of 50–70 (Lutz and Bujard 1997). In each strain, a second plasmid, pUHA1, carried the wild-type lac repressor gene, allowing IPTG control of CI expression. Western blots (data not shown) and gel shift assays (summarized in Fig. 4B) showed that pZC320cI gave good control of CI expression, ranging from about 0.06 CI wild-type lysogenic units in the absence of IPTG to ∼3 CI WLU at 300 μM IPTG, with the lysogenic CI concentration produced at 60 μM IPTG. In contrast, Western blots (data not shown) indicated that plasmid pZE15cI could be induced to produce at least 20 CI WLU.
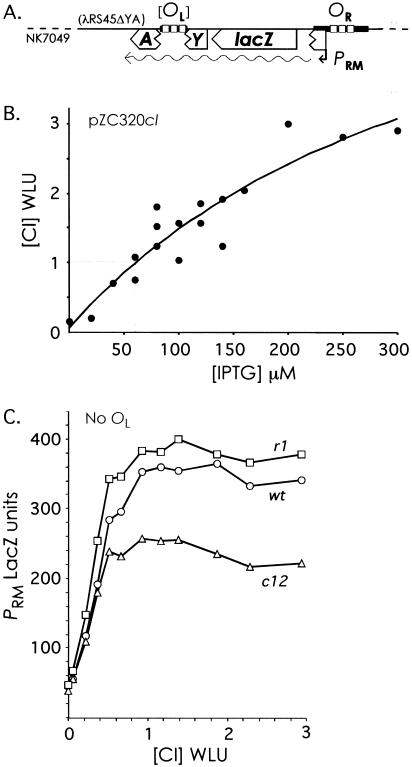
Figure 4.
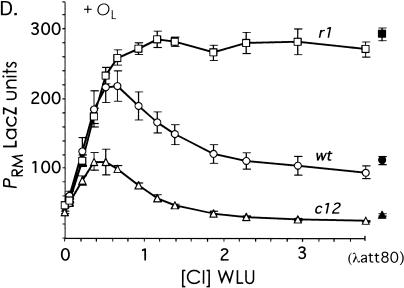
The effect of OR3 mutations on CI regulation of PRM in the absence and presence of OL. (A) The PRM::lacZ reporter system. The PRM::lacZ operon fusion was present on a single copy λimm21 prophage in NK7049. The lacY and lacA genes were removed by deletion. OL, when present, was inserted at the deletion point and was 3.8 kb downstream of OR. All strains in (B) to (D) were NK7049, carrying a reporter prophage and plasmid pUHA1 (makes Lac repressor) and the Plac CI expression plasmid pZC320cI or its cI-less parent, pZC320. (B) CI levels produced by the pZC320cI plasmid. Extracts of (λRS45ΔYA-PRMwt::lacZ) with pZC320cI, grown with the indicated IPTG concentrations, were analyzed for CI concentration relative to the wild-type lysogenic level by the gel shift assay. Each point represents the mean of at least two determinations of CI levels from one extract. The curve is the best fit to a rectangular hyperbola (Sigmaplot). (WLU) Wild-type lysogenic units. (C) Activity of wt and mutant PRM promoters in the absence of OL. LacZ activities of (λRS45ΔYA-PRM::lacZ) pZC320cI versus CI concentration produced by IPTG induction according to the relationship of (B). Points are the means of three experiments. The zero CI points are the basal activities of the wt and mutant PRM promoters (in the presence of pZC320 and no IPTG). (D) As (C) but in the presence of OL, that is, with (λRS45ΔYAOL-PRM::lacZ). Points are the means of 4–10 experiments; the error bars show the 95% confidence limits. The solid symbols to the right of the curves are the mean LacZ activities of the PRM reporters in the presence of lysogenic CI levels produced by a λatt80 prophage (with pZC320 and IPTG concentrations ranging from 50 to 500 μM).
Figure 4C shows the activity of the wt, r1, and c12 PRM promoters in the presence of increasing IPTG induction of CI expression from pZC320cI. The IPTG concentrations have been converted to CI concentrations using the empirical relationship of Figure 4B. The mutations did not strongly affect the basal activity of PRM. Over the range of CI concentrations produced by pZC320cI, the wild-type and mutant PRM promoters were activated but not repressed. All the promoters reached maximal activity at about the lysogenic CI concentration, but increasing CI levels produced no reduction in activity, even in the c12 mutant (Fig. 4C). These data support the observation of Maurer et al. (1980) that very little or no repression of PRM occurs at the lysogenic CI level. In accord with Maurer et al. (1980), we did see repression of wt and c12 PRM when we supplied very high levels of CI with pZE15cI (data not shown). The c12 mutation improved the efficiency of repression of PRM, consistent with its expected effect on CI binding at OR3. However, the r1 mutant PRM remained at maximal activity even at the very high CI concentrations, confirming that the r1 mutation eliminates repression of PRM.
Judging from the maximal PRM activities under conditions in which there was no apparent occupation of OR3 (Fig. 4C), the mutations appear also to affect the ability of PRM to be activated by CI. The r1 mutation improved maximal PRM activity by ∼9%, whereas the c12 mutation reduced it by ∼30%. Thus, the r1 mutation, at the –17 position of PRM, seems to slightly improve the ability of RNA polymerase at PRM to be activated by CI bound at OR2, whereas the c12 mutation, which alters the −25 position of PRM, may interfere with the response of RNA polymerase to CI at OR2.
The regulation of PRM by CI seen in our lacZ reporter assays in the absence of OL confirms the discrepancy with our measurements of lysogenic CI levels. The λr1 prophage produced a CI level that was 2.3–2.8-fold that of the λwt prophage, yet the activity of PRM–r1 at 2.3–2.8 CI WLU was no more than 1.1 times that of PRM–wt at 1 CI WLU. This small increase in PRM activity due to the r1 mutation is unable to explain the large increase in CI concentration in the λr1 lysogen.
CI transcriptional autoregulation in the presence of OL
We reasoned that if the CI-mediated DNA loop between OR and OL (Révet et al. 1999) occurs in the prophage DNA, then OL may affect CI regulation of PRM in the lysogenic state. We therefore introduced OL downstream of lacZ in our reporter constructs (Fig. 4A). The OL operators were placed in the same orientation relative to the OR operators as they are in the phage, but OL and OR were spaced 3.8 kb apart rather than the natural 2.8-kb distance. Like Révet et al. (1999), we mutationally inactivated the PL promoter on the OL fragment, but we included all three OL operators, rather than just OL1 and OL2.
The PRM activities versus CI concentration in the presence of OL are shown in Figure 4D. Repression of the wild-type and c12 PRM promoters was much more efficient in the presence of OL (Fig. 4, cf. C and D). At the lysogenic CI concentration (1 CI WLU), wild-type PRM activity was reduced 16% from its maximal level and the activity of c12 PRM was reduced 36% from its maximal level. However, even in the presence of OL, PRM in the r1 mutant remained resistant to repression. This shows that the improvement of repression caused by OL requires an intact OR3, indicating that OL is acting by increasing CI occupation of OR3. The magnitude of the effect of OR3 on PRM repression can be seen by comparing wt and r1 PRM. At the lysogenic CI concentration, the activity of wild-type PRM-wt was 34% lower than the activity seen in the r1 mutant. Thus, assuming that OR3 is completely unoccupied in the r1 mutant, and allowing for the 9% improvement of PRM activation by CI caused by the r1 mutation, we can estimate that OR3 is occupied 26% of the time in the lysogen.
The negative autoregulation by CI in the presence of OL goes some way to explaining the difference in CI concentration between the λwt and λr1 lysogens. From the data of Figure 4D, the expected activity of PRM–r1 in the λr1 lysogen (at 2.3–2.8 CI WLU) is ∼1.5 times the expected activity of PRM-wt in the λwt lysogen (at 1 CI WLU). Assuming that CI production is proportional to PRM activity quantitated by LacZ units, we would therefore predict the concentration of CI in the λr1 lysogen to be 1.5 times that of the λwt lysogen, a value ∼60% of the measured CI level. A similar argument predicts the CI concentration in the λc12 lysogen to be 0.6 that of wild type, matching the measurement of CI levels for the λc12 lysogen (0.6 CI WLU).
As a check on our determination that the wild-type lysogenic CI level is produced from pZC320cI at 60 μM IPTG (Fig. 4B), we supplied the lysogenic CI level to the reporter constructs from a single-copy λ prophage (λatt80) integrated at the φ80 chromosomal attachment site. The activities of the wt and mutant PRM reporters in the presence of this prophage are shown to the right of the curves in Figure 4D. Surprisingly, the activities of the PRM-wt and PRM-c12 promoters were considerably lower than those obtained with 1 CI WLU from pZC320cI. In fact, all three promoters showed activities similar to those seen at 2 CI WLU with pZC320cI. Yet the activity of CI in the λatt80 lysogen, as measured by the gel shift assay, was similar to that produced from pZC320cI at 60 μM IPTG (measurements of two extracts gave values of 1.0 and 1.1 CI WLU). Maurer et al. (1980) reported a similar anomaly between CI activity in lacZ reporter assays, depending on whether CI was produced from a plasmid or a prophage. We believe that the anomaly is due to the plasmid-produced CI level being prone to cell-to-cell variability. Large cell-to-cell variation can occur in levels of proteins produced from expression plasmids and this variation has the effect of smoothing activity versus concentration curves, such as those in Figure 4, C and D (Cluzel et al. 2000). Thus, the culture with pZC320cI at 60 μM IPTG has the same average CI content as a lysogen, but the population may be a mix of cells: those with less CI and a higher-than-lysogenic PRM activity and those with more CI and similar-to-lysogenic PRM activity. The average PRM activity of such a population would overestimate the true activity at the lysogenic concentration. In contrast, we expect the native λ cI gene on the λatt80 prophage to produce a more consistent CI level. This is because negative autoregulation by a protein has the effect of reducing fluctuation around its steady state concentration (see Discussion), as demonstrated experimentally by Becskei and Serrano (2000). We therefore expect that the PRM activities in the presence of the λatt80 prophage more truly reflect the lysogenic state.
In support of this, a lower activity of wt PRM in the lysogenic state resolves the discrepancy between the reporter data and the lysogenic CI concentrations. The expected activity of PRM-r1 in the λr1 lysogen becomes ∼2.4 times the expected activity of PRM-wt in the λwt lysogen, which matches the observed 2.3–2.8-fold difference in CI concentration. The predicted CI concentration in the λc12 lysogen becomes ∼0.5 times that of wild type and thus remains consistent with measurement.
On this basis, CI negative autoregulation in the lysogenic state is more severe: in the presence of the λatt80 prophage, PRM-wt activity was reduced 58% from the activity of PRM-r1 (Fig. 4D), giving an estimate of OR3 occupation in the wild-type lysogen of 53% (see above).
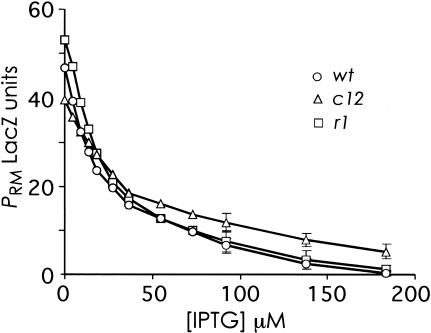
The effect of the OR3 mutants on Cro repression of PRM
The effects of the OR3 mutations on the efficiency of prophage induction can be explained by their effects on the CI concentration in the lysogenic state. However, the mutations may alter prophage induction for a different reason: by changing the ability of PRM to be repressed by the early lytic protein Cro. Cro binds more tightly to OR3 than to the other λ operators (Takeda et al. 1989; Darling et al. 2000), and it has been suggested that Cro produced early in prophage induction binds to OR3 to repress PRM, preventing further CI synthesis and thus aiding CI removal (Johnson et al. 1981; Ptashne 1992). By this reasoning, the c12 mutation, which should reduce Cro binding, should cause an impairment of prophage induction rather than the improvement that we saw. Nevertheless, we wished to check whether differences in Cro repression of PRM might have impact on the differences in prophage induction found between the wild-type and two mutant phages. The cro gene was therefore cloned into the high-copy pZE15 expression vector and the effect of Cro on basal PRM transcription in the presence of OL was examined (Fig. 5). Wild-type PRM was effectively repressed by Cro. The r1 mutation had no significant effect, whereas the c12 mutation caused a slight reduction in the efficiency of Cro repression of PRM. It is clear, then, that the effects of the two mutations on prophage induction cannot be explained by differences in Cro repression of PRM. Friedman and Court (2001) have suggested that Cro repression of PRM might be affected by OL, but no significant changes were seen on repeating the experiment in the absence of OL (data not shown).
Figure 5.
The effect of OR3 mutations on Cro repression of PRM. Activity of wt or mutant PRM promoters with IPTG induction of Cro expression in NK7049(λRS45ΔYAOL-PRM::lacZ) with pUHA1 and pZE15cro. Points are the means of four or five experiments; the 95% confidence limits for the last three highest [IPTG] are shown.
Discussion
We have shown that the efficiency of CI repression of PRM is increased by OL to the point that PRM is ∼50% repressed at the lysogenic CI concentration. The r1 mutation in OR3 abolishes this negative autoregulation, resulting in a 2.3–2.8-fold increase in the CI level in the lysogen and causing a strong defect in prophage induction. These findings have important implications for the role and structure of the OR–CI–OL DNA loop and for the function of OR3 and OL3, and illustrate the utility of negative autoregulation in allowing efficient switching between developmental states.
The CI-mediated OL–OR DNA loop
As pointed out by Bell et al. (2000), the experiments of Révet et al. (1999) do not show that the CI-mediated OL–OR loop is important in phage development. Our results show that the OL–OR interaction, presumably via the DNA loop, plays a physiologically relevant role in the phage. The large increase in lysogenic CI concentration caused by the r1 mutation can only be rationalized if there is substantial repression of PRM by CI in the wild-type prophage, and our reporter studies show that such repression requires OL. In the absence of OL, we saw no repression of PRM by CI at concentrations close to the lysogenic level, whereas, in the presence of OL, CI supplied from the λatt80 prophage reduced PRM activity 47% from its maximal observed value. The r1 mutation abolished this repression, showing that the OL effect is mediated through OR3. Comparison with the activity of PRM-r1 indicated that OR3 is 53% occupied by CI in the wild-type lysogen.
Révet et al. (1999) showed that four operators were needed for the OL–OR interaction in vivo, suggesting that the loop was mediated by a CI octamer. CI is known to associate to octamers in solution (Senear et al. 1993), and the recently described structure of a tetramer of the CI C-terminal domain revealed surfaces that could provide for such a tetramer:tetramer interaction (Bell et al. 2000). The simplest model to explain our results is that a CI octamer linking its preferred binding sites, OL1, OL2, OR1, and OR2 (see Fig. 1), may juxtapose OL3 and OR3 such that they can be linked by a CI tetramer. Such an interaction between CI dimers bound to OL3 and OR3 is consistent with the ability of CI dimers to interact across a short DNA loop (Hochschild and Ptashne 1986). In this model, OL3 would play a central role in CI negative autoregulation, and we predict that phage defective in CI binding at OL3 will behave like the λr1 mutant. Experiments to test this prediction are in progress.
The OL–OR interaction is a striking example of gene regulation by a distal site. As far as we are aware, a DNA loop of 2.4 kb is the longest natural, repressive DNA loop known in prokaryotes. We expect that a stable loop of this size requires the large free energy change due to CI octamerization (Senear et al. 1993). Also, repression is completely dependent on the distal site—we saw no repression of PRM at physiological CI concentrations in the absence of OL. In the absence of cooperative interactions supplied by OL, the affinity of CI for OR3 is presumably too low for it to compete with RNA polymerase.
Our results support the idea that the fourfold enhancement of CI repression of PR by OL seen by Révet et al. (1999) and, presumably, a reciprocal enhancement of PL repression by OR, will occur in the phage context. The linking of OL3 and OR3 by a CI tetramer should help to stabilize the OL–OR loop and give an even greater improvement of repression of the lytic promoters. We suspect that the effect of the OL–OR loop on repression of the lytic promoters is physiologically significant. To stably maintain the lysogenic state, CI must repress PR tightly to prevent production of Cro; otherwise Cro would bind to OR3 and repress PRM, turning off CI production. Reinitz and Vaisnys (1990) measured the efficiency of Cro production from PR in the absence of CI and calculated that the activity of CI in the lysogenic state, on the basis of the data of Maurer et al. (1980), would be too low to repress Cro production sufficiently. Our data and that of Révet et al. (1999) show that OL increases the activity of CI at OR, helping to explain how CI can maintain effective repression of PR. Ptashne (1992) has argued that CI cooperativity is critical for switching because it provides for large changes in PR activity (and PRM activity) in response to relatively small changes in CI concentration. The extra level of cooperativity introduced by CI octamerization may add significantly to this effect.
It is clear that OL has a strong effect on CI repression of PRM. Does OL also affect CI activation of PRM? Friedman and Court (2001) suggested that activation of PRM by CI at OR2 might be improved by octamerization of CI through interaction with OL. We found the opposite effect. The r1 mutation eliminates repression of PRM, and therefore reveals an effect of OL on activation of PRM. We found a drop of ∼28% in maximal PRM-r1 activity when OL was introduced (Fig. 4C and D), indicating that OL reduces activation of PRM. This effect is similar to, but much weaker than, the disruption to PRM activation seen previously when CI at OR2 interacts with a CI dimer located 6 or 7 DNA turns further upstream (Hochschild and Ptashne 1988).
The role of OR3 in switching from lysogeny to lytic development
Johnson et al. (1981) and Ptashne (1992) postulated that the binding of Cro to OR3 is important in efficient switching from the lysogenic to the lytic state. The idea is that Cro produced during the early stages of prophage induction could bind to OR3 and repress PRM, preventing further synthesis of CI and thus aiding its removal. This proposal was based on experiments described by Johnson (1980) that used a λ phage carrying two mutations in OR3: the c12 mutation and the r1 mutation. It was shown that this λOR3–c12r1 mutant was strongly defective in UV induction of the prophage. Because it was believed from the results of Maurer et al. (1980) that any effect of the r1 mutation on CI repression of PRM was not significant in the lysogen, the prophage induction defect was attributed to the c12 mutation and a lack of Cro repression of PRM. However, our results show that the r1 mutation strongly impairs prophage induction without affecting Cro repression of PRM, whereas the c12 mutation, which slightly impairs Cro repression of PRM, gives an improvement in prophage induction. Experiments with our own λr1c12 double mutant indicate a similar prophage induction phenotype to the λr1 single mutant (data not shown). Therefore, the idea that Cro binding to OR3 is important in prophage induction, although plausible, lacks any supportive evidence. We are currently constructing phage mutants defective in Cro binding at OR3 but with normal CI binding in order to test this proposal.
The effects of the OR3 mutations on switching from lysogeny are instead readily explained by the degree to which CI is able to limit its own concentration in the lysogenic state; the inducibility of the phage decreased with increasing lysogenic CI concentration. This is not surprising, because alterations in CI levels will affect the duration and intensity of RecA co-protease activity (dependent on the dose of DNA damage) that is needed to reduce the CI concentration to the lytic transition threshold (Bailone et al. 1979).
Negative autoregulation
An alternative way for λ to limit the CI concentration in the lysogen would be by reducing the degree of activation of PRM by CI. What advantage is there, then, in using negative autoregulation? λ lysogeny is extremely stable. The apparent rate of spontaneous transition from the lysogenic state to the lytic state (in a recA– host) is of the order of 4 × 10−7 per generation (Little et al. 1999), or about once every 160 yr for a single cell. Despite this stability, lysogeny is also sensitive. Transient SOS-inducing signals are able to cause the prophage to efficiently and rapidly switch to lytic development. We believe that negative autoregulation allows λ to limit the lysogenic CI concentration to provide sensitivity without overly compromising the stability of the lysogen.
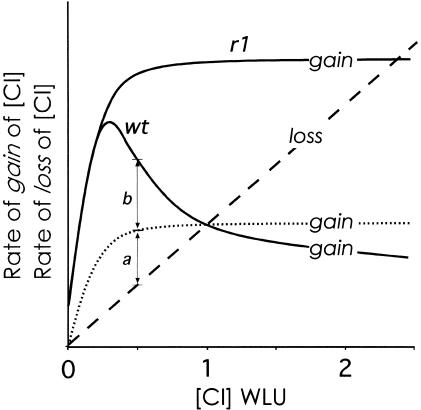
The regulatory consequences of negative autoregulation are illustrated in Figure 6, which shows how the CI concentration affects both the rate of gain of CI concentration and the rate of loss of CI concentration. CI has a half-life of >10 h in the cell (Parsell et al. 1990); the loss of CI concentration is primarily by dilution due to cell growth. Exponential cell growth means that CI concentration decays with a constant half-life, that is, that the rate of loss of CI concentration is first order, or proportional to the CI concentration (shown in Fig. 6 as a dashed line). Assuming that our measured PRM activities are proportional to the rate of gain of CI concentration, we can plot gain curves for CI either with (wt) or without (r1) negative autoregulation (Fig. 6, solid lines). Changes in the concentration of CI are dictated by the relative rates of gain and loss of the protein: if gain is greater than loss, the CI concentration will increase; if gain is less than loss, the CI concentration will decrease. The lysogenic CI concentration in each case is the stable state of this regulatory system, where gain of CI balances loss of CI. With the removal of negative autoregulation (r1), the stable state occurs at a much higher CI concentration, as observed. However, negative autoregulation is not the only way to achieve the wild-type lysogenic CI concentration—a reduction in the degree of positive autoregulation would have the same effect (Fig. 6, dotted curve). The advantage of negative autoregulation is that it provides extra stabilization of the lysogenic CI concentration against fluctuations (Becskei and Serrano 2000). From Figure 6, it can be seen that any downward fluctuation in CI concentration will be resisted more strongly in the presence of negative autoregulation because the resulting net rate of gain of CI (the difference between its rates of gain and loss; a + b in Fig. 6) will be greater than in the absence of negative autoregulation (a in Fig. 6). This noise reduction system makes it less likely that random fluctuations will lower CI concentrations to a point where lysogeny cannot be maintained.
Figure 6.
The function of negative autoregulation. The curves show the rate of gain of CI concentration versus its concentration for λwt and λr1 (solid lines). The first order rate of loss of CI concentration is plotted on the same axes as a straight line passing through the origin (dashed line). Note that the y-axis represents the rate of gain or the rate of loss of CI concentration, as indicated on the curves. The stable steady state CI concentrations in the λwt and λr1 lysogens (1 and 2.4 CI wild-type lysogenic units [WLU]) occur where the rates of CI gain and loss are equal. The dotted gain curve shows that the wild-type CI concentration could be achieved without negative autoregulation, but this would result in a less stable lysogenic CI concentration. Without negative autoregulation, a downward fluctuation in the CI concentration (e.g., to 0.5 WLU) causes the system to respond with a small net gain of CI (gain − loss = a) and therefore a fairly weak restoration of the CI concentration. With negative autoregulation (wt), the same fluctuation causes the system to produce a larger net gain of CI (gain − loss = a + b) due to derepression of PRM. This gives a stronger restoration of the CI concentration.
We expect that the ability of negative autoregulation to confer stability and sensitivity will be used in the genetic switches that underlie development in more complex organisms.
Materials and methods
Strains and media
C600 was used as a λ indicator. NK7049 (ΔlacIZYA)X74 galOP308 StrR Su– from R. Simons (Simons et al. 1987) was the host for all LacZ assays. DH5α was used for recombinant DNA work. DH5αZ1 (Lutz and Bujard 1997) was the lacIq host for construction of the cI and cro gene expression plasmids. λ+ and 434 were obtained from the laboratory of Dale Kaiser at Stanford University. λatt80 was λimmλhφ80attφ80 from Gary Gussin (University of Iowa, Iowa City). λRS45 (Simons et al. 1987) was modified for lac operon fusions. λ lysogens were tested for monolysogeny by the polymerase chain reaction (PCR) method of Powell et al. (1994); the λatt80 lysogens were checked by an analogous test. Cells were grown in LB (Miller 1972) with addition of ampicillin (30 μg/mL for pZC320, 100 μg/mL for pZE15) and kanamycin (50 μg/mL for pUHA1).
Construction of λ OR3 mutants
The r1 and c12 mutations were introduced by the Quickchange oligonucleotide-directed method (Stratagene) into pDT843, a pACYC184 derivative carrying the λ+ BclI:37352–EcoRV:39355 fragment. The sequence of the BclI:37352–BstXI:38292 region was then confirmed and this fragment was used to replace the equivalent wild-type portion of pAP831, a pACYC184 derivative carrying the λ+NheI:34678–EcoRV:39355 fragment. The wild-type and mutant sequences were crossed onto the λ chromosome by infecting C600 cells carrying pAP831 or its mutant derivative with λimm434 and selecting immλ recombinants among the progeny as plaques on C600(434). Recombination leads to replacement of the imm434 region with λ sequences. The sequence of the BclI:37352–BstXI:38292 region of the λ isolates was confirmed.
Phage infection and prophage induction
For phage infection, log phase cultures in LBM (LB, 10 mM MgSO4) plus 0.2% maltose were concentrated fivefold by centrifugation and infected at a multiplicity of addition ∼0.1 at 37°C for 10 min. The culture was diluted at least 100-fold in LB, incubated with shaking at 37°C, and plaque-forming units (PFU) assayed over time by mixing dilutions with indicator bacteria (C600 in LBM) and 3 mL molten top agar (0.7% agar, 10 mM MgSO4) and pouring onto TB plates (1% Bacto-tryptone [Difco], 0.5% NaCl, 1.5% Bacto-agar). PFU/mL were normalized to preinfection colony forming units (CFU)/mL, assayed by spreading on TB plates. For UV irradiation, log phase cultures in LB were diluted in M9 salts (Miller 1972) and irradiated with a germicidal UV lamp, all manipulations being performed under yellow lighting at 37°C. The kinetics of phage production after UV were assayed as for infection. To measure the fraction of lysogens induced after UV, we assayed aliquots of the cultures for PFU with NK7049 in LBM on LB plates. Preirradiation CFU/mL were assayed by pouring onto LB plates with top agar.
Immunoquantitation of CI
Gel electrophoresis and Western blotting were essentially as described by Sambrook et al. (1989). Cells were grown at 37°C in LB, plus ampicillin, plus kanamycin (plus IPTG, as appropriate) to O.D.600 ∼ 0.6, washed, resuspended in 1/200th volume (adjusted for slight deviations from O.D.600 = 0.6) in MinA (Miller 1972), sonicated, diluted 1:1 with 2× SDS sample buffer, and heated to 95°C for 2 min. After SDS-PAGE (10% polyacrylamide), samples were electrotransferred to PVDF membranes (0.1 μm pore, MSI, Westborough, MA). Membranes were blocked and washed in 100 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5% low fat milk, and 0.1% Tween 20. Primary rabbit antibody to λ CI was from Invitrogen and in some experiments was preadsorbed to nonlysogenic cell extract in blocking buffer. HRP-conjugated goat anti-rabbit secondary antibody and SuperSignal reagents for chemiluminesence were from Pierce. A range of exposures of Kodak Max film were scanned by laser densitometry and analyzed using NIH Image 1.62.
Gel shift quantitation of CI
Extracts were prepared from 80 mL of log phase culture at 37°C in LB, plus ampicillin, plus kanamycin (plus IPTG, as appropriate) at O.D.600 ∼ 0.6, washed, and concentrated 100-fold in TEG150 (50 mM Tris-HCl at pH 7.5, 0.1 mM EDTA, 150 mM NaCl, 10% glycerol), followed by sonication and centrifugation at 5000g to remove debris. For the gel mobility shift reactions, extracts were diluted equally in TEG150 and then the CI-containing extracts were diluted to differing extents using diluted extract from nonlysogenic cells, that is, all reactions contained the same amount of cellular extract but different amounts of CI. The DNA probe was a gel-purified 154 bp PCR-generated OLPL– fragment from pRS308ΔYAOL, labeled by treatment of one primer with [γ-32P]ATP and T4 polynucleotide kinase. Binding reactions (10 μL) were in TEG150 at 0°C with ∼150 pM probe DNA and 4 ng/μL sheared salmon sperm DNA as competitor. After 30–50 min, 4 μL of each reaction was loaded onto a 6% polyacrylamide, 10% glycerol, 1× TBE gel at 4°C. Loading and running were done at 22 mA/cm2; the running buffer was 1× TBE (Sambrook et al. 1989). After drying of the gel, the total and unbound DNA in each lane were quantitated using storage phosphor technology (Molecular Dynamics) and the fraction of DNA bound calculated as 1 − unbound/total.
Construction of chromosomal PRM::lacZ reporter fusions
The 37880–38064 PRM region of λ was amplified by PCR with primers flanked by HindIII and XhoI sites from pDT843wt or its r1 and c12 derivatives, and inserted into the polylinker of the lacZ reporter plasmid pTL61T (Linn and St. Pierre 1990). The PRM::lacZ fusions were then recombined in vivo onto λRS45ΔYA and λRS45ΔYAOL (see below) by infection of plasmid-containing strains followed by screening for blue plaque forming phage on plates containing X-gal (Simons et al.1987). The resulting λRS45ΔYA-PRM::lacZ and λRS45ΔYAOL-PRM::lacZ phage (wt, rl, and c12) were used to make monolysogens of NK7049.
The modifed lacZ reporter phage λRS45ΔYA and λRS45ΔYAOL used earlier were derived from the lac reporter λRS45 (Simons et al. 1987) by deleting the DNA between the SgrAI site (in lacY) and the BsrGI site (in lacA) to inactivate these genes (Fig. 4A). This was done to avoid IPTG-controlled expression of CI being affected by lacY expression from the reporter, because the LacY permease aids uptake of IPTG by the cell. The ΔYA deletion was first made in pRS308 (Simons et al. 1987) by endfilling and religating the SgrAI and the BsrGI sites and was then recombined in vivo onto λRS45Ptet::lacZ to obtain λRS45ΔYA. Recombinant phage were isolated as white plaque formers on X-gal (λRS45Ptet::lacZ gives blue plaques) and then checked for the presence of ΔYA using PCR. To make λRS45ΔYAOL, a synthetic (sticky-ended) λOLPL– sequence containing the OL1, OL2, and OL3 operators (underlined) and mutations to inactivate PL (lowercase), GTAcaTATCACCGCCAGTGGTATTTActaCAACACCGCCAGAGATAATTTATC ACCGCAGATGGTTATCGCCGG, was inserted into the SgrAI and BsrGI sites (right end toward lacZ) in pRS308 and recombined onto λRS45Ptet::lacZ as for λRS45ΔYA. The relevant sequences of λRS45ΔYA and λRS45ΔYAOL were confirmed.
Construction of CI expression plasmids
The wild-type λ cI gene (9 bp upstream of start codon to the stop codon) was amplified by PCR using primers with flanking PstI and HindIII sites and inserted into pBS-SK+ (Stratagene) to give pBS-SK+cI. To make pZC320cI, the PstI–cI–BamHI fragment from pBS-SK+cI was inserted downstream of Plac+; between the NsiI and BamHI sites of the mini-F based, ampicillin-resistant plasmid pZC320 (Shi and Biek 1995). The colE1, ampicillin-resistant expression plasmid pZE15 was derived by inserting Plac+ from pZC320 (on an endfilled BsaBI–BamHI fragment) into the endfilled XhoI and XbaI sites of pZE12-luc (Lutz and Bujard 1997), replacing the PLlacO-1 promoter and luc gene. The pZE15cI or pZE15cro expression plasmids were made by inserting the cI or cro genes between the NsiI and BamHI sites downstream of Plac+ in pZE15: cI by inserting the PstI I–BamHI fragment from pBS-SK+cI; cro on a PCR-generated fragment (16 bp upstream of start codon to the stop codon with flanking PstI and BamHI sites). The sequences of the cloned cI and cro genes were confirmed. Lac repressor was supplied to the expression plasmids by pUHA1, a p15A plasmid encoding kanamycin resistance and carrying the wild-type lacI gene and promoter, obtained from H. Bujard.
LacZ assays
Kinetic LacZ assays were done in 96-well microtiter plates by an extensively modified method of Miller (1972). Fresh colonies on selective LB plates were resuspended in LB and used to inoculate 200 μL of LB +antibiotic +IPTG. Dishes were sealed and incubated for ∼16 h without shaking. These cultures were subcultured by diluting 2 μL into 98 μL fresh medium and incubated with rotation to an O.D.600 of 0.2–1.2 (log phase). OD600 was measured using a Labsystems Multiskan Ascent plate reader with a 620-nm filter; the O.D.620 values were converted to O.D.600 (1-cm path length) values using an empirically derived relationship and adjusted for light-scattering nonlinearity according to Bipatnath et al. (1998). Cells were chilled and then permeabilized with polymyxin B (Schupp et al. 1995) by adding 25 μL of culture plus 25 μL of LB to 150 μL of lysis buffer in a microtiter dish. Lysis buffer was TZ8 (100 mM Tris-HCl at pH 8.0, 1 mM MgSO4, 10 mM KCl) + 2.7 μL/mL 2-mercaptoethanol and 50 μg/mL polymyxin B. The presence of detergents and chelating agents (Schupp et al. 1995) did not improve the assay. Use of pH 8 for the assay buffer rather than the pH 7 used by Miller (1972) improved display of o-nitrophenol in the absence of Na2CO3 added to stop the reaction. Assays were at 28°C and were begun by addition of 40 μL of o-nitrophenyl-β-D galactoside (4 mg/mL in TZ8). The plate reader was used to incubate the reactions and take A414 readings every 2 min for 1 h. Enzyme activity was determined as the slope of the line of best fit of A414 versus time (readings with A414>2.5 were ignored). Enzyme activity was found to be directly proportional to the O.D.600 of the culture and the volume of culture added to the assay (V, in μL). LacZ units were calculated as 200,000 × (A414/min)/(A600 x V) and were roughly equivalent to standard Miller units.
Acknowledgments
We thank Donald Biek, Herman Bujard, Gary Gussin, Thomas Linn, and Bob Simons for gifts of plasmids, phage, and bacterial strains; Don Court, Alex Gann, John Little, Mark Ptashne, John Reinitz, and Kim Sneppen for discussions and comments on the manuscript; Keith Shearwin and other members of the Egan lab for advice and support; and John Little for helping us correct one of our constructs. Research in the Egan lab is funded by the Australian Research Council. This paper is dedicated to Mark Ptashne and his colleagues, whose seminal work on λ has been an inspiration.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ian.dodd@adelaide.edu.au; FAX 61-8-8303-4348.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.937301.
References
- Bailone A, Levine A, Devoret R. Inactivation of prophage λ repressor in vivo. J Mol Biol. 1979;131:553–572. doi: 10.1016/0022-2836(79)90007-x. [DOI] [PubMed] [Google Scholar]
- Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- Bell CE, Frescura P, Hochschild A, Lewis M. Crystal structure of the λ repressor C-terminal domain provides a model for cooperative operator binding. Cell. 2000;101:801–811. doi: 10.1016/s0092-8674(00)80891-0. [DOI] [PubMed] [Google Scholar]
- Bipatnath M, Dennis P, Bremer H. Initiation and velocity of chromosome replication in Escherichia coli B/r and K-12. J Bacteriol. 1998;180:265–273. doi: 10.1128/jb.180.2.265-273.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenowitz M, Senear DF, Shea MA, Ackers GK. Quantitative DNase footprint titration: A method for studying protein–DNA interactions. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- Cluzel P, Surette M, Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- Darling PJ, Holt JM, Ackers GK. Coupled energetics of λ cro repressor self-assembly and site-specific DNA operator binding II: Cooperative interactions of cro dimers. J Mol Biol. 2000;302:625–638. doi: 10.1006/jmbi.2000.4050. [DOI] [PubMed] [Google Scholar]
- Friedman DI, Court DL. Bacteriophage λ: Alive and well and still doing its thing. Curr Opin Microbiol. 2001;4:201–207. doi: 10.1016/s1369-5274(00)00189-2. [DOI] [PubMed] [Google Scholar]
- Hasty J, McMillen D, Isaacs F, Collins JJ. Computational studies of gene regulatory networks: in numero molecular biology. Nat Rev Genet. 2001;2:268–279. doi: 10.1038/35066056. [DOI] [PubMed] [Google Scholar]
- Hochschild A, Ptashne M. Cooperative binding of λ repressors to sites separated by integral turns of the DNA helix. Cell. 1986;44:681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- ————— Interaction at a distance between λ repressors disrupts gene activation. Nature. 1988;336:353–357. doi: 10.1038/336353a0. [DOI] [PubMed] [Google Scholar]
- Johnson AD. “Mechanism of action of the λ cro protein.” Ph.D. thesis. Cambridge, MA: Harvard University; 1980. [Google Scholar]
- Johnson AD, Poteete AR, Lauer G, Sauer RT, Ackers GK, Ptashne M. λ repressor and cro-components of an efficient molecular switch. Nature. 1981;294:217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- Koudelka GB. Cooperativity: Action at a distance in a classic system. Curr Biol. 2000;10:R704–R707. doi: 10.1016/s0960-9822(00)00710-7. [DOI] [PubMed] [Google Scholar]
- Linn T, St. Pierre R. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J Bacteriol. 1990;172:1077–1084. doi: 10.1128/jb.172.2.1077-1084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JW. Autodigestion of lexA and phage λ repressors. Proc Natl Acad Sci. 1984;81:1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JW, Shepley DP, Wert DW. Robustness of a gene regulatory circuit. EMBO J. 1999;18:4299–4307. doi: 10.1093/emboj/18.15.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer R, Meyer BJ, Ptashne M. Gene regulation at the right operator (OR) of bacteriophage λ. I. OR3 and autogenous negative control by repressor. J Mol Biol. 1980;139:147–161. doi: 10.1016/0022-2836(80)90302-2. [DOI] [PubMed] [Google Scholar]
- Meyer BJ, Ptashne M. Gene regulation at the right operator (OR) of bacteriophage λ. III. λ repressor directly activates gene transcription. J Mol Biol. 1980;139:195–205. doi: 10.1016/0022-2836(80)90304-6. [DOI] [PubMed] [Google Scholar]
- Meyer BJ, Maurer R, Ptashne M. Gene regulation at the right operator (OR) of bacteriophage λ. II. OR1, OR2 and OR3: Their roles in mediating the effects of repressor and cro. J Mol Biol. 1980;139:163–194. doi: 10.1016/0022-2836(80)90303-4. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Parsell DA, Silber KR, Sauer RT. Carboxy-terminal determinants of intracellular protein degradation. Genes & Dev. 1990;4:277–286. doi: 10.1101/gad.4.2.277. [DOI] [PubMed] [Google Scholar]
- Powell BS, Court DL, Nakamura Y, Rivas MP, Turnbough CL. Rapid confirmation of single copy λ prophage integration by PCR. Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. A genetic switch. Cambridge, MA: Cell Press, Blackwell Scientific Publications; 1992. [Google Scholar]
- Reinitz J, Vaisnys JR. Theoretical and experimental analysis of the phage λ switch implies missing levels of cooperativity. J Theor Biol. 1990;145:295–318. doi: 10.1016/s0022-5193(05)80111-0. [DOI] [PubMed] [Google Scholar]
- Révet B, von Wilcken-Bergmann B, Bessert H, Barker A, Müller-Hill B. Four dimers of λ repressor bound to two suitably spaced pairs of λ operators form octamers and DNA loops over large distances. Curr Biol. 1999;9:151–154. doi: 10.1016/s0960-9822(99)80069-4. [DOI] [PubMed] [Google Scholar]
- Roberts JW, Devoret R. Lysogenic induction. In: Hendrix RW, Roberts JW, Stahl FW, Weisburg RA, editors. Lambda II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1983. pp. 123–144. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sarai A, Takeda Y. λ repressor recognizes the approximately 2-fold symmetric half-operator sequences asymmetrically. Proc Natl Acad Sci. 1989;86:6513–6517. doi: 10.1073/pnas.86.17.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupp JM, Travis SE, Price LB, Shand RF, Keim P. Rapid bacterial permeabilization reagent useful for enzyme assays. Biotechniques. 1995;19:18–20. [PubMed] [Google Scholar]
- Senear DF, Laue TM, Ross JBA, Waxman E, Eaton S, Rusinova E. The primary self-assembly reaction of bacteriophage λ cI repressor dimers is to octamer. Biochemistry. 1993;32:6179–6189. doi: 10.1021/bi00075a010. [DOI] [PubMed] [Google Scholar]
- Shi J, Biek DP. A versatile low-copy-number cloning vector derived from plasmid F. Gene. 1995;164:55–58. doi: 10.1016/0378-1119(95)00419-7. [DOI] [PubMed] [Google Scholar]
- Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Takeda Y, Sarai A, Rivera VM. Analysis of the sequence-specific interactions between Cro repressor and operator DNA by systematic base substitution experiments. Proc Natl Acad Sci. 1989;86:439–443. doi: 10.1073/pnas.86.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]