Abstract
Studies of embryonic stem cells (ESCs) reveal that these cell lines can be derived from differing stages of embryonic development. We analyzed common changes in the expression of microRNAs (miRNAs) and mRNAs in 9 different human ESC (hESC) lines during early commitment and further examined the expression of key ESC-enriched miRNAs in earlier developmental states in several species. We show that several previously defined hESC-enriched miRNA groups (the miR-302, −17, and −515 families, and the miR-371–373 cluster) and several other hESC-enriched miRNAs are down-regulated rapidly in response to differentiation. We further found that mRNAs up-regulated upon differentiation are enriched in potential target sites for these hESC-enriched miRNAs. Interestingly, we also observed that the expression of ESC-enriched miRNAs bearing identical seed sequences changed dynamically while the cells transitioned through early embryonic states. In human and monkey ESCs, as well as human-induced pluripotent stem cells (iPSCs), the miR-371–373 cluster was consistently up-regulated, while the miR-302 family was mildly down-regulated when the cells were chemically treated to regress to an earlier developmental state. Similarly, miR-302b, but not mmu-miR-295, was expressed at higher levels in murine epiblast stem cells (mEpiSC) as compared with an earlier developmental state, mouse ESCs. These results raise the possibility that the relative expression of related miRNAs might serve as diagnostic indicators in defining the developmental state of embryonic cells and other stem cell lines, such as iPSCs. These data also raise the possibility that miRNAs bearing identical seed sequences could have specific functions during separable stages of early embryonic development.
Introduction
The isolation of embryonic stem cells (ESCs) from the inner cell mass of murine and human embryos and the subsequent derivation of cell lines from these pluripotent cells provided researchers with powerful tools for developmental studies and for potential cellular therapeutics [1,2]. Embryonic stem cells are capable of self-renewal and possess the ability to differentiate into a broad array of cell types. Numerous studies have demonstrated the ability to direct the differentiation of ESCs into various lineages [3,4]. As such, the potential applications of ESCs in both basic and clinical research have been highly touted [3].
Recently, several groups reported that the forced expression of select transcription factors facilitated the reprogramming of adult somatic cells into pluripotent, ES-like cells [5–9]. The creation of these induced pluripotent stem (iPS) cells brings with it the exciting possibility that pluripotent donor-specific stem cell lines can be created to treat a host of maladies. Although excellent progress has been made, further studies aimed at understanding similarities and differences between hESC and iPS are still required.
In the past decade, miRNAs have been identified as an important class of genes involved in regulating organismal development and differentiation of specific cell types [10–12]. These endogenous, small (∼22 nucleotide) RNAs regulate the stability and translation of mRNAs bearing partially complementary sequences in their 3′ untranslated regions (UTRs). Computational and functional studies have identified nucleotides 2–8, or the “seed region,” as the critical portion of the miRNA for base pairing interactions with target mRNAs [11,13]. Families of miRNAs with highly conserved sequences are found in many organisms, and additionally, clusters of miRNAs located in discrete genomic loci can be coordinately expressed. These clusters and families of miRNAs are ideal molecular tools for regulating developmental processes, as the miRNAs can be expressed in a temporal manner to quickly regulate the expression of a host of genes. Studies have shown that miRNAs play critical roles in the maintenance and differentiation of various populations of mammalian stem cells; however, careful examination of miRNAs and their function in hESCs is still in early stages [14–19].
Previously, miRNA expression has been examined in murine and human ES cells using cloning, microarray, quantitative polymerase chain reaction (PCR), and deep sequencing technologies on selected cell lines [20–29]. These studies typically involve embryoid body (EB) formation as an intermediate step, and the differentiation protocols extend for several weeks. In order to examine the miRNAs responding rapidly to early differentiation cues, we analyzed 9 NIH-approved hESCs 4 days after the initiation of serum-enforced differentiation. In corroboration with other studies, we find that members of several previously described miRNA families and clusters are enriched in hESCs. Additionally, we observe that the expression of miRNAs not previously associated with hESCs, as well as several recently discovered miRNAs and miRNA families. These miRNAs responded dramatically upon initiation of differentiation, suggesting that they may perform highly ES-specific roles. Finally, we examined the expression of the hESC-enriched miRNAs in early developmental stages in several species and find that the expression of miRNAs bearing identical seed sequences (miR-302b and hsa-miR-372/mmu-miR-295) correlates with the state of the ESC, suggesting that relative expression levels of these microRNAs might serve as diagnostic indicators of the stem cell state.
Materials and Methods
hESC cells and culture techniques
For this study, the human ESC lines H1 (WA01), H7 (WA07), H9 (WA09), H13 (WA13), H14 (WA14), HSF-6, BG01, BG02, and BG03, as well as the embryonal carcinoma cell line, NTera-2, were used [30,31]. hESCs were maintained as previously described [31]. In brief, hESCs were initially cultured on monolayers of γ-irradiated primary mouse embryonic fibroblasts (MEFs) with Dulbecco's modified Eagle's medium/Ham's F-12 medium (DMEM/F12) containing glutamax with 20% knockout serum replacer (SR), 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 50 U/mL penicillin, 50 μg/mL streptomycin (all from Invitrogen, Carlsbad, CA), 0.1 mM β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO), and 2 ng/mL basic fibroblast growth factor (FGF; Peprotech, Rocky Hill, NJ). Cells were passaged using 1.2 U/mL dispase dissolved in phosphate-buffered saline (PBS) containing 10% ES-qualified fetal bovine serum (FBS; all from Invitrogen). Cells were grown for 3 passages in feeder-free conditions prior to differentiation. For feeder-free conditions, hESCs were grown on plates coated with Matrigel (BD Biosciences, San Jose, CA) in MEF-conditioned media, supplemented with 2 ng/mL FGF [32]. For butyrate-treatment studies, the hESCs were grown for at least 3 passages in non-MEF treated, non-FGF hESC media containing 0.2 mM sodium butyrate (Sigma), as previously described [33].
For differentiation studies, hESCs were grown in feeder-free conditions in DMEM/F12 containing glutamax with 20% ES-qualified FBS, 50 U/mL penicillin, and 50 μg/mL streptomycin (all from Invitrogen) for 4 days. Differentiation of NTera-2 cells was performed using the same conditions with the exception that 5 μM retinoic acid (Sigma-Aldrich, St. Louis, MO) was also included in the media, because NTera-2 cells are otherwise refractory to differentiation. Retinoic acid treatment biases the NTera-2 differentiation to an ectodermal lineage. Media changes were performed daily for all cells used in these analyses.
Murine embryonic stem cells (mESCs) were treated as previously described [33]. Murine epiblast stem cells (mEpiSC #5) were the kind gift of Dr. Paul J. Tesar and were cultured in hESC culture conditions as described [34]. The tissue culture-derived mEpiSCs were obtained by passaging R1 mESCs with 20 ng/mL activin A (HumanZyme) and 10 ng/mL bFGF (Invitrogen, Carlsbad, CA) as described [35].
Non-human primate (nhp) ESCs were the gift of Tom Burbacher and Eric Hayes, Washington National Primate Research Center). The MF-1 cell line was generated from a male Macaca fascicularis embryo and had a normal G-banded karyotype on arrival at passage 56. These cells were grown on a feeder layer of γ-irradiated (3,000 rads) primary MEF from passage 59 to passage 72. Base culture medium consisted of DMEM/F12 containing GlutaMax™ supplemented with 20% SR, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 50 U/mL penicillin, 50 mg/mL streptomycin (all from Invitrogen), 0.1 mM β-mercaptoethanol (Sigma, St. Louis, MO). MF-1 were grown on feeders in base medium supplemented either with 4 ng/mL FGF2 (Invitrogen) to approximate standard hESC/epiblast stage culture, in the presence of 0.1 mM sodium butyrate (Sigma) + 200 nM suberoylanilide hydroxamic acid (Vorinostat, Cayman Chemical) + 10 ng/mL human LIF (Millipore, Temecula, CA) to induce a reversion to an earlier developmental stage between embryonic equivalent epiblast and inner cell mass [33].
Establishment of induced pluripotent stem (iPS) cell lines
iPS cells were generated from human fibroblasts using either a previously described 4-component cocktail (Oct4, Sox2, Nanog, and Lin28; OSLN) [8] or 4/3-component cocktails (Oct4, Sox2, Klf4 with or without c-Myc; OSKM or OSK and M83.9) [36].
Human myocardial fibroblasts (HMF2) and human foreskin fibroblasts (HFF1) were infected by lentiviruses expressing Oct4, Sox2, Nanog, and LIN28 and Moloney murine leukemia retroviruses expressing Oct4, Sox2, KLF4, and c-Myc, respectively. Viral transductions were performed in the presence of polybrene (4 μg/mL). Following 1- or 2-day incubation, cells were trypsinized, transferred to 10-cm dishes, seeded with irradiated MEF, and cultured in hESC medium. For the generation of 3 factor-iPS cells, hESC medium was supplemented with 0.5 mM valproic acid during the first 10 days.
To establish iPS cell lines, iPS colonies were picked ∼1 month post-infection based on hESC-like colony morphology. The selected colonies were subsequently expanded and maintained on irradiated MEF in hESC media. Butyrate treatment of induced pluripotent stem cells (iPSCs) was as described earlier for hESCs.
Reverse transcription and quantitative polymerase chain reaction (qPCR)
Total RNA was extracted from cells using Trizol (Invitrogen, Carlsbad, CA), according to manufacturer's protocol. The RNA was then treated with DNase I (Fermentas) to remove any contaminating genomic DNA. Reverse transcription was performed using Omniscript (Qiagen, Valencia, CA) according to manufacturer's protocol. Reactions were carried out at 37°C for 60 min. Quantitative PCR was performed using Power Sybr® Green PCR Master Mix (Applied Biosystems, Foster City, CA) using an Applied Biosystems 7300 Real-Time PCR System. All primer sets were validated for use in qPCR by several methods, including agarose gel analysis, dissociation curve analysis, and template dilution studies. Reactions were carried out using the following conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s steps. All qPCRs were normalized using β-actin as an internal control. Primer sequences are as follows: β-actin fwd-5′-TCAAGATCATTGCTCCTCCTGAG-3′, β-actin rev-5′-ACATCTGCTGGAAGGTGGACA-3′, Nanog fwd-5′-CCTGTGATTTGTGGGCCTG-3′, Nanog rev-5′-GACAGTCTCCGTGTGAGGCAT-3′, Oct4 fwd-5′-GTGGAGGAAG CTGACAACAA-3′, Oct4 rev-5′-ATTCTCCAGGTTGCCTCT CA-3′, Sox2 fwd-5′-GTATCAGGAGTTGTCAAGGCAGAG-3′, Sox2-rev-5′-TCCTAGTCTTAAAGAGGCAGCAAAC-3′.
All miRNA qPCR assays were performed using TaqMan miRNA assays (Applied Biosystems), according to the manufacturer's protocol.
Immunofluorescence
hESCs were grown on Matrigel-treated chamber slides (Nalge Nunc Intl.) under conditions previously described. Following the 4 days of CM or differentiation media treatment, the cells were fixed with 4% paraformaldehyde. Cells stained for intracellular Oct4 were permeablized using Triton-X (0.1%). The following primary antibodies were used at the indicated dilutions: Oct 3/4 (R&D Systems, Minneapolis, MN; 1:100), SSEA-4, and Tra-1–60 (Chemicon Intl., Temecula, CA; 1:200). Secondary antibodies and dilutions were as follows: Alexa-568 goat α-mouse and Alexa-488 donkey α-goat (Molecular Probes, Eugene, OR; 1:1,000). Cells were further treated with the nuclear stain 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (1 μg/mL; Sigma-Aldrich, St. Louis, MO) and visualized using confocal microscopy (Lecia SPE5).
miRNA Microarray
Total RNA for miRNA microarray analysis was extracted using Trizol (Invitrogen, Carlsbad, CA), according to manufacturer's protocol. RNA fractionation, labeling, hybridization, and detection were performed as previously described using Agilent Technologies single-color, miRNA microarrays [37]. The microarray platform contains probes for detection of 470 annotated human miRNAs and the sensitivity and dynamic range of the assay allows for reliable detection of miRNAs expressed at low levels. Background-subtracted gene signals were normalized to the 75th percentile of signal intensity for each chip. Placental RNA samples, provided by the microarray manufacturer, were used as internal control samples for the microarray slides. All miRNA microarray data analysis was performed using MultiExperiment Viewer (MeV, version 4.3) [38]. Hierarchical clustering analysis using the log of ratio (differentiated expression/undifferentiated expression) following normalization was performed and Euclidian Distance and average linkage for similarity measurements were utilized.
For comparison of miRNA expression in the BG02-CM and BG02-butyrate cells, statistical analyses were performed using the freely available statistical programming and graphics environment R and using the Limma bioconductor package. After general log2 transformation, the background-corrected data was quantile normalized. A linear model was fitted for each gene and an empirical Bayesian approach was used to calculate standard errors and to rank genes using a combination of magnitude and consistency of differential expression. Differentially expressed genes were defined as those with a B statistic > 0 [39].
mRNA microarray and hexamer seed-match analysis
Total RNA for mRNA microarray analysis was isolated using RNeasy kit (Qiagen, Valencia, CA) as per manufacturer's protocol. Samples were amplified and labeled using a custom-automated version of the RT/IVT protocol and reagents provided by Affymetrix. Hybridization, labeling, and scanning were completed following the manufacturer's recommendations (Affymetrix). Sample amplification, labeling, and microarray processing were performed by the Rosetta Inpharmatics Gene Expression Laboratory in Seattle, WA. Two-way hierarchical clustering of whole genome microarray expression (Affymetrix) and Rosetta error modeling was used to find groups of genes whose expression changed at least 1.5-fold and had a P value <0.01 in all 9 hESCs [40]. GO term annotation and 3′-UTR hexamer analysis of gene sets and were performed as previously described [41].
Chromatin immunoprecipitation (ChIP) and real-time PCR
ChIP assays were performed based on a modification of previously published methods [42]. In brief, cells (1 × 106) were treated with 1% formaldehyde for 8 min to cross-link histones to DNA. After washing with cold PBS, the cell pellets are resuspended in lysis buffer (150 mM NaCl/25 mM Tris–Cl, pH 7.5/5 mM EDTA/1% Triton X-100/0.1% SDS/0.5% sodium deoxycholate) and sonicated 7 times for 8 s. The lysate was then divided into 3 fractions. The first lysate was incubated with 10 μL of either anti-K9 acetylated histone H3 (H3K9Ac; Upstate Biotechnology, Charlottesville, VA) or anti-histone H3 (Abcam, Cambridge, UK) antibodies at 4°C overnight. The second lysate was incubated with TE buffer (10 mM Tris/1 mM EDTA, pH 8.0, 10 μL) at 4°C overnight as a negative control. The third lysate (2% of total) was used for input control. To collect the immunoprecipitated complexes, protein G-Sepharose beads (GE Healthcare, Fairfield, CT) were added and incubated for 1 h at 4°C. After washing, the beads were treated with RNase (50 μg/mL) for 30 min at 37°C and then proteinase K overnight. The cross-links were then reversed by heating the sample at 65°C for 6 h. DNA was extracted by the phenol/chloroform method, ethanol-precipitated, and resuspended in water.
For genome-wide analysis of H3K9Ac modification, the ChIP products were labeled with cy-5 (red) and input with cy-3 (green) using a random primed Klenow polymerase reaction (Invitrogen, Carlsbad, CA) at 37°C for 3 h. Labeled samples were then hybridized to the 88K human promoter array (Agilent Technologies, Santa Clara, CA) in the presence of human Cot-1 DNA for 40 h at 65°C. After washing the array according to the manufacturer's protocol, arrays were scanned on an Agilent scanner and analyzed using Feature Extraction software (Agilent Technologies). Results were analyzed using the neighborhood error model in ChIP Analytics software (version 1.1; Agilent Technologies) [43].
For qPCR analysis, the immunoprecipitated DNA was quantified by real-time PCR using Taqman approach (Applied Biosystems, Foster City, CA). Two sets of primers located at either putative core promoter or mature miR-371 regions were used for miR-371–373 cluster [44]. In addition, a PCR primer pair specific for the miR-302 region was used. To correct for differences in nucleosome density, the percentage of input that was bound to H3K9AC was normalized to those of total histone H3. Primer sequences are as follows: miR-371-Forward-5′-TCAGCCTGTGGCACTCAAAC-3′, miR-371-Reverse-5′ AGTCTTCTCAAGCGGTAACACTC-3′, miR-371 Probe-5′-TCTGCTCTCTGGTGAAAGTGCCGCC-3′, miR-371–373-Forward-5′-GTCTGACTAAGGCAAGCTAGGATC-3′, miR-371–373-Reverse-5′CCCTTCCCACCCTCTCATTCC-3′,miR-371–373 Probe-5′-CTCCGCCCCAAGCCACCCTGC-3′, miR-302-Forward-5′-GCTGTTAACATTGACATCTGTATAC-3′, miR-302-Reverse-5′ GGACTTCAGCCACTTCTATTTATAC-3′, miR-302 Probe-5′ TCCAGACCCACCCAGGATCATA CAT-3′
Karyotype analysis
G-band karyotyping was performed by the Cytogenetics Laboratory at the University of Washington.
Results
Examining early commitment of hESCs
To explore the involvement of miRNAs in hESC maintenance and early differentiation, we subjected 9 NIH-approved hESC lines to a serum-enforced differentiation scheme. Karyotypically normal hESC lines were grown in MEF feeder-free conditions and maintained in an undifferentiated state in MEF-conditioned media (CM). For induction of differentiation, hESCs were removed from conditioned medium and exposed to medium containing 20% FBS (Fig. 1A) [31]. This method was chosen in order to analyze the rapid shift from pluripotency toward multilineage differentiation events and it is in contrast to many published differentiation protocols, in which the switch from pluripotency is slow due to a reliance on removal of pluripotency maintenance conditions prior to exposure to defined growth factors.
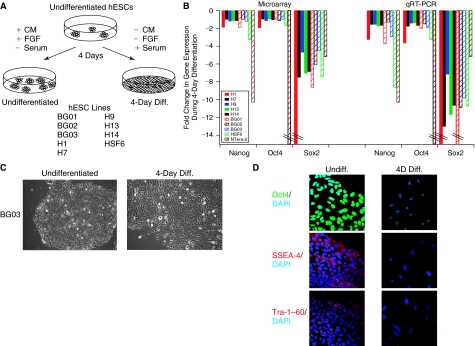
FIG. 1.
Assessing undirected differentiation in human embryonic stem cells (hESCs). (A) Undifferentiated hESCs were treated with either mouse embryonic fibroblast (MEF)-conditioned media to maintain an undifferentiated state or serum-containing media to induce undirected differentiation over a 4-day period. Total RNA was extracted from cells and used for qRT-PCR, microRNA (miRNA), and mRNA microarray analysis. (B) Stem cell marker expression levels at Day 4 of the differentiation scheme were assessed by both qRT-PCR and microarray in all undifferentiated and differentiated hESC lines. qRT-PCR analysis was performed as described in B, and both microarray and qRT-PCR data are expressed as the change in the level of gene expression during 4-day differentiation. (C) Phase-contrast microscopy was used to assess morphology of hESC colonies at Day 3 of the differentiation scheme. All images shown were captured using 10× magnification. (D) Immunofluorescent detection of the stem cell markers Oct4, SSEA-4, and Tra-1–60 demonstrates a strong reduction in stem cell markers in the differentiating H1 hESCs. Images were captured using confocal microscopy at 40× magnification.
Typical hESC differentiation protocols entail exposure of hESCs to differentiation stimuli for prolonged periods of time (>1 week). In contrast, we determined that several key stem cell markers were already significantly down-regulated on both the RNA and protein levels following 4 days of serum-induced differentiation (Fig. 1B and 1D; Supplementary Fig. 1; Supplementary materials are available online at www.liebertonline.com/scd). Immunofluorescent staining of the stem cell markers Oct4, SSEA-4, and Tra-1–60 demonstrate a strong reduction consistent with the morphological changes observed in the cells after 4-day serum-induced differentiation (Fig. 1C–1D; Supplementary Fig. 2). Microarray analyses also indicate that the 4-day serum-induced differentiation protocol leads to a concomitant induction of germ layer markers in the hESC lines used in this study (Supplementary Fig. 3).
Hierarchical clustering of miRNA expression levels reveal hESC-enriched miRNAs
To gain a full perspective on miRNA expression in hESCs, we measured global miRNA expression levels in all 9 NIH-approved hESC lines in both an undifferentiated state and early committed state. Additionally, we examined the NTera-2 embryonal carcinoma cell line, because these cells have been reported to share hESC characteristics, such as the formation of teratomas in vivo and the ability to differentiate into multiple tissue types upon differentiation in cell culture [30,45].
Microarray-based hybridizations were utilized to detect miRNA expression in the samples and the changes in miRNA expression in each cell line (log ratio of differentiated signal/undifferentiated signal) were further analyzed using unsupervised, hierarchical clustering. This analysis revealed a high degree of similarity in the changes in miRNA expression induced by the serum-induced differentiation protocol for the 9 hESC cell lines examined (Fig. 2). We observed that many of the miRNAs that are highly expressed in hESCs, including members of the miR-302 cluster and the miR-17 family, were consistently down-regulated in all lines analyzed in response to early differentiation cues, while the previously identified hESC-enriched miR-371–373 cluster was down-regulated in 6 of the 9 hESC lines (Fig. 2 and Table 1).In analyzing these miRNAs that respond most acutely to the differentiation scheme, we find only modest correlation between the hESCs and NTera-2 (Supplementary Table 1 and Supplementary Fig. 4).
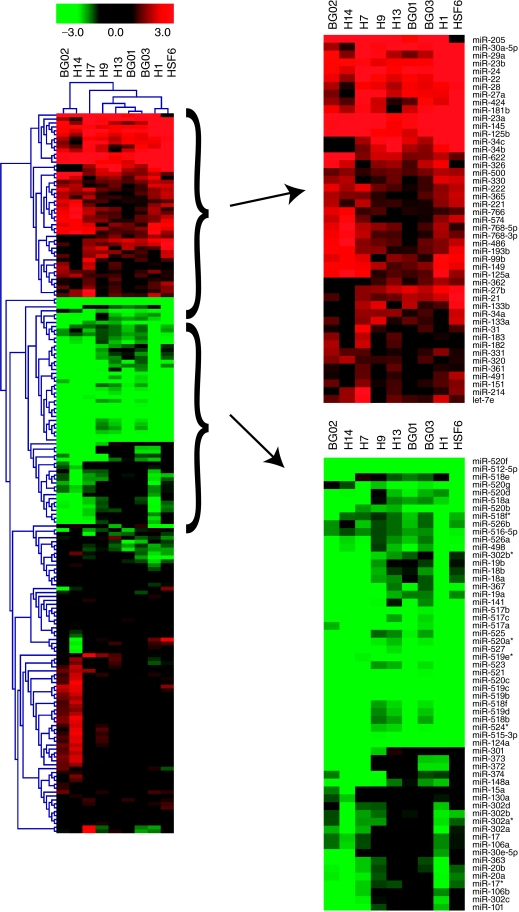
FIG. 2.
Clustering analysis of microRNA (miRNA) expression reveals human embryonic stem cell (hESC)-enriched miRNAs. Single-color, miRNA microarrays (Agilent Technologies, Santa Clara, CA) were used to analyze miRNA expression in 9 hESC lines. Of the 470 miRNAs detected by the array, 184 were consistently expressed above background levels in the samples and were included in the clustering analysis. The log2 ratio of differentiated/undifferentiated signal (fold change) for each cell line was subjected to hierarchical clustering by Euclidean distance metric using average linkage. miRNAs that are down-regulated upon differentiation are indicated by green color in the cluster and up-regulated miRNAs are indicated by red.
Table 1.
miRNAs Enriched in hESCs
| Lakshmipathy and colleagues, Stem Cells Dev (2007) [24] | Morin and colleagues, Genome Res (2008) [25] | Laurent and colleagues, Stem Cells (2008) [27] | Bar and colleagues, Stem Cells (2008) [29] | Tzur and colleagues, PLoS One (2008) [21] | Ren and colleagues, J Transl Med (2009) [28] | This study | |
|---|---|---|---|---|---|---|---|
| hESC Lines | CyT25, CyT203, HES-2, HES-3, HES-4, HUES-20, HUES-21, BG01v | H9 | H1, H9, HUES7, HUES13, HSF-6 | H1 | HES-1, HES-2 | H9,16, BG01v | H1, H7, H9, H13, H14, BG01, BG02, BG03, HSF-6 |
| Differentiation protocol | 12-day diff. to EB (CyT25 and 203, HES-2, −3, and −4 only) | 14-day diff. to EB | Diff. to extraembryonic endoderm (H9 only) | 10–14-day undirected diff. | 7–14-day directed endodermal diff. | 14-day diff. to EB | 4-day serum-induced diff. |
| Platform | Microarray | Sequencing | Microarray | Sequencing | Microarray | Microarray | Microarray |
| hESC-enriched miRNAs |
miR-302 family |
miR-302 family |
miR-302 family |
miR-302 family |
miR-302 family |
miR-302 family |
miR-302 family |
| miR-371–373 cluster | miR-92 | miR-17 family | miR-17 family | miR-17 family | miR-372 | miR-17 family | |
| miR-21 | miR-1 | miR-371–373 cluster | C19MC miRNAs | miR-520f | C19MC miRNAs | miR-371–373 cluster | |
| miR-29a | miR-21 | C19MC miRNAs | miR-135a | miR-107 | miR-141 | C19MC miRNAs | |
| miR-29c | miR-25 | miR-187 | miR-124 | miR-200c | miR-130 family | ||
| miR-143 | miR-184 | miR-324-3p | miR-130a | miR-15a | |||
| miR-154 | miR-221 | miR-766 | miR-142-3p | miR-30e-5p | |||
| miR-200c | miR-222 | miR-187 | miR-101 | ||||
| miR-222 | miR-320 | miR-200c | miR-124a | ||||
| miR-296 | miR-340 | miR-301a | miR-141 | ||||
| miR-494 | miR-423 | miR-409-3p | miR-148a | ||||
| let-7a | miR-594 | miR-423-5p | miR-374 | ||||
| miR-744 | miR-485-3p | ||||||
| let-7a | miR-654-3p | ||||||
| let-7c | let-7i | ||||||
| let-7i |
Data procured from several studies of human embryonic stem cell (hESC)-enriched miRNAs are compared with those found in this study.
Brief descriptions of the differentiation protocols, cell lines examined, and profiling platforms used in each analysis are also indicated. Members of microRNA (miRNA) families and related clusters are highlighted (miR-302 cluster—red, miR-17 family—blue, miR-371–373 cluster—green, C19MC cluster—brown).
The microarray analysis also detected the enrichment of multiple members of a recently described, large cluster of miRNAs on chromosome 19 (henceforth called C19MC for chromosome 19 miRNA cluster) in undifferentiated hESCs (Fig. 2) [27,29,46,47]. This large cluster of highly related miR-NAs is largely comprised of members of the miR-515 and miR-506 family, and was detected previously in placental tissue [46–48]. Across the undifferentiated hESC lines, we uniformly observed low levels of expression of these family members, as well as miR-498, which is also contained in the C19MC. This cluster of miRNAs also demonstrates a key discrepancy between the hESCs and NTera-2 cells, because unlike the highly expressed miRNAs from the miR-302 and miR-17 families, we did not detect significant differential expression of many miRNAs from C19MC in the NTera-2 cells (Supplementary Fig. 4). Indeed, expression of the C19MC miRNAs was extremely low in the NTera-2 cells and no members of C19MC were amongst the top 20 differentially expressed (as assessed by fold change) miRNAs in these cells (Supplementary Table 1).
In addition to the hESC-enriched miRNAs found in the miR-302 family, miR-17 family, and C19MC, we also observed that several other miRNAs not commonly described as hESC-enriched appeared to change expression rapidly in response to the loss of pluripotency. These include miRs-301, −101, −141, −148a, and −374 (Fig. 2, Table 1).Further studies will be necessary to determine the role of these miRNAs in hESCs.
Several of these newly described hESC-enriched miRNAs, as well as many members of C19MC were lowly or moderately expressed in the undifferentiated hESCs. To further validate the expression of these low to moderately expressed miR-NAs, we performed replicate miRNA microarrays on new differentiation samples, as well as miRNA qPCR analysis for specific miRNAs. Examination of some of these early-responder miRNAs from the new samples corroborated the data from the initial arrays (Supplementary Fig. 5). These data demonstrate the reproducibility of the microarray platform, as well as confirm the rapid down-regulation of these low to moderately expressed miRNAs detected upon differentiation. Additionally, the expression profiling data for the hESCs was subjected to a one-class, T-test t to determine significantly regulated genes. Reassuringly, this technique derived a nearly identical set of significantly regulated miRNAs as was observed by hierarchical clustering and analysis of fold change in gene expression (Supplementary Fig. 6).
miRNAs induced upon early commitment
In examining those miRNAs whose expression was up-regulated upon early differentiation of the ES cell lines, we again saw highly consistent results across the 9 hESCs (Fig. 2). It is important to note that our differentiation protocol does not intentionally direct the ES cells into a specific cell type or lineage, although initial induction of mesoderm and endoderm was observed (Supplementary Fig. 3). Instead, removal of feeder influence and addition of FBS to the ESC cultures produces cell types of various morphologies and each plate of cells likely contains an assortment of cell types. Therefore, this protocol does not aim to determine miRNAs critical for specific lineage pathways. Rather, the miRNAs up-regulated across multiple cell lines during differentiation may more accurately reflect a general set of miRNAs whose expression is normally suppressed in order to maintain pluripotency. In support of this idea, we find that miR-145, a miRNA that was recently identified as a repressor of several pluripotency factors in hESCs, is consistently up-regulated during our early differentiation scheme [49].
In analyzing the miRNAs whose expression was most increased during differentiation, we again found that several of these miRNAs were located in specific chromosomal clusters (miR-24/miR-27a/miR-23a) indicating that they are coordinately regulated (Fig. 2; Supplementary Table 1).Additionally, our analysis revealed that miR-21, a miRNA previously reported as being suppressed in murine ES cells, becomes highly up-regulated upon differentiation of hESCs [50]. Further analyses of these miRNAs are needed to determine if they, like miR-145, act as repressors of pluripotency or whether they act in the early mesodermal or endodermal induction observed in this differentiation paradigm.
Properties of hESC-enriched miRNAs
We compared our miRNA profiling data with several other recent hESC miRNA profiling efforts, in order to determine those miRNAs uniformly described as hESC-enriched (Table 1).From our analysis of miRNAs consistently down-regulated during early commitment of 9 hESC lines, we saw 5 distinct groups of miRNAs: miRNAs from the miR-302 cluster, miRNAs from the miR-17 family, miRNAs from the miR-371–373 cluster, miRNAs from C19MC, the miR-130 family, and a group of unrelated, nonclustered miRNAs (Fig. 2 and Table 1).Consistent with these findings, nearly all of the other recent reports observe the highest levels of differential expression in the members of the miR-302 cluster and the miR-17 family, while some also describe the miR-371–373 cluster and C19MC miRNAs as being hESC-enriched [21,24,25,27–29]. We only observed significant changes in expression of members of the miR-371–373 cluster in 6 of 9 hESC lines, possibly because of their already modest expression levels. Our study also highlights the significant, early changes in expression of members of the C19MC cluster, as well as other clustered and nonclustered miRNAs (miRs-101, −124a, −130a, −141, −148a, −301, −374), which may serve as important early-response mediators of commitment [27,29].
Strikingly, closer inspection of the hESC-enriched miRNAs found in this study reveals that their seed sequences show a high level of similarity. Table 2 displays the seed sequences (nucleotides 2–8) for members of hESC-enriched miRNA families and clusters (as defined by miRBase, release 14) found in our analysis. Importantly, numerous members of the newly identified hESC-specific C19MC miRNAs contain seed sequences that are highly similar to the seed sequences from members of the miR-302, hsa-miR-371–373, and miR-17 family, which have previously been identified as hESC-enriched [27,29]. This redundancy in seed sequence amongst the hESC-enriched miRNAs suggests that either a group of highly related miRNAs work in concert to maintain hESCs and coordinately suppress the expression of mRNAs containing similar target sequences or that subtle, nonseed sequence differences between the miRNAs are critical to their regulatory functions [27].
Table 2.
hESC-Enriched miRNA Families
| Family/miRNA | Chromosome | Seed Seq. | Family/miRNA | Chromosome | Seed Seq. |
|---|---|---|---|---|---|
| miR-302 Cluster | C19MC Cluster | ||||
| miR-302a | 4 | aagugcu | miR-512-5p | 19 | acucagc |
| miR-302a* | 4 | cuuaaac | miR-498 | 19 | uucaagc |
| miR-302b | 4 | aagugcu | miR-515-3p | 19 | agugccu |
| miR-302b* | 4 | cuuuaac | miR-519e* | 19 | ucuccaa |
| miR-302c | 4 | aagugcu | miR-520f | 19 | agugcuu |
| miR-302d | 4 | aagugcu | miR-519c | 19 | ucuagag |
| miR-367 | 4 | auugcac | miR-520a-3p | 19 | aagugcu |
| miR-526b | 19 | ucuugag | |||
| miR-519b | 19 | ucuagag | |||
| miR-371–373 Cluster | miR-525 | 19 | uccagag | ||
| miR-372 | 19 | aagugcu | miR-523 | 19 | aacgcgc |
| miR-373 | 19 | aagugcu | miR-518f | 19 | aaagcgc |
| miR-518f* | 19 | ucuagag | |||
| miR-520b | 19 | aagugcu | |||
| miR-17 Family | miR-518b | 19 | aaagcgc | ||
| miR-17 | 13 | aaagugc | miR-526a | 19 | ucuagag |
| miR-17* | 13 | cugcagu | miR-520c | 19 | ucuagag |
| miR-18a | 13 | aaggugc | miR-524-3p | 19 | aaggcgc |
| miR-19a | 13 | gugcaaa | miR-517a | 19 | ucgugca |
| miR-20a | 13 | aaagugc | miR-519d | 19 | aaagugc |
| miR-19b-1 | 13 | gugcaaa | miR-521 | 19 | acgcacu |
| miR-520d | 19 | uacaaag | |||
| miR-106a | X | aaagugc | miR-517b | 19 | cgugcau |
| miR-18b | X | aaggugc | miR-520g | 19 | caaagug |
| miR-20b | X | aaagugc | miR-518e | 19 | aagcgcu |
| miR-19b-2 | X | gugcaaa | miR-518a | 19 | ugcaaag |
| miR-363 | X | auugcac | miR-517c | 19 | ucgugca |
| miR-527 | 19 | ugcaaag | |||
| miR-106b | 7 | aaagugc | miR-516a-5p | 19 | ucucgag |
| miR-130 Family | |||||
| miR-130a | 11 | agugcaa | |||
| miR-301 | 22 | agugcaa | |||
Members of microRNA (miRNA) families enriched in human embryonic stem cells (hESCs) as determined in our microarray profiling analyses are further annotated to describe in which chromosome they reside and their seed sequence (nucleotides 2–8 of miRNA).
Highly similar seed sequences are similarly colored. All miRNA annotation data are obtained from miRBase, Release 14.0, September 2009 (http://microrna.sanger.ac.uk/).
Enrichment of hESC-specific miRNA target sequences in genes regulated during loss of pluripotency
In addition to the miRNA profiling analysis, we also examined mRNA gene expression in the differentiated and undifferentiated hESC samples using Affymetrix single-color microarrays. Other studies have examined gene expression profiles of differentiating human ES cells, but few have looked at very early time points in the differentiation process. We analyzed the ratio of undifferentiated sample to differentiated sample for each cell line and used 2-way hierarchical clustering along with the Rosetta error model to find genes that are differentially expressed [40]. To meet the specific criteria for differential gene expression between the undifferentiated and differentiated samples, genes must have at least 1.5-fold change in expression and a P value ≤0.01 in all 9 hESC lines (Fig. 3A). This analysis generated a group of 1,263 genes down-regulated (blue) and 1,509 genes up-regulated (pink) from the 9 hESC lines during differentiation.
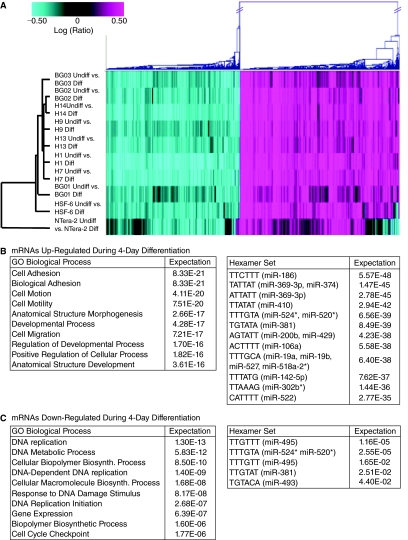
FIG. 3.
Genome-wide mRNA profiling and differential gene expression analysis in human embryonic stem cell (hESC) lines. (A) Two-way hierarchical clustering of whole genome microarray expression (Affymetryx) was used to find groups of genes whose expression changed at least 1.5-fold and had a P value <0.01 in all 9 hESCs. This analysis generated a group of 1,263 genes down-regulated (blue) and 1,509 genes up-regulated (pink). (B) The gene set of up-regulated mRNAs generated from the clustering analysis was analyzed for enrichment of biological function using gene ontology (GO) biological process functional categories and analyzed for enrichment of hexamer sequences in their 3′-UTRs. (C) The gene set of down-regulated mRNAs generated from the clustering analysis was annotated by GO biological process functional categories and analyzed for enrichment of hexamer sequences in their 3′-UTRs as described in B.
To gain a better understanding of the cellular processes that change upon differentiation, we categorized the differentially expressed genes using gene ontology (GO) biological process annotation. In the group of genes up-regulated upon differentiation (Fig. 3B), we found enrichment for genes associated with developmental pathways, locomotion, and cellular structural features, as would be expected for cells undergoing early differentiation. The possibility that some of the up-regulated genes result from specific mesodermal or endodermal differentiation cannot be excluded, since the serum-enforced differentiation showed a bias toward these germ layers (Supplementary Fig. 3). In contrast, the down-regulated genes were enriched for several cellular processes, including nucleic acid biogenesis, DNA replication, and cell cycle (Fig. 3C). This GO term enrichment likely reflects differences in metabolic processes inherent in the pluripotent ESCs as compared with differentiated, multipotent, or unipotent cell lineages.
To determine whether the 4-day differentiation protocol utilized in our study led to similar definitions of hESC-specific genes as has previously been demonstrated in hESC profiling studies, we directly compared our data with the microarray profiling data reported by Sperger and colleagues [51]. Our differential gene expression data were overlapped with the lists of hESC-specific and EC-specific genes in the Sperger and colleagues study using Fisher's exact test to generate P values for the probability of these gene sets overlapping randomly (Supplementary Fig. 7). The resultant high degree of correlation between these data sets suggests that the differentiation protocols used in this study elicit data that corroborate previous, comparative differentiation studies. Additionally, our mRNA profiling data is in agreement with previous microarray-based expression analyses of hESCs, which demonstrated that EC lines expressed a similar, but not identical, mRNA profile to hESCs (Fig. 3A) [23,51].
In an attempt to integrate the miRNA and mRNA profiling data from our studies, we used a hexamer-enrichment algorithm to search the 3′-UTR regions of the up-regulated and down-regulated gene sets [41]. Enrichment of particular hexamer sequences in a gene set may indicate regulation by miRNAs complementary to these sequences. More specifically, we looked for an intersection of miRNAs that are down-regulated upon hESC differentiation, with those genes that are up-regulated during differentiation and bear complementary sequences to the miRNAs in their 3′-UTRs, and vice versa. This analysis demonstrated that the genes down-regulated during differentiation were not significantly enriched for miRNA hexamers (Fig. 3C). Of the top 5 enriched hexamers, none corresponded to miRNAs found to be significantly up-regulated during differentiation (Fig. 2). In contrast, hexamer analysis of the 3′-UTRs of the up-regulated genes revealed enrichment for several ES-enriched miRNAs (Fig. 3B). Specifically, we found that mRNAs with 3′-UTRs bearing hexamer seed matches for miRs-374, −524*, −520a*, −106a, −19a/b, −518a*, −302b*, and −522 were significantly up-regulated amongst this gene set. This positive correlation between down-regulated miRNAs with up-regulated mRNAs containing matches for hESC-enriched miRNAs suggests that some of these miRNAs may function to down-regulate expression of genes associated with differentiation. These putative hESC-enriched miRNA/target mRNA relationships represent potential starting points for examining the roles of specific miRNAs in maintenance and differentiation of hESCs.
miRNA expression delineates separable embryonic states
We further wished to determine if the ESC-enriched miRNAs are also expressed at earlier developmental stages. In order to further delineate the precise stages of expression of the hESC-enriched miRNAs, we took advantage of a recently published method involving the use of sodium butyrate, a histone deacetylase (HDAC) inhibitor, to induce hESCs regression to an earlier developmental stage [33]. Cells treated with a low concentration of sodium butyrate, but not MEF-CM with FGF, still displayed the normal characteristics of hESCs, including the ability to self-renew and form teratomas in vivo. However, expression profiling data and the lack of X chromosome inactivation indicated that these cells were more similar to a mouse ES cell state than human ES cell state. Interestingly, murine embryonic stem cells (mESCs) are proposed to reflect an earlier stage of embryonic development (approximately embryonic day (E)3.5) than human ES cells, which are believed to be of an origin more closely related to murine epiblast stem cells (∼E5.75) [34,52]. Therefore, examination of the butyrate-treated hESCs allows for investigation of a cellular state approximating earlier embryonic cells.
To investigate the behavior of hESC-enriched miRNAs in this early stem cell state, we compared the miRNA expression profile of CM-treated, undifferentiated BG02 cells and sodium butyrate-treated BG02 cells. The log-transformed expression data was fit to a linear model and a Bayesian approach was used to determine significant differential expression between the samples (Fig. 4A). Interestingly, we find that the hESC-specific miR-371–373 cluster is highly up-regulated in the butyrate-treated cells, while several members of the miR-302 family are slightly down-regulated. To validate these findings, we performed qPCR analysis for miR-372 and miR-302b on several independent sets of CM- and butyrate-treated cells (H1, H13, BG02; Fig. 4B). Similar to the microarray analysis, the qPCR data demonstrates a dramatic up-regulation for miR-372 and a modest down-regulation for miR-302b in all 3 butyrate-treated cell lines, as assessed by ΔCt value and average fold change in expression for each set of CM- and butyrate-treated hES cell lines (Fig. 4B). To corroborate the butyrate-treatment studies of hESCs, we also tested the response of miR-372 to butyrate treatment in an ES cell line derived from monkeys, MF-1. Using qPCR, we again observed that miR-372 was up-regulated in response to butyrate in comparison with cells treated with standard ES cell media (Fig. 4C). We also found that the level of miR-302b was unaffected following butyrate treatment, further confirming our hESC findings in a nonhuman primate ESC line.
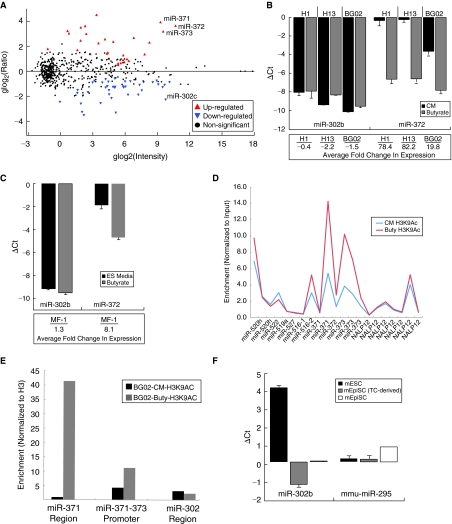
FIG. 4.
Relative expression of microRNAs (miRNAs) with similar seeds delineates separable embryonic states. (A) Microarray data for a BG02 human embryonic stem cell (hESC) line cultured in conditioned media (CM) or low-level sodium butyrate media (butyrate) as depicted by scatter plot. A Bayesian approach was utilized to determine significant genes as described in Materials and Methods, and the data are plotted as the log ratio (Buty/CM) versus the log average intensity (Buty+CM) for each miRNA gene. (B) qPCR data for the miR-302b and miR-372 are shown for hESC lines cultured in CM or butyrate media. Two independent pairs of samples are shown for both the BG02 and H13 hESC lines, as indicated by the dashed error bars. Data are represented as ΔCt (average Ct of miRNA – average Ct of RNU66 control RNA), and fold changes (butyrate/CM) in expression are indicated below each pair of samples. (C) Nonhuman primate MF-1 cells were maintained on feeder cells in ES media or exposed to sodium butyrate-containing media and qPCR analysis for miR-302b and miR-372 was performed. Data are represented as ΔCt (miRNA-RNU66 control RNA) and fold changes (butyrate/ES media) in expression are indicated below the samples. (D) ChIP assays were performed in conjunction with DNA microarray analysis to determine promoter regions enriched for acetylated histone, H3K9, a mark of active chromatin. The region of chromosome 19 containing the hsa-miR-371–373 locus is displayed and a fold enrichment of the ac-H9K3 antibody in comparison with the input fractions for both CM-treated and butyrate-treated cells are shown. (E) ChIP using an antibody specific for acetylated H3K9 was performed and subsequent qPCR for specific miR-371–373 promoter and mature miRNA regions (both miR-372 and miR-302) were performed from both CM and butyrate-treated extracts. Data are normalized to control samples obtained from ChIP with a histone H3 antibody. (F) qPCR analysis of murine mmu-miR-295 and miR-302b are shown for mESC and mEpiSC. Both naturally derived mEpiSC and chemically induced EpiSC cell lines were analyzed. The data are represented as ΔCt (miRNA-snoRNA202).
To further explore the stage-specific expression of the miR-371–373 cluster, we assessed the epigenetic state of this genomic locus by performing chromatin immunoprecipitation (ChIP) using an antibody specific for acetylated histone H3K9, a marker of active transcription. Microarray analysis in conjunction with ChIP revealed that the miR-371–373 promoters are hyperacetylated in the butyrate-treated cells, but not the CM-treated cells (Fig. 4D). Furthermore, ChIP-qPCR analysis of the mature miR-371 sequence and the miR-371–373 promoter region confirmed that this locus, but not the miR-302 locus, was specifically enriched for ace-tylated H3K9 in the butyrate-treated cells (Fig. 4E). These data support the findings that the miR-371–373 cluster is expressed at higher levels in earlier developmental stages than hESCs, and show that epigenetic modifications correlate with the stage-specific transcription of the miR-371–373 locus.
Our finding that the miR-371–373 cluster is expressed at higher levels in the butyrate-treated cells indicates that these miRNAs may play important roles at earlier stages of embryonic development than is reflected by hESCs. Interestingly, it has been shown that the murine homologs of human miR-371–373 cluster (the mmu-290–295 cluster) are highly expressed in murine ES cells, which emanate from an earlier embryonic stage than hESCs. Additionally, the miR-302 family, which is highly expressed in hESCs, is less expressed in mESCs [20,22]. In order to determine if our observed pattern of expression of these ES-enriched miRNAs in early primate stem cell populations also occurred in mouse, we examined the expression levels of mmu-miR-295 and miR-302b in mESCs and in both naturally derived murine epiblast stem cells (mEpiSC) and mEpiSCs derived in tissue culture from mESCs [34,35]. We found that mmu-miR-295 has only a subtle change in expression level between mESC and the mEpiSCs, as indicated by the ΔCt values. However, the expression level of a member of the miR-302 family, miR-302b, was considerably higher in the mEpiSCs than the mESC counterparts, in corroboration with a previous study of mouse embryos (Fig. 4F) [53]. Similar to hESCs, murine ES cells down-regulate expression of both the mmu-miR-290–295 cluster and miR-302 family upon differentiation [22,54].
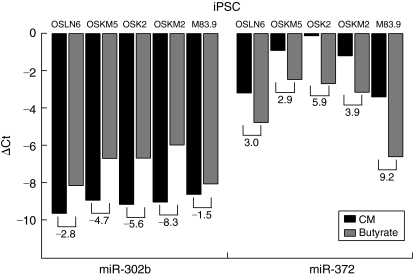
A recent report indicated that some hESC-enriched miR-NAs, including the miR-371–373 cluster, are differentially expressed in iPSCs as compared with hESCs [55]. We tested whether butyrate treatment can similarly up-regulate expression of the miR-371–373 cluster in iPSC lines. We created 5 individual iPSC lines using different cocktails of transcription factors (Supplementary Figs. 8 and 9). We then grew the iPSC lines in CM or butyrate-treated media to monitor the expression of miR-302b and miR-372. Similar to hESCs, the butyrate-treated iPS cells exhibited an increased expression of miR-372 (3–9-fold) as compared with the CM-treated cells, while miR-302b levels decreased (Fig. 5).
FIG. 5.
Similar to human embryonic stem cells (hESCs) regressed to earlier stem cell states, induced pluripotent stem cells (iPSCs) show increased miR-372 levels upon butyrate treatment. Five distinct iPSC clonal cell lines were generated and validated as described in Materials and Methods. qPCR data for the miR-302b and miR-372 are shown for these iPSC lines cultured in conditioned media (CM) or butyrate-containing media (butyrate). Data are represented as ΔCt (miRNA-RNU66 control RNA), and fold changes (butyrate/CM) in expression are indicated below each pair of samples.
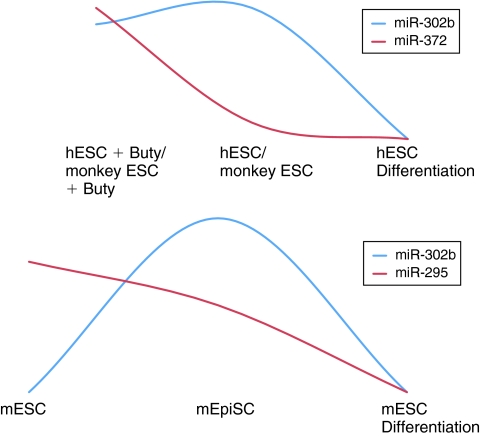
In our studies of hESC-enriched miRNAs, we consistently observe differential expression of highly similar miRNAs at separable stem cell stages. Since the miR-302 family and hsa-miR-372/mmu-miR-295 share an identical seed (Table 2), our data from primate and murine ES cells, as well as human iPSCs, imply that ES-enriched miRNAs with the same seed sequence may have critical roles at differing stages of embryonic development in both human and mice (Fig. 6).
FIG. 6.
MicroRNAs (miRNAs) with identical seed sequences are differentially expressed at separable stages of embryonic development. Summary of the observed changes in miR-302b and hsa-miR-372/mmu-miR-295 expression in early embryonic stages and in response to differentiation. The highest levels of miR-372/mmu-miR-295 are found at the earliest stages in butyrate-treated primate embryonic stem cells (ES cells) and in murine ES cells, while miR-302b levels increase at a later embryonic stage represented by primate ES cells and murine epiSCs.
Discussion
Previously, others have examined miRNA and/or mRNA expression profiles in individual or several hESC lines, while this study encompasses the largest number of NIH-approved hESC lines during early differentiation [21,23–26, 51, 56, 57]. We describe a core group of early-response miRNAs, including the miR-302 family, the miR-17 family, the miR-371–373 cluster, and a cluster of miRNAs located on chromosome 19, C19MC, as being hESC-enriched (Fig. 2 and Table 1).The C19MC cluster is restricted to primates, and heretofore, expression of the C19MC miRNAs has been detected only in placental tissue and very recently in hESCs [27,29,47,48, 58]. Although not as highly expressed as other ES-enriched miRNAs, we consistently observed expression of members of C19MC in undifferentiated hESCs but not in the differentiated cells. We also found several additional low to moderately expressed miRNAs (miRs-101, −124a, −130a, −141, −148a, −301, −374), whose expression appears to be enriched in hESCs, as these miRNA genes are down-regulated rapidly upon early differentiation. The high similarity in differential expression patterns across the hESC lines suggests that expression of a core group of miRNAs may be critical for hESC maintenance, as these 9 hESC lines represent 9 distinct individuals.
Due to the nature of the differentiation scheme used in these studies, the miRNA identified as hESC-enriched represent the early-response genes upon loss of hESC pluripotency. As such, these miRNAs are likely to regulate key differentiation-associated genes. Alternatively, these miRNAs might regulate cellular processes that are inherently different in hESCs as compared with their differentiated progeny. In accordance with this idea, a recent study found that miR-302a can regulate expression of cyclin D1 in hESCs [53]. In contrast, those miRNAs whose expression is acutely up-regulated in response to differentiation cues may be key regulators of stem cell-specific genes or processes, as has already been observed for miR-145 [49].
Intriguingly, many members of these ES-enriched miRNA families and clusters bear similar seed sequences (Table 2).The origin of these similar miRNA clusters and a determination of whether they act in a functionally redundant fashion to target the same or related mRNAs in hESCs await further analyses. It is possible that these related miRNAs act coordinately to suppress the expression of related genes involved in differentiation or in specific developmental stages of hESCs.
Finally, in order to gain a greater perspective of hESC-specific miRNAs, we sought to examine miRNA expression of an embryonic state that precedes hESCs. A previous study of mouse embryos describes differential expression of the miR-302 family at successive embryonic stages [53]. In our study, using embryonic cell lines from several species, we similarly observe that individual ES-enriched miRNAs can change expression dramatically in separate embryonic cell states. In human and monkey ES cells, we find that treatment with a low concentration of butyrate to induce an earlier embryonic state results in a large up-regulation in expression of the miR-371–373 cluster, while the highly expressed miR-302 family is largely unaffected (Fig. 4B–4E). A similar expression pattern is observed for these miRNA families and clusters when examining the mEpiSCs in comparison with their embryonic predecessors, the mESCs, which have previously been demonstrated to contain a high level of expression of the homologs of the miR-371–373 cluster (mmu-miR-290–295; Fig. 4F) [20,22]. We additionally found that butyrate treatment of human iPSCs increased the level of expression of a member of the miR-371–373 cluster, miR-372 (Fig. 5). Our results reinforce the idea that butyrate treatment of hESCs and iPSCs drives the cells toward an earlier embryonic state and demonstrate that hESC-enriched miRNAs are differentially expressed at specific embryonic stages.
Interestingly, the miRNAs from the miR-302 family and hsa-miR-371–373/mmu-miR-290–295 clusters share identical or highly similar seed sequences, as discussed earlier. Since these miRNAs show high expression at separable embryonic stages, one implication is that the “non-seed” portions of the miRNAs are likely important in influencing stage-specific regulatory functions. Alternatively, these miRNAs may have overlapping functions in embryonic stages and have differential expression caused by epigenetic changes in the local chromatin structure of these miRNAs at different embryonic stages or by stage-specific expression of transcription factors.
In collaboration with the serum-enforced differentiation studies, our hESC butyrate-treatment studies provide us with a window to observe changes in miRNA expression in developmental stages both preceding and following the ICM-derived hESC stage. As such, we were able to determine miRNAs enriched in hESCs, as well as those expressed in earlier developmental stages. As the research and clinical applications for iPSCs expand, it is critical to characterize the molecular factors responsible for stem cell plasticity or “stemness.” As important regulators of development, miR-NAs are attractive genes to codify early embryonic stages. Interestingly, a recent study demonstrated that enforced expression of some member of the murine miR-290–295 cluster and miR-302 family improved the efficiency and kinetics of iPSC formation [59]. These findings raise the possibility that hESC-enriched miRNAs could serve as important diagnostic indicators of stem cell state, and correspondingly, as indicators for their potential utility in research and clinical applications.
Supplementary Material
Acknowledgments
This work was supported by the Tietze Award for B.S. and grants from NIH for C.T.B., C.W. and H.R.B., MOD for H.R.B., and CCEH for B.S. and H.R.B. Microarray data is MIAME compliant and is stored at the GEO database (accession numbers GSE14473 and GSE14389).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Thomson JA. Itskovitz-Eldor J. Shapiro SS. Waknitz MA. Swiergiel JJ. Marshall VS. Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Evans MJ. Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 3.Klimanskaya I. Rosenthal N. Lanza R. Derive and conquer: sourcing and differentiating stem cells for therapeutic applications. Nat Rev Drug Discov. 2008;7:131–142. doi: 10.1038/nrd2403. [DOI] [PubMed] [Google Scholar]
- 4.Spagnoli FM. Hemmati-Brivanlou A. Guiding embryonic stem cells towards differentiation: lessons from molecular embryology. Curr Opin Genet Dev. 2006;16:469–475. doi: 10.1016/j.gde.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Maherali N. Sridharan R. Xie W. Utikal J. Eminli S. Arnold K. Stadtfeld M. Yachechko R. Tchieu J. Jaenisch R. Plath K. Hochedlinger K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K. Tanabe K. Ohnuki M. Narita M. Ichisaka T. Tomoda K. Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K. Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Yu J. Vodyanik MA. Smuga-Otto K. Antosiewicz-Bourget J. Frane JL. Tian S. Nie J. Jonsdottir GA. Ruotti V. Stewart R. Slukvin II. Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 9.Park IH. Zhao R. West JA. Yabuuchi A. Huo H. Ince TA. Lerou PH. Lensch MW. Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y. Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Plasterk RH. Micro RNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 12.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 13.Grimson A. Farh KK. Johnston WK. Garrett-Engele P. Lim LP. Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanellopoulou C. Muljo SA. Kung AL. Ganesan S. Drapkin R. Jenuwein T. Livingston DM. Rajewsky K. Dicerdeficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murchison EP. Partridge JF. Tam OH. Cheloufi S. Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y. Medvid R. Melton C. Jaenisch R. Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stadler BM. Ruohola-Baker H. Small RNAs: keeping stem cells in line. Cell. 2008;132:563–566. doi: 10.1016/j.cell.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sengupta S. Nie J. Wagner RJ. Yang C. Stewart R. Thomson JA. MicroRNA 92b controls the G1/S checkpoint gene p57 in human embryonic stem cells. Stem Cells. 2009;27:1524–1528. doi: 10.1002/stem.84. [DOI] [PubMed] [Google Scholar]
- 19.Qi J. Yu JY. Shcherbata HR. Mathieu J. Wang AJ. Seal S. Zhou W. Stadler BM. Bourgin D. Wang L. Nelson A. Ware C. Raymond C. Lim LP. Magnus J. Ivanovska I. Diaz R. Ball A. Cleary MA. Ruohola-Baker H. microRNAs regulate human embryonic stem cell division. Cell Cycle. 2009;8:3729–3741. doi: 10.4161/cc.8.22.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calabrese JM. Seila AC. Yeo GW. Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tzur G. Levy A. Meiri E. Barad O. Spector Y. Bentwich Z. Mizrahi L. Katzenellenbogen M. Ben-Shushan E. Reubinoff BE. Galun E. MicroRNA expression patterns and function in endodermal differentiation of human embryonic stem cells. PLoS ONE. 2008;3:e3726. doi: 10.1371/journal.pone.0003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houbaviy HB. Murray MF. Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 23.Josephson R. Ording CJ. Liu Y. Shin S. Lakshmipathy U. Toumadje A. Love B. Chesnut JD. Andrews PW. Rao MS. Auerbach JM. Qualification of embryonal carcinoma 2102Ep as a reference for human embryonic stem cell research. Stem Cells. 2007;25:437–446. doi: 10.1634/stemcells.2006-0236. [DOI] [PubMed] [Google Scholar]
- 24.Lakshmipathy U. Love B. Goff LA. Jornsten R. Graichen R. Hart RP. Chesnut JD. MicroRNA Expression Pattern of Undifferentiated and Differentiated Human Embryonic Stem Cells. Stem Cells Dev. 2007;16:1003–1016. doi: 10.1089/scd.2007.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morin RD. O'Connor MD. Griffith M. Kuchenbauer F. Delaney A. Prabhu AL. Zhao Y. McDonald H. Zeng T. Hirst M. Eaves CJ. Marra MA. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suh MR. Lee Y. Kim JY. Kim SK. Moon SH. Lee JY. Cha KY. Chung HM. Yoon HS. Moon SY. Kim VN. Kim KS. Human embryonic stem cells express a unique set of microR-NAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 27.Laurent LC. Chen J. Ulitsky I. Mueller FJ. Lu C. Shamir R. Fan JB. Loring JF. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells. 2008;26:1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- 28.Ren J. Jin P. Wang E. Marincola FM. Stroncek DF. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J Transl Med. 2009;7:20. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bar M. Wyman SK. Fritz BR. Qi J. Garg KS. Parkin RK. Kroh EM. Bendoraite A. Mitchell PS. Nelson AM. Ruzzo WL. Ware C. Radich JP. Gentleman R. Ruohola-Baker H. Tewari M. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26:2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrews PW. Retinoic acid induces neuronal differentiation of a cloned human embryonal carcinoma cell line in vitro. Dev Biol. 1984;103:285–293. doi: 10.1016/0012-1606(84)90316-6. [DOI] [PubMed] [Google Scholar]
- 31.Ware CB. Nelson AM. Blau CA. A comparison of NIH-approved human ESC lines. Stem Cells. 2006;24:2677–2684. doi: 10.1634/stemcells.2005-0452. [DOI] [PubMed] [Google Scholar]
- 32.Xu C. Inokuma MS. Denham J. Golds K. Kundu P. Gold JD. Carpenter MK. Feeder-free growth of undiff-erentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 33.Ware CB. Wang L. Mecham BH. Shen L. Nelson AM. Bar M. Lamba DA. Dauphin DS. Buckingham B. Askari B. Lim R. Tewari M. Gartler SM. Issa JP. Pavlidis P. Duan Z. Blau CA. Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell. 2009;4:359–369. doi: 10.1016/j.stem.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tesar PJ. Chenoweth JG. Brook FA. Davies TJ. Evans EP. Mack DL. Gardner RL. McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 35.Guo G. Yang J. Nichols J. Hall JS. Eyres I. Mansfield W. Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi K. Okita K. Nakagawa M. Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc. 2007;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- 37.Wang H. Ach RA. Curry B. Direct and sensitive miRNA profiling from low-input total RNA. RNA. 2007;13:151–159. doi: 10.1261/rna.234507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saeed AI. Sharov V. White J. Li J. Liang W. Bhagabati N. Braisted J. Klapa M. Currier T. Thiagarajan M. Sturn A. Snuffin M. Rezantsev A. Popov D. Ryltsov A. Kostukovich E. Borisovsky I. Liu Z. Vinsavich A. Trush V. Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 39.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 40.Weng L. Dai H. Zhan Y. He Y. Stepaniants SB. Bassett DE. Rosetta error model for gene expression analysis. Bioinformatics. 2006;22:1111–1121. doi: 10.1093/bioinformatics/btl045. [DOI] [PubMed] [Google Scholar]
- 41.Linsley PS. Schelter J. Burchard J. Kibukawa M. Martin MM. Bartz SR. Johnson JM. Cummins JM. Raymond CK. Dai H. Chau N. Cleary M. Jackson AL. Carleton M. Lim L. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kondo Y. Shen L. Yan PS. Huang TH. Issa JP. Chromatin immunoprecipitation microarrays for identification of genes silenced by histone H3 lysine 9 methylation. Proc Natl Acad Sci USA. 2004;101:7398–7403. doi: 10.1073/pnas.0306641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kondo Y. Shen L. Cheng AS. Ahmed S. Boumber Y. Charo C. Yamochi T. Urano T. Furukawa K. Kwabi-Addo B. Gold DL. Sekido Y. Huang TH. Issa JP. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 44.Houbaviy HB. Dennis L. Jaenisch R. Sharp PA. Characterization of a highly variable eutherian microRNA gene. RNA. 2005;11:1245–1257. doi: 10.1261/rna.2890305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andrews PW. Damjanov I. Simon D. Banting GS. Carlin C. Dracopoli NC. Føgh J. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab Invest. 1984;50:147–162. [PubMed] [Google Scholar]
- 46.Borchert GM. Lanier W. Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 47.Bentwich I. Avniel A. Karov Y. Aharonov R. Gilad S. Barad O. Barzilai A. Einat P. Einav U. Meiri E. Sharon E. Spector Y. Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 48.Liang Y. Ridzon D. Wong L. Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu N. Papagiannakopoulos T. Pan G. Thomson JA. Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 50.Singh SK. Kagalwala MN. Parker-Thornburg J. Adams H. Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sperger JM. Chen X. Draper JS. Antosiewicz JE. Chon CH. Jones SB. Brooks JD. Andrews PW. Brown PO. Thomson JA. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci USA. 2003;100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brons IG. Smithers LE. Trotter MW. Rugg-Gunn P. Sun B. Chuva de Sousa Lopes SM. Howlett SK. Clarkson A. Ahrlund-Richter L. Pedersen RA. Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 53.Card DA. Hebbar PB. Li L. Trotter KW. Komatsu Y. Mishina Y. Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen C. Ridzon D. Lee CT. Blake J. Sun Y. Strauss WM. Defining embryonic stem cell identity using differentiation-related microRNAs and their potential targets. Mamm Genome. 2007;18:316–327. doi: 10.1007/s00335-007-9032-6. [DOI] [PubMed] [Google Scholar]
- 55.Wilson KD. Venkatasubrahmanyam S. Jia F. Sun N. Butte AJ. Wu JC. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18:749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adewumi O. Aflatoonian B. Ahrlund-Richter L. Amit M. Andrews PW. Beighton G. Bello PA. Benvenisty N. Berry LS. Bevan S. Blum B. Brooking J. Chen KG. Choo AB. Churchill GA. Corbel M. Damjanov I. Draper JS. Dvorak P. Emanuelsson K. Fleck RA. Ford A. Gertow K. Gertsenstein M. Gokhale PJ. Hamilton RS. Hampl A. Healy LE. Hovatta O. Hyllner J. Imreh MP. Itskovitz-Eldor J. Jackson J. Johnson JL. Jones M. Kee K. King BL. Knowles BB. Lako M. Lebrin F. Mallon BS. Manning D. Mayshar Y. McKay RD. Michalska AE. Mikkola M. Mileikovsky M. Minger SL. Moore HD. Mummery CL. Nagy A. Nakatsuji N. O'Brien CM. Oh SK. Olsson C. Otonkoski T. Park KY. Passier R. Patel H. Patel M. Pedersen R. Pera MF. Piekarczyk MS. Pera RA. Reubinoff BE. Robins AJ. Rossant J. Rugg-Gunn P. Schulz TC. Semb H. Sherrer ES. Siemen H. Stacey GN. Stojkovic M. Suemori H. Szatkiewicz J. Turetsky T. Tuuri T. van den Brink S. Vintersten K. Vuoristo S. Ward D. Weaver TA. Young LA. Zhang W International Stem Cell Initiative. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 57.Ma L. Sun B. Hood L. Tian Q. Molecular profiling of stem cells. Clin Chim Acta. 2007;378:24–32. doi: 10.1016/j.cca.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 58.Cao H. Yang CS. Rana TM. Evolutionary emergence of microRNAs in human embryonic stem cells. PLoS ONE. 2008;3:e2820. doi: 10.1371/journal.pone.0002820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Judson RL. Babiarz JE. Venere M. Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.