Abstract
The eubacterial species Streptomyces coelicolor proceeds through a complex growth cycle in which morphological differentiation/development is associated with a transition from primary to secondary metabolism and the production of antibiotics. We used DNA microarrays and mutational analysis to investigate the expression of individual genes and multigene antibiotic biosynthetic pathways during these events. We identified expression patterns in biosynthetic, regulatory, and ribosomal protein genes that were associated highly specifically with particular stages of development. A knowledge-based algorithm that correlates temporal changes in expression with chromosomal position identified groups of contiguous genes expressed at discrete stages of morphological development, inferred the boundaries of known antibiotic synthesis gene loci, and revealed novel physical clusters of coordinately regulated genes. Microarray analysis of RNA from cells mutated in genes regulating synthesis of the antibiotics actinorhodin (Act) and undecylprodigiosin (Red) identified proximate and distant sites that contain putative ABC transporter and two-component system genes expressed coordinately with genes of specific biosynthetic pathways and indicated the existence of two functionally and physically discrete regulons in the Red pathway.
Keywords: Microarray, antibiotic, pathway, cluster, regulation, transcription
Members of the Gram-positive eubacterial genus Streptomyces initiate vegetative growth as a tangle of multinucleate substrate mycelia. Later, aerial mycelia extend upward and develop cross walls that become boundary lines for spores; during this process, a biochemical transition from primary to secondary metabolism occurs (Chater 1993). The production of these compounds and other secondary metabolites used in medicine and agriculture generally coincides with the onset of morphological differentiation in surface-grown cultures (Chater and Bibb 1997). Most antibiotics are produced by complex biosynthetic pathways encoded by physically clustered genes located contiguously on the chromosome. These clusters usually contain pathway-specific transcriptional activators that are themselves subject to control by pleiotropic regulatory genes (Hopwood et al. 1995; Kieser et al. 2000).
Streptomyces coelicolor A3(2) has been used extensively for the study of morphological and physiological development and for investigation of the genetic control of antibiotic production (Hopwood et al. 1995). The chemically diverse antibiotics made by S. coelicolor A3(2) include the red-pigmented tripyrrole undecylprodigiosin (Red), the lipopeptide calcium-dependent antibiotic (CDA), and the deep blue pigmented polyketide actinorhodin (Act), whose color is responsible for the S. coelicolor name (Hopwood 1999).
Most of what is known currently about the regulation of antibiotic biosynthetic pathways has been gleaned through investigations of individual genes in specific pathways. However, the availability of the 8.7 Mb S. coelicolor chromosomal DNA sequence from the Sanger Centre (http://www.sanger.ac.uk/Projects/S_coelicolor/), and the development of efficient methods for genome-wide analysis of expression profiles using DNA microarrays (DeRisi et al. 1997) enabled us to simultaneously and globally assess factors that affect the transcription of Streptomyces genes and regulatory pathways. We used DNA microarrays, together with mutations in S. coelicolor genes, to identify alterations in gene expression that occur during S. coelicolor growth, and particularly during the transition from primary to secondary metabolism, and to elucidate the regulation of key antibiotic biosynthetic pathways in S. coelicolor. Analysis of microarray results using knowledge-based algorithms has facilitated delineation of the physical boundaries of biosynthetic clusters, defined chromosomally dispersed groupings of coordinately regulated genes, and identified novel regulons.
Results
Global gene expression during the growth of S. coelicolor M145
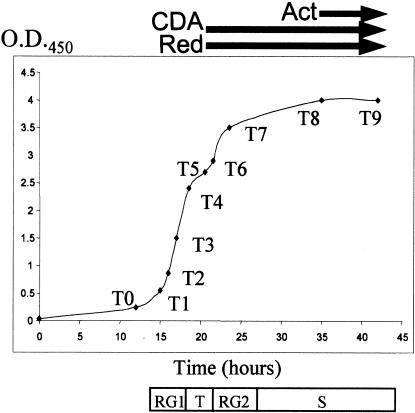
Pregerminated spores from wild-type S. coelicolor strain M145 were synchronized and inoculated into modified liquid R5 medium, where four distinct stages of growth were observed (Fig. 1), as reported previously (Leskiw et al. 1993; Puglia et al. 1995; Vohradsky et al. 2000). The antibiotics CDA and Red were first detected during T phase, whereas Act production was not detectable until S phase (Fig. 1). RNA isolated prior to logarithmic cell growth (timepoint 0), was used as template for synthesis of Cy3-labeled cDNA; this was mixed with Cy5-labeled cDNA corresponding to RNA from other timepoints and hybridized with PCR-amplified sequences corresponding to 4960 known genes or putative open reading frames (ORFs) of S. coelicolor A3(2) M145 arrayed on glass slides. The fluorescence intensity ratio of these two dyes in individual spots was analyzed by a hierarchical clustering algorithm (Eisen et al. 1998) and/or by supervised learning algorithms that create gene groupings according to their ability to satisfy specified conditions (Genetic Analysis By Rules Incorporating Expert Logic [GABRIEL]; K.-H. Pan, C.-J. Lih, and S.N. Cohen, in prep.).
Figure 1.
Growth of Streptomyces coelicolor A3(2) strain M145 in liquid R5− medium. Arrows indicate when antibiotics were detected in cultures; T0–T9 indicates times (timepoints 0 to 9) when cultures were harvested for isolation of total RNA. Stages of growth were defined by change in the rate of increase in cell density at OD450. An initial period of rapid growth (RG1) beginning ∼14 h after inoculation of liquid cultures was followed by a brief growth slowdown starting ∼18.5 h after inoculation (transition [T] phase). After ∼2–3 h, cultures briefly resumed another rapid growth phase (RG2) before entering stationary (S) phase.
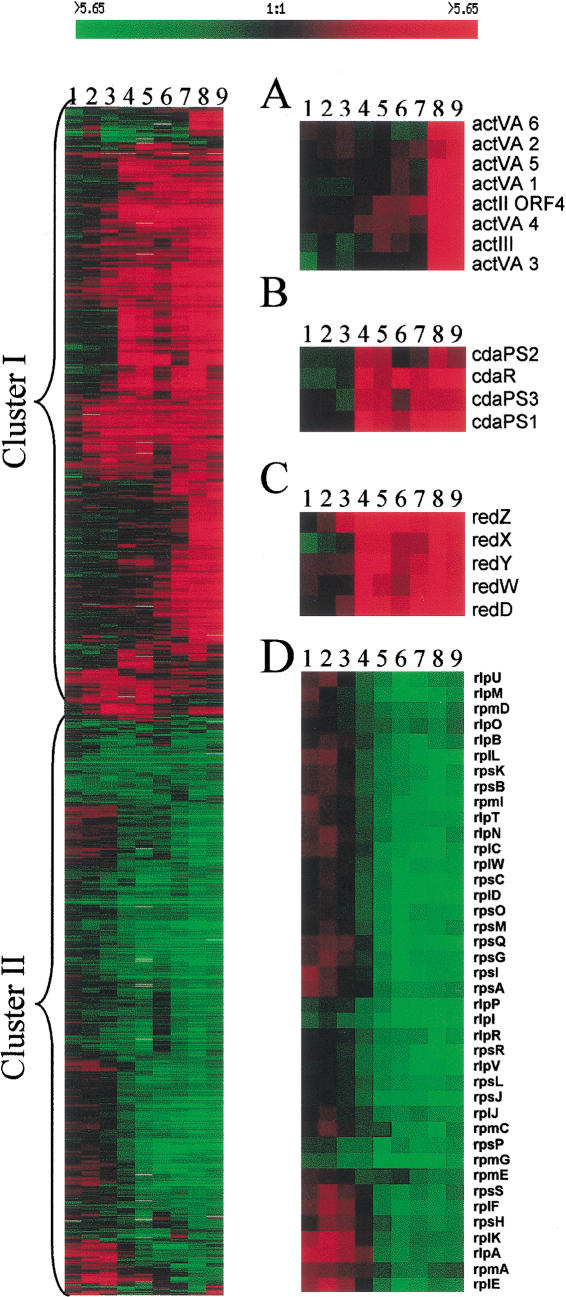
Approximately 1100 genes showed a two-fold or greater change in expression at two or more time points; the expression profiles generated by hierarchical clustering (Fig. 2, left) indicated that transcription of ∼200 of these genes was significantly increased or decreased during vegetative growth (timepoints 1–3) whereas others showed altered transcription during the transition stage (timepoints 4 and 5) or in posttransition samples (timepoints 6–9). Almost equal numbers of genes were up- or down-regulated in the posttransition period and these formed two large expression clusters (Fig. 2, left, cluster I and II). Most known genes in antibiotic biosynthetic pathways and some genes associated with morphological or physiological differentiation were up-regulated during or after the transition from primary to secondary metabolism (Fig. 2A–C, cluster I), whereas genes encoding ribosomal proteins, factors involved in translation initiation or peptide elongation, and tRNA synthetases were down-regulated (Fig. 2D, cluster II).
Figure 2.
Expression profiles of genes differentially expressed during growth of Streptomyces coelicolor A3(2). Profiles indicate the extent of gene expression relative to timepoint 0. Genes were clustered hierarchically (Eisen et al. 1998) according to similarity in the expression profile. Consistency in the amount of cDNA used for analysis at different timepoints was inferred from our finding that 80% of the genes analyzed showed no change in expression during cell growth and that the remaining 20% was equally divided into up-regulated and down-regulated genes. Normalization thus assumed that the total amount of RNA present in samples obtained at different timepoints was sufficiently constant to preclude effects on gene groupings. cDNA ratios are represented in tabular form according to the color scale shown at top; rows correspond to individual genes and columns correspond to successive timepoints. Red shades represent an increase and green shades represent a decrease in hybridizing cDNA. Black indicates no detectable change in transcript level and gray represents the absence of data. (A,B,C) Expression profiles of named members of the Act, CDA, and Red biosynthetic pathways, respectively, that were present as targets on arrays. (D) Expression profile of ribosomal protein genes. The results shown are from one of two duplicate experiments that yielded similar gene expression profiles. Lanes 1–9, represent timepoints 1 to 9 from Figure 1.
Approximately 15% of the annotated known or putative regulatory genes on the DNA microarrays we analyzed showed altered transcription during S. coelicolor growth. Not surprisingly, genes whose transcription changed prominently included those encoding transcriptional regulatory proteins, two-component regulators, kinases, and sigma factors, (e.g., glkA, whiB, afsQ2, and cprB [down-regulated] and abaA, afsS, absA2, sigE, bldN, whiK/bldM, and ramR [up-regulated]) (Fig. 3).
Figure 3.
Some known and putative regulatory genes showing growth-phase dependent expression. Data analysis and format of presentation are as described in Figure 2.
Delineation of biosynthetic gene clusters by expressional and positional analysis
Transcripts of known genes in the Red (redD,X,W,Y and redZ) and CDA (cdaPS1,2,3 and cdaR) pathways increased in abundance immediately prior to the appearance of these antibiotics in the culture medium (Fig. 1, timepoint 4; Fig. 2B,C). RNA encoded by redZ, which has been proposed as the initiator of Red pathway activity (White and Bibb 1997) was elevated slightly earlier than transcripts of other Red pathway genes and prior to emergence of cells from exponential growth. Elevation of RNA encoded by redD, a redZ-dependent, pathway-specific secondary regulator (Takano et al. 1992; White and Bibb 1997) and of previously named Red biosynthetic genes (redX,W,Y) followed. In contrast, Act pathway gene transcripts did not show a major increase under our experimental conditions until stationary phase (Fig. 2A), which was found previously by S1 mapping to be associated with elevated transcription of the Act pathway regulator, actII–ORF4, and the actIII biosynthetic gene (Strauch et al. 1991; Floriano and Bibb 1996).
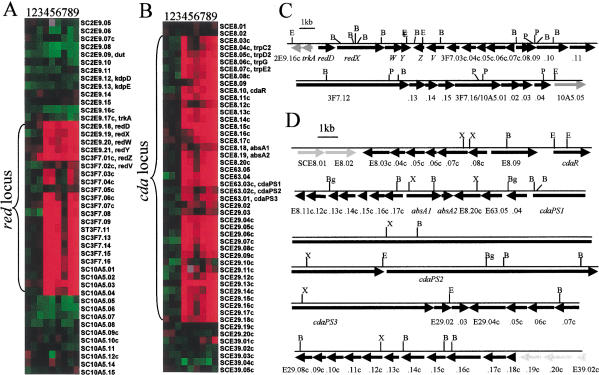
Functionally related genes commonly are arranged contiguously on the chromosomes of bacteria (Blattner et al. 1997; Cole et al. 1998). In Streptomyces, genes encoding secondary metabolites, especially antibiotic biosynthetic pathways, are clustered together on the chromosome with pathway-specific regulatory genes (Feitelson et al. 1985; Martin and Liras 1989; Fernandez-Moreno et al. 1991). The frequent occurrence of functionally related Streptomyces genes in physical contiguity enabled us to use microarray expression profiles to discover and delineate groupings of genes having potentially related functions and to infer the boundaries of previously identified biosynthetic pathways. These tasks were accomplished by a GABRIEL algorithm that relates the profile at various timepoints to chromosomal position by examining the expression profiles of contiguous genes (Fig. 4A,B). Using this approach, we identified dispersed loci that contain sets of coordinately regulated contiguous genes (i.e., SCJ1.11∼SCJ1.30c, SCJ12.10c∼SCJ12.32 and SCF62.07∼SCF62.28c; http://www.sanger.ac.uk/Projects/S_coelicolor/; see website http://sncohenlab.stanford.edu/streptomyces for other groupings). We suggest that the components of these and other similarly identified loci, some of which appear from sequence analysis to be comprised of polycistronic operons, may carry out related functions.
Figure 4.
Chromosomal boundaries of red and cda gene clusters. (A,B) Profiles of genes and open reading frames (ORFs) present in the chromosome regions associated with production of the antibiotics undecylprodigiosin and CDA, respectively. The order of genes listed reflects position on the chromosome. Contiguous genes whose expression is coordinately regulated throughout growth as determined by GABRIEL proband-based and continuity/gap rules are indicated as red and cda clusters, and these are displayed on linear maps (C, red) and (D, cda) constructed from sequence data available from the Sanger Centre and previous studies (Malpartida et al. 1990; White and Bibb 1997; Chong et al. 1998; Anderson et al. 2001). Arrows indicate the extent and direction of putative ORFs. Numbers are listed at some locations without cosmid designations, which are identified in A and B. Black arrows indicate ORFs of antibiotic locus genes; gray arrows indicate nonlocus genes. Cleavage sites for restriction endonucleases are designated (B, BamHI; Bg, BglII; E, EcoRI; P, PstI; X, XhoI). The Sanger Centre designations SC3F.16 and SC10A5.01 refer to a single putative red ORF. SC3F.10 and SC3F.12 were not present as targets in these arrays and their presence in the red locus is inferred. Lanes 1–9, represent timepoints 1 to 9 from Figure 1. The unavailability of sequence information for genes surrounding Act genes at the time of preparation of arrays used in this analysis precluded the use of GABRIEL to define Act cluster boundaries, as was done for Red and CDA.
Temporal coordination of expression of unnamed ORFs present on a 35.7-kb DNA fragment that encodes the undecylprodigiosin pathway (Malpartida et al. 1990) with expression of known Red pathway genes (i.e., redD,X,W,Y and redZ) identified other ORFs on this fragment as components of the red locus and suggested that the locus extends for ∼33 kb through 23 putative ORFs (Fig. 4A,C); this conclusion, which was supported by the effects of redD mutations (Fig. 6I, see below), recently has been deduced independently by other methods of analysis (Cerdeno et al. 2001). Transcription of trkA, which is included in the 35.7-kb S. coelicolor DNA fragment that confers Red function, was not coordinated temporally with expression of known red genes (Fig. 4A,C), implying that it is not a red locus component (cf., Cerdeno et al. 2001). Similar analysis suggests that the cda locus, which previously was not fully defined, extends for ∼83 kb from SCE8.03c to SCE29.18c and includes 40 putative ORFs (Fig. 4B,D). Interestingly, transcripts of most constituents of both the cda and red loci were transiently, but distinctly, decreased during the RG2 phase, suggesting that growth rate per se may affect their expression.
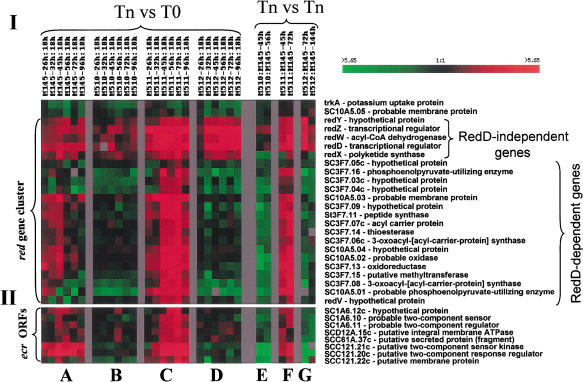
Figure 6.
Effects of mutations on red genes and ecr open reading frames (ORFs). Array experiments and data analysis were carried out as indicated in Figure 5 (I). Effects of mutations on transcription profiles of contiguous ORFs in the chromosomal DNA region containing the red locus. SC3F.10 and SC3F.12 were not present as targets in these arrays and their presence in the red locus is inferred (Fig. 4). (II). Effects of mutations on transcription profiles of ecr ORFs. GenBank accession numbers for ecr ORFs are AF425991–AF425994.
Microarray analysis of mutants in the Act or Red pathways
The pathway-specific regulatory genes redD and actII–ORF4 play pivotal roles in the biosynthesis of Red and Act, respectively (Bibb 1996), and ActII–ORF4 has been shown by transcriptional analysis and DNase I footprinting assays to interact with intergenic sites identified in the act locus (Arias et al. 1999). To identify possible additional genes regulated by redD and actII–ORF4—both within and external to their respective biosynthetic loci—we analyzed transcription in bacteria mutated in these genes (M510, ΔredD; M511, ΔactII–ORF4]; and M512, ΔredD ΔactII–ORF4) (Floriano and Bibb 1996) using solid media to allow cells to fully differentiate morphologically and proceed to sporulation. We compared gene expression at timepoint 0 (T0) with expression during various stages of growth (Tn versus T0, Figs. 5I and 6, panels A–D) and mutant versus wild-type expression at various time points (Tn versus Tn, Figs. 5I and 6, panels E–G) using 6740 targets amplified from M145 chromosomal DNA. Patterns of gene expression were analyzed by both hierarchical clustering and GABRIEL.
Figure 5.
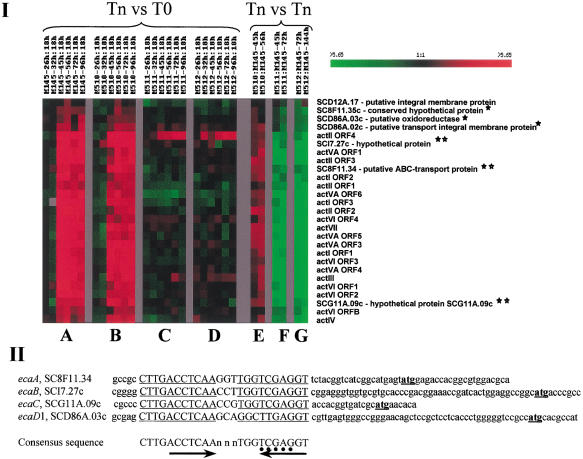
Distant genes coregulated with Act locus during growth on solid media. (I) Effects of mutations. cDNAs derived from RNA isolated from M145 (wt), M510 (ΔredD), M511 (ΔactII–ORF4), and M512 (ΔredD ΔactII–ORF4) (Floriano and Bibb 1996) at the indicated timepoints (Tn) during growth on solid media were compared. T0 RNA was isolated 18 h after inoculation (timepoint 0). Tn vs. Tn indicates comparisons between mutant and wild-type cells harvested at the same timepoint. T0 cDNA was labeled with Cy3 (green) dye and compared with Tn cDNA labeled with Cy5 (red) dye (A–D). In Tn vs. Tn experiments, cDNA from mutants was labeled with Cy5 and from M145 cells with Cy3 (E–G). Data were analyzed by hierarchical clustering (HC) (Eisen et al. 1998) and knowledge-based GABRIEL algorithms (K.-H. Pan, C.-J. Lili, and S.N. Cohen, in prep.). For HC, genes were ordered according to the extent of similarity of expression at each timepoint and groups were delineated based on the resulting dendrogram. GABRIEL analyses employed a proband-based algorithm that tested the correlation of expression for various genes using a modified zero-based Pearson correlation coefficient. Rows correspond to individual genes and columns to different experiments, as indicated. The change in transcript abundance for each gene is displayed by means of a color scale, in which color saturation represents the magnitude of the difference. Black color indicates no detectable change in transcript level and gray represents the absence of data. Asterisks indicate eca open reading frames (ORFs). Double asterisks indicate >90% correlation with Act locus expression. (II). Sequences 5′ to ORFs of eca genes. The underlined capital letters indicate a consensus sequence identified 5′ to the putative ORFs of eca genes and show its distance relative to the proposed translational start site (underlined lowercase letters). Arrowheads indicate inverted repeats within each sequence. The dots indicate a possible ActII–ORF4 binding site (Arias et al. 1999). GenBank accession numbers for eca ORFs are AF425987–AF425990.
The transcription profile of act locus genes (Fig. 5I) showed that genes in this biosynthetic pathway were tightly coordinated temporally, and on solid media were up-regulated 2- to 17-fold at later stages of cell growth, as had been observed also in liquid media. In actII–ORF4 mutants, transcripts of Act biosynthetic genes failed to increase during stationary phase (Fig. 5I, panels C,D,F,G). However, transcripts of actII–ORF4 itself, which contained an in-frame deletion, responded to growth stage control, indicating that the upstream signals that turn on the Act biosynthetic pathway are intact.
Identification of distant ORFs whose expression is temporally coordinated with Act locus genes
Expression of six putative ORFs located at distant sites was highly coordinated with act gene expression (correlation coefficient >0.8) (Fig. 5I). These ORFs are here named ecaA-E (expression coordinated with act). ecaD1 and ecaD2 are adjacent ORFs in the same orientation and may belong to a single operon. ecaA and ecaE are also adjacent to each other but are transcribed convergently according to their Sanger Centre ORF assignments. Expression of ecaA, B, and C had a correlation coefficient with known Act pathway genes of >0.9 and the regions 5′ to these ORFs, as well as to ecaD1, showed a consensus sequence that contains the inverted repeat, CTTGACCTCAAnnnTGGTCGAGGT (Fig. 5II) located 14 to 48 nucleotides 5′ to the ORF. Within the consensus sequence is 5′-TCGAG-3′, which was shown by DNase I footprinting assays to be an act locus binding site for the Act II–ORF4 protein (Arias et al. 1999). The protein-coding sequences of ecaD1 and ecaD2 suggest that they may function as an oxidoreductase and a transport integral membrane protein respectively (Sanger Centre annotation), whereas ecaA is suggested to be an ATP-binding cassette (ABC) transporter. ecaB, ecaC, and ecaE are hypothetical proteins.
Identification of red-associated genes by transcriptional analysis
The effects of a redD mutation on expression of the physically contiguous genes in the red locus defined two distinct subgroups (Fig. 6I). Elevated transcription of one subgroup during and following the transition required the function of redD, which earlier work has shown is a pathway-specific transcriptional activator (Narva and Feitelson 1990). However, transcripts of redZ, redD, and redX,W,Y were not suppressed in redD mutants. Transcription of RedD-independent and RedD-dependent constituents of the red locus was enhanced in bacteria mutated in actII–ORF4 (M511; Fig. 6I, panels C,F) and RedD-independent transcription was increased also in the redD/actII–ORF4 double mutant strain (M512; Fig. 6I, panels D,G). Transcription of trkA and SC10A5.05 was not elevated during growth of wild-type or actII–ORF4 mutants on solid modified R5 media, consistent with our conclusion that these ORFs are not components of the red locus.
We found several ecr (expression coordinated with red; shown in Fig. 6II) ORFs that mapped at a distance from the red locus. Among these were ecrA1/A2 (SCC121.21c/20c) and ecrE1/E2 (SCIA6.10/11), which encode respectively the sensor kinases and regulators of putative two-component systems, and ecrB (SCC121.22c), which encodes a putative membrane protein (Fig. 6II). Other ORFs whose transcription was coordinated with red locus expression are ecrC (SCD12A.15c) and ecrD (SCC61A.37c), which respectively are proposed to encode an integral membrane ATPase and a secreted protein of unknown function, and ecrF (SC1A6.12c), which is a hypothetical protein.
Discussion
Antibiotic production in Streptomyces species generally is dependent on growth phase and involves the expression of physically clustered regulatory and biosynthetic genes. Our data show that antibiotic pathway genes are coordinately regulated at the level of transcription during S. coelicolor growth, and this mode of regulation has helped to identify the physical boundaries of biosynthetic loci. By combining microarray data with information obtained by mapping, annotation, and sequencing of the S. coelicolor chromosome, we inferred the extent of known antibiotic biosynthetic clusters and also discovered new groupings of physically contiguous and coordinately expressed genes that may have related functions. Additionally, our mutational analysis identified chromosomally dispersed sets of ORFs whose expression was coordinated temporally with expression of known antibiotic biosynthetic pathway genes and which appear to be regulated in common with these genes (for example, the eca ORFs). The ability to identify such ORFs now provides a tool for investigating possible mechanisms responsible for coordinated expression, including coexpression under control of a pleiotropic regulator, regulation by gene(s) in the biosynthetic pathway, and indirect effects of antibiotic production.
Expression of eca and ecr ORFs was generally coordinated with the expression of genes in the Act and Red pathways in liquid media as well as on solid media; however, some variation in the precise timing of expression occurred between certain eca or ecr ORFs and the respective biosynthetic pathway genes. Some dispersed ORFs coregulated with Act shared, 5′ to the predicted ORF, a common sequence that includes a stretch (TCGAG) identical to one proposed as a DNA binding site for ActII–ORF4 in the act locus, suggesting a possible mechanism by which ActII–ORF4 may coregulate eca ORFs and Act biosynthetic genes. No sequence commonality among the ecr ORFs was identified.
One of the eca ORFs, ecaA, encodes a putative ABC transporter. Several previously identified ABC transporters of Streptomyces spp. maintain self-resistance to, or accomplish the secretion of, antibiotics produced by those species (Linton et al. 1994; Barrasa et al. 1995; Olano et al. 1995; Fernandez et al. 1996; Ikeno et al. 2000), and can be found outside the biosynthetic cluster encoding the antibiotic (Rodriguez et al. 1993; Aguirrezabalaga et al. 2000). Whether ecaA has a role in Act resistance or the secretion of actinorhodin has not been determined.
Earlier results have suggested that undecylprodigiosin production is controlled in S. coelicolor by a linear pathway in which the autoregulated redZ gene activates another positive regulatory gene, redD, which in turn promotes the transcription of Red biosynthetic genes (White and Bibb 1997; Guthrie et al. 1998). Consistent with the previously determined relationship of redZ to redD, our microarray data show that transcription of these genes is independent of RedD function. Surprisingly, however, we also found that the transcription of redX,W,Y also does not depend on RedD function, whereas other genes within the Red biosynthetic locus (from redV to SC10A5.04) are RedD-dependent. Northern blotting experiments we have recently carried out indicate that transcription of redX,W,Y does not occur in the redZ deletion strain, M550 (data not shown). Consistent with the notion that these genes may be controlled directly by redZ is evidence that redD overexpression in M550 (ΔredZ) does not restore the production of undecylprodiosin (White and Bibb 1997).
Genetic studies have mapped the locus encoding the lipopeptide CDA to a region later deduced to correspond to an ∼88 kb region of the S. coelicolor chromosome (Hopwood and Wright 1983; Chong et al. 1998). An ∼35-kb segment of this region has been shown be essential for biosynthesis of CDA (Chong et al. 1998); however, the full extent of the CDA locus has not been defined previously. Our analysis of transcription of genes within the CDA contig, which includes four overlapping cosmids sequenced by the Sanger Centre, has identified a continuous stretch of the S. coelicolor chromosome whose expression is temporally coordinated with known CDA genes (Fig. 4B). This stretch includes the region suggested to contain the CDA locus. Sequence-derived information about the putative function of ORFs in this region supports our inference from microarray analysis that the CDA locus extends for ∼83 kb and includes 40 ORFs, from ORF SCE8.03c to SCE29.18c. We find that expression of previously unidentified genes within this region shows a high degree of temporal correlation with known CDA genes (see website, http://sncohenlab.stanford.edu/streptomyces); however, the correlation coefficient drops sharply to <0.2 for SC8.02 and SCE29.19, which bracket the continuous stretch that our data suggest comprises the CDA locus. The presence within the region encompassed by the CDA locus of absA1 and absA2, which encode proteins of a two-component regulatory system and which modulate the biosynthesis of Act and Red in addition to CDA biosynthesis, has also been observed recently by others (Anderson et al. 2001).
Whereas transcription of genes encoding antibiotic biosynthetic pathways increased during and following the transition from primary to secondary metabolism, genes encoding ribosomal proteins were globally down-regulated (Fig. 2D). Two dimensional gel electrophoresis has shown that production of ribosomal proteins in S. coelicolor varies with the phase of cell growth (Blanco et al. 1994). Evidence that activity of the promoter for the rplJL operon, which encodes ribosomal proteins L10 and L7/L12, is growth-phase dependent (Blanco et al. 2001), provides a possible mechanistic basis for the observed decrease in ribosomal proteins and transcripts.
Elevation of transcripts encoding some sigma factors and other pleiotropic regulatory proteins during the later stages of S. coelicolor growth did not require redD or actII–ORF4, but in some instances was affected by mutation of these genes. The pleiotropic regulatory protein afsR2/afsS (Vogtli et al. 1994; Matsumoto 1995) was more abundant in cells mutated in the actII–ORF4 gene (data shown on website, http://sncohenlab.stanford.edu/streptomyces), implying that the Act biosynthetic pathway, which is regulated by afsR2/afsS (Kim et al. 2001) may exercise feedback control over transcription of this multipathway regulator. Additionally, Red pathway RNAs were more abundant in cells blocked in the Act pathway (actII–ORF4 mutant, M511), as compared to wild-type cells (Fig. 6I, panel F); this differential expression was observed at the time that Act pathway genes normally are up-regulated. Expression of four of the eight ORFs that comprise the whiE locus also showed a greater increase in posttransition redD mutant cells than in wild-type bacteria (data shown on website). Such findings are consistent with the notion that pathways of antibiotic production and spore pigment production in S. coelicolor are competitive or may be balanced by an interactive regulatory network.
Materials and methods
Strains, growth conditions, and assay of antibiotics
S. coelicolor A3(2), wild-type strain M145 and in-frame deletion mutants M510 (ΔredD), M511 (ΔactII–ORF4), and M512 (ΔredD ΔactII–ORF4) (Floriano and Bibb 1996) were used in these studies. Fresh spores were collected and pregerminated as described previously (Kieser et al. 2000), and then inoculated into modified R5 liquid medium (R5−), pH7.2, lacking additional KH2PO4, CaCl2, and L-proline. CaCl2 was specifically omitted as we found that calcium can delay onset of Act production in M145 and reduce the yield of Red and Act. 6% PEG 8000 was added to enhance cell dispersal. Medium was filter-sterilized and antifoam 289 was added (0.05% v/v) before use. Mycelia were collected for RNA isolation at the zero timepoint when OD450 was 0.25.
In preliminary experiments using various solid media we found that growth, antibiotic production, and sporulation occurred most synchronously on modified R5 medium and used cellophane membranes placed on R5− plates for RNA isolation from surface grown cells (Aceti and Champness 1998).
Act and Red were detected spectrophotometrically (Kieser et al. 2000). CDA production on solid media was assayed as described previously (Anderson et al. 2001); for liquid cultures, small filter paper discs containing droplets of supernatant taken from cultures at different timepoints were used to assay inhibition of growth of Staphylococcus aureus.
Construction and use of S. coelicolor microarrays
DNA sequences from 5000 to 7100 S. coelicolor M145 ORFs (ftp://ftp.sanger.ac.uk/pub/S_coelicolor/sequences/) were used to design primer pairs that amplified 50- to 2400-bp internal fragments of putative ORFs (protocol details are available at http://sncohenlab.stanford.edu/streptomyces). Primer design was automated using PRIMER 3 (Rozen and Skaletsky 2000) and primers were grouped according to their melting temperatures. To reduce the selection as targets of redundant ORFs having different names and sequences present in multiple ORFs, we carried out a BLAST search using the e-12 as the cut-off value.
HotstartTaq and a standard protocol (QIAGEN) were used for PCR amplification and products were verified by gel electrophoresis and printed onto glass slides coated with poly-L-lysine (details about protocol used are available at http://cmgm.stanford.edu/pbrown/mguide/index.html). Microarrays used to analyze expression during liquid growth contained PCR products for the 4960 putative ORFs available at the time these arrays were prepared; slides used for mutational analysis and other experiment involving growth on solid media contained 6740 putative ORFs. About 10% of ORFs on arrays were duplicates or overlapping sequences. Total RNA was isolated and purified by the modified Kirby mix procedure as described (Kieser et al. 2000) and used as template for incorporation of fluorescent nucleotide analogs during a first-strand reverse transcription reaction. Ten to fifteen micrograms of total RNA was denatured in the presence of 5–6 μg of 72%-GC-content hexamers (total 13 μL) at 75°C for 10– 15 min and snap-cooled in ice water before addition of the remaining reaction components: 3 μL of Cy3–dCTP or Cy5–dCTP (Amersham Pharmacia Biotech) and a 14 μL of cocktail that included 6 μL 5× superscriptII Buffer, 3 μL of DTT (0.1 M), 3 μL of dNTP (4 mM dATP, 4 mM dTTP, 10 mM dGTP, and 0.5 mM dCTP), and 2 μL of SuperscriptII (GIBCO BRL). The reverse transcriptase reaction was carried out for 10 min at 25°C followed by 2 h at 42°C. After labeling, purification of cDNA, hybridization, and washing were as described previously (Wilson et al. 1999; DeRisi et al. 1997).
Data analysis
Analysis of data was performed with the software available at http://genome-www4.stanford.edu/MicroArray/SMD/restech.html and http://rana.stanford.edu or by the GABRIEL program (http://gabriel.stanford.edu), a knowledge-based machine learning software system (K-H. Pan, C-J. Lih, and S.N. Cohen, in prep.). Identification of ORFs coregulated with genes of antibiotic biosynthetic pathways were identified by a GABRIEL proband-based rule that classifies ORFs according to the extent of similarity of expression to a proband pattern or a composite pattern representing multiple probands. Boundaries of coordinately expressed loci were identified using a GABRIEL continuity/gap rule that determines whether correlation between expression of each succeeding gene along the chromosome exceeds a defined threshold, which in these studies involved a correlation coefficient >0.75. Random permutation of values was used to calculate the false discovery rate due to random occurrence, and signal-to-noise ratio analysis determined the statistical significance of alterations in expression.
Acknowledgments
These investigations were supported by grant AI08619 from the NIAID and by DARPA grant MDA972-00-1-0032 to SNC. We acknowledge the contributions to this project of the Streptomyces Genome Sequencing Project carried out in the U.K. at the Sanger Centre and the John Innes Centre. We thank J. Lam for technical assistance, D. Hopwood, M. Bibb, C. Kao, W. Champness, J. Ryding, J. Bernstein, K. Bao, C. Miller, and Y. Lu for strains, helpful discussions, and advice.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sncohen@stanford.edu; FAX (650) 725-1536.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.943401.
References
- Aceti DJ, Champness WC. Transcriptional regulation of Streptomyces coelicolor pathway-specific antibiotic regulators by the absA and absB loci. J Bacteriol. 1998;180:3100–3106. doi: 10.1128/jb.180.12.3100-3106.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirrezabalaga I, Olano C, Allende N, Rodriguez L, Brana AF, Mendez C, Salas JA. Identification and expression of genes involved in biosynthesis of L-oleandrose and its intermediate L-olivose in the oleandomycin producer Streptomyces antibioticus. Antimicrob Agents Chemother. 2000;44:1266–1275. doi: 10.1128/aac.44.5.1266-1275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson T, Brian P, Champness W. Genetic and transcriptional analysis of absA, an antibiotic gene cluster-linked two-component system that regulates multiple antibiotics in Streptomyces coelicolor. Mol Microbiol. 2001;39:553–566. doi: 10.1046/j.1365-2958.2001.02240.x. [DOI] [PubMed] [Google Scholar]
- Arias P, Fernandez-Moreno MA, Malpartida F. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J Bacteriol. 1999;181:6958–6968. doi: 10.1128/jb.181.22.6958-6968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrasa MI, Tercero JA, Lacalle RA, Jimenez A. The ard1 gene from Streptomyces capreolus encodes a polypeptide of the ABC-transporters superfamily which confers resistance to the aminonucleoside antibiotic A201A. Eur J Biochem. 1995;228:562–569. doi: 10.1111/j.1432-1033.1995.tb20295.x. [DOI] [PubMed] [Google Scholar]
- Bibb M. 1995 Colworth Prize Lecture. The regulation of antibiotic production in Streptomyces coelicolor A3(2) Microbiology. 1996;142:1335–1344. doi: 10.1099/13500872-142-6-1335. [DOI] [PubMed] [Google Scholar]
- Blanco G, Rodicio MR, Puglia AM, Mendez C, Thompson CJ, Salas JA. Synthesis of ribosomal proteins during growth of Streptomyces coelicolor. Mol Microbiol. 1994;12:375–385. doi: 10.1111/j.1365-2958.1994.tb01027.x. [DOI] [PubMed] [Google Scholar]
- Blanco G, Sanchez C, Rodicio MR, Mendez C, Salas JA. Identification of a growth phase-dependent promoter in the rplJL operon of Streptomyces coelicolor A3(2) Biochim Biophys Acta. 2001;1517:243–249. doi: 10.1016/s0167-4781(00)00280-3. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Cerdeno AM, Bibb MJ, Challis GL. Analysis of the prodiginine biosynthesis gene cluster of Streptomyces coelicolor A3(2): New mechanisms for chain initiation and termination in modular multienzymes. Chem Biol. 2001;8:817–829. doi: 10.1016/s1074-5521(01)00054-0. [DOI] [PubMed] [Google Scholar]
- Chater KF. Genetics of differentiation in Streptomyces. Annu Rev Microbiol. 1993;47:685–713. doi: 10.1146/annurev.mi.47.100193.003345. [DOI] [PubMed] [Google Scholar]
- Chater KF, Bibb MJ. Regulation of bacterial antibiotic production. Weinheim, Germany: VCH; 1997. [Google Scholar]
- Chong PP, Podmore SM, Kieser HM, Redenbach M, Turgay K, Marahiel M, Hopwood DA, Smith CP. Physical identification of a chromosomal locus encoding biosynthetic genes for the lipopeptide calcium-dependent antibiotic (CDA) of Streptomyces coelicolor A3(2) Microbiology. 1998;144:193–199. doi: 10.1099/00221287-144-1-193. [DOI] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feitelson JS, Malpartida F, Hopwood DA. Genetic and biochemical characterization of the red gene cluster of Streptomyces coelicolor A3(2) J Gen Microbiol. 1985;131:2431–2441. doi: 10.1099/00221287-131-9-2431. [DOI] [PubMed] [Google Scholar]
- Fernandez E, Lombo F, Mendez C, Salas JA. An ABC transporter is essential for resistance to the antitumor agent mithramycin in the producer Streptomyces argillaceus. Mol Gen Genet. 1996;251:692–698. doi: 10.1007/BF02174118. [DOI] [PubMed] [Google Scholar]
- Fernandez-Moreno MA, Caballero JL, Hopwood DA, Malpartida F. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell. 1991;66:769–780. doi: 10.1016/0092-8674(91)90120-n. [DOI] [PubMed] [Google Scholar]
- Floriano B, Bibb M. afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2) Mol Microbiol. 1996;21:385–396. doi: 10.1046/j.1365-2958.1996.6491364.x. [DOI] [PubMed] [Google Scholar]
- Guthrie EP, Flaxman CS, White J, Hodgson DA, Bibb MJ, Chater KF. A response-regulator-like activator of antibiotic synthesis from Streptomyces coelicolor A3(2) with an amino-terminal domain that lacks a phosphorylation pocket. Microbiology. 1998;144:727–738. doi: 10.1099/00221287-144-3-727. [DOI] [PubMed] [Google Scholar]
- Hopwood DA. Forty years of genetics with Streptomyces: From in vivo through in vitro to in silico. Microbiology. 1999;145:2183–2202. doi: 10.1099/00221287-145-9-2183. [DOI] [PubMed] [Google Scholar]
- Hopwood DA, Wright HM. CDA is a new chromosomally-determined antibiotic from Streptomyces coelicolor A3(2) J Gen Microbiol. 1983;129:3575–3579. doi: 10.1099/00221287-129-12-3575. [DOI] [PubMed] [Google Scholar]
- Hopwood DA, Chater KF, Bibb MJ. Genetics of antibiotic production in Streptomyces coelicolor A3(2), a model streptomycete. Biotechnology. 1995;28:65–102. doi: 10.1016/b978-0-7506-9095-9.50009-5. [DOI] [PubMed] [Google Scholar]
- Ikeno S, Yamane Y, Ohishi Y, Kinoshita N, Hamada M, Tsuchiya KS, Hori M. ABC transporter genes, kasKLM, responsible for self-resistance of a kasugamycin producer strain. J Antibiot (Tokyo) 2000;53:373–384. doi: 10.7164/antibiotics.53.373. [DOI] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Chater KF, Hopwood DA. Practical Streptomyces genetics. Norwich, United Kingdom: John Innes Foundation; 2000. [Google Scholar]
- Kim ES, Hong HJ, Choi CY, Cohen SN. Modulation of actinorhodin biosynthesis in Streptomyces lividans by glucose repression of afsR2 gene transcription. J Bacteriol. 2001;183:2198–2203. doi: 10.1128/JB.183.7.2198-2203.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskiw BK, Mah R, Lawlor EJ, Chater KF. Accumulation of bldA-specified tRNA is temporally regulated in Streptomyces coelicolor A3(2) J Bacteriol. 1993;175:1995–2005. doi: 10.1128/jb.175.7.1995-2005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton KJ, Cooper HN, Hunter IS, Leadlay PF. An ABC-transporter from Streptomyces longisporoflavus confers resistance to the polyether-ionophore antibiotic tetronasin. Mol Microbiol. 1994;11:777–785. doi: 10.1111/j.1365-2958.1994.tb00355.x. [DOI] [PubMed] [Google Scholar]
- Malpartida F, Niemi J, Navarrete R, Hopwood DA. Cloning and expression in a heterologous host of the complete set of genes for biosynthesis of the Streptomyces coelicolor antibiotic undecylprodigiosin. Gene. 1990;93:91–99. doi: 10.1016/0378-1119(90)90141-d. [DOI] [PubMed] [Google Scholar]
- Martin JF, Liras P. Enzymes involved in penicillin, cephalosporin and cephamycin biosynthesis. Adv Biochem Eng Biotechnol. 1989;39:153–187. doi: 10.1007/BFb0051954. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Ishizukz H, Beppu T, Horinouchi S. Involvement of a small ORF downstream of the afsR gene in the regulation of secondary metabolism in Streptomyces coelicolor A3(2) Actinomycetologica. 1995;9:37–43. [Google Scholar]
- Narva KE, Feitelson JS. Nucleotide sequence and transcriptional analysis of the redD locus of Streptomyces coelicolor A3(2) J Bacteriol. 1990;172:326–333. doi: 10.1128/jb.172.1.326-333.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olano C, Rodriguez AM, Mendez C, Salas JA. A second ABC transporter is involved in oleandomycin resistance and its secretion by Streptomyces antibioticus. Mol Microbiol. 1995;16:333–343. doi: 10.1111/j.1365-2958.1995.tb02305.x. [DOI] [PubMed] [Google Scholar]
- Puglia AM, Vohradsky J, Thompson CJ. Developmental control of the heat-shock stress regulon in Streptomyces coelicolor. Mol Microbiol. 1995;17:737–746. doi: 10.1111/j.1365-2958.1995.mmi_17040737.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez AM, Olano C, Vilches C, Mendez C, Salas JA. Streptomyces antibioticus contains at least three oleandomycin-resistance determinants, one of which shows similarity with proteins of the ABC-transporter superfamily. Mol Microbiol. 1993;8:571–582. doi: 10.1111/j.1365-2958.1993.tb01601.x. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Strauch E, Takano E, Baylis HA, Bibb MJ. The stringent response in Streptomyces coelicolor A3(2) Mol Microbiol. 1991;5:289–298. doi: 10.1111/j.1365-2958.1991.tb02109.x. [DOI] [PubMed] [Google Scholar]
- Takano E, Gramajo HC, Strauch E, Andres N, White J, Bibb MJ. Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase-dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2) Mol Microbiol. 1992;6:2797–2804. doi: 10.1111/j.1365-2958.1992.tb01459.x. [DOI] [PubMed] [Google Scholar]
- Vogtli M, Chang PC, Cohen SN. afsR2: A previously undetected gene encoding a 63-amino-acid protein that stimulates antibiotic production in Streptomyces lividans. Mol Microbiol. 1994;14:643–653. doi: 10.1111/j.1365-2958.1994.tb01303.x. [DOI] [PubMed] [Google Scholar]
- Vohradsky J, Li XM, Dale G, Folcher M, Nguyen L, Viollier PH, Thompson CJ. Developmental control of stress stimulons in Streptomyces coelicolor revealed by statistical analyses of global gene expression patterns. J Bacteriol. 2000;182:4979–4986. doi: 10.1128/jb.182.17.4979-4986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Bibb M. bldA dependence of undecylprodigiosin production in Streptomyces coelicolor A3(2) involves a pathway-specific regulatory cascade. J Bacteriol. 1997;179:627–633. doi: 10.1128/jb.179.3.627-633.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M, DeRisi J, Kristensen HH, Imboden P, Rane S, Brown PO, Schoolnik GK. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc Natl Acad Sci. 1999;96:12833–12838. doi: 10.1073/pnas.96.22.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]