Abstract
Salt and drought stress signal transduction consists of ionic and osmotic homeostasis signaling pathways, detoxification (i.e., damage control and repair) response pathways, and pathways for growth regulation. The ionic aspect of salt stress is signaled via the SOS pathway where a calcium-responsive SOS3-SOS2 protein kinase complex controls the expression and activity of ion transporters such as SOS1. Osmotic stress activates several protein kinases including mitogen-activated kinases, which may mediate osmotic homeostasis and/or detoxification responses. A number of phospholipid systems are activated by osmotic stress, generating a diverse array of messenger molecules, some of which may function upstream of the osmotic stress–activated protein kinases. Abscisic acid biosynthesis is regulated by osmotic stress at multiple steps. Both ABA-dependent and -independent osmotic stress signaling first modify constitutively expressed transcription factors, leading to the expression of early response transcriptional activators, which then activate downstream stress tolerance effector genes.
Keywords: osmotic stress, abscisic acid, SOS, protein kinase, phospholipid, calcium signaling, gene expression
INTRODUCTION
Modern-day plants are products of eons of evolution from primal living organisms in response to abiotic and biotic environmental changes. Among the abiotic factors that have shaped and continue shaping plant evolution, water availability is the most important. Water stress in its broadest sense encompasses both drought and salt stress. Because cell signaling controls plant responses and adaptation, it is probably not an exaggeration to state that water stress signaling has in large part shaped the flora on earth. Drought and salt stress, together with low temperature, are the major problems for agriculture because these adverse environmental factors prevent plants from realizing their full genetic potential.
Salt stress afflicts plant agriculture in many parts of the world, particularly irrigated land (16a). Compared to salt stress, the problem of drought is even more pervasive and economically damaging (5a). In this regard, drought stress signaling certainly merits separate treatment. Nevertheless, most studies on water stress signaling have focused on salt stress primarily because plant responses to salt and drought are closely related and the mechanisms overlap. From a practical point, salt stress can be imposed more easily and precisely in laboratory settings. Although the importance of salt and drought stress signaling was recognized long ago, few molecular components were known until recently. As such, salt and drought stress signaling was reviewed only as part of salt and drought stress tolerance (24, 31), and it has not been reviewed as a separate subject in this series.
This review focuses on general cell-based salt and drought stress signaling. Long-distance signaling within a plant and even interplant signaling are important for plant adaptation, but little mechanistic information is available. In drought stress responses, guard cell signaling is of critical importance because it is a key denominator within the plant water budget. Much effort has been justifiably dedicated to guard cell signaling and substantial advances have been made. Several excellent reviews are devoted to this subject, including Schroeder et al. (85) and Leung & Giraudat (51). Despite the fact that guard cells are specialized structures, most of what is learned there is probably applicable to other cells as well. Therefore, some of the knowledge of general cell signaling presented here has roots in guard cell studies.
INPUTS AND OUTPUTS: MAKING SENSE OF SALT AND DROUGHT STRESS SIGNALING PATHWAYS
Salt and drought stresses affect virtually every aspect of plant physiology and metabolism. Numerous changes that occur under these stresses have been documented. Although some of the changes are clearly adaptive, many may simply be pathological consequences of stress injury. True, there may be active signaling to activate pathological responses. Knowledge of them is just as important because they represent suitable targets for genetic suppression to improve salt and drought stress tolerance. In nature, for a plant to sacrifice a part of its structure constitutes an adaptive strategy to survive a stress episode.
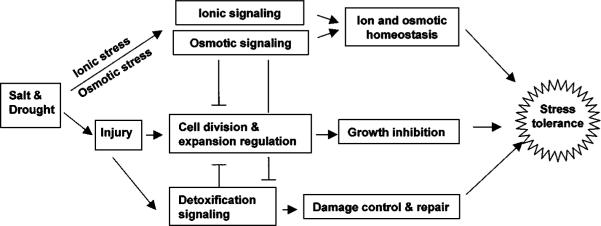
For adaptive or presumed adaptive responses, it may be helpful to conceptually group them into three aspects: (a) homeostasis that includes ion homeostasis, which is mainly relevant to salt stress, and osmotic homeostasis or osmotic adjustment; (b) stress damage control and repair, or detoxification; and (c) growth control (119). Accordingly, salt and drought stress signaling can be divided into three functional categories: ionic and osmotic stress signaling for the reestablishment of cellular homeostasis under stress conditions, detoxification signaling to control and repair stress damages, and signaling to coordinate cell division and expansion to levels suitable for the particular stress conditions (Figure 1). Homeostasis signaling negatively regulates detoxification responses because, once cellular homeostasis is reestablished, stress injury would be reduced, and failure to reestablish homeostasis would aggravate stress injury. Homeostasis and detoxification signaling lead to stress tolerance and are expected to negatively regulate the growth inhibition response, i.e., to relieve growth inhibition.
Figure 1.
Functional demarcation of salt and drought stress signaling pathways. The inputs for ionic and osmotic signaling pathways are ionic (excess Na+) and osmotic (e.g., turgor) changes. The output of ionic and osmotic signaling is cellular and plant homeostasis. Direct input signals for detoxification signaling are derived stresses (i.e., injury), and the signaling output is damage control and repair (e.g., activation of dehydration tolerance genes). Interactions between the homeostasis, growth regulation, and detoxification pathways are indicated.
It is not enough to know only the components or elements of signaling pathways (113). Good comprehension requires knowledge of precise inputs and outputs of the pathways. It is here that water stress signaling is most poorly understood. Just because certain changes occur upon drought stress treatment, one cannot assume that drought is the direct input signal, nor can one assume that the output is any of the adaptive responses. For the ionic aspect of salt stress, a signaling pathway based on the SOS (Salt Overly Sensitive) genes has been established. The input of the SOS pathway is likely excess intracellular or extracelluar Na+, which somehow triggers a cytoplasmic Ca2+ signal (117). The outputs are expression and activity changes of transporters for ions such as Na+, K+, and H+. The input for osmotic stress signaling is likely a change in turgor. Several osmotic stress–activated, SOS-independent protein kinases probably mediate osmotic stress signaling, but no output is known (119). Possible outputs of osmotic signaling pathways include gene expression and/or activation of osmolyte biosynthesis enzymes as well as water and osmolyte transport systems. Most of the other changes induced by salt or drought stress can be considered as part of detoxification signaling. These include (a) phospholipid hydrolysis; (b) changes in the expression of LEA/dehydrin-type genes, molecular chaperones, and proteinases that remove denatured proteins; and (c) activation of enzymes involved in the generation and removal of reactive oxygen species and other detoxification proteins. The input signal(s) for the detoxification pathways is mostly likely not an ionic or osmotic change, but a product of stress injury, e.g., reactive oxygen species or protein denaturation.
Water stress generally inhibits plant growth, and indirect evidence suggests active signaling to cell division and expansion machineries (119). Slower cell division under water stress is probably a result of reduced cyclin-dependent kinase (CDK) activity (87). Reduced CDK activity may be a result of combined effects of transcription suppression of cyclins and CDKs (7) and induction of CDK inhibitors (106). The direct input signal(s) for the CDK regulation is unclear but can be a product of stress injury or any of the primary or intermediary signals involved in the homeostasis and detoxification pathways.
THE SOS REGULATORY PATHWAY FOR ION HOMEOSTASIS AND SALT TOLERANCE
Although a number of possible pathways for salt, drought, or cold signaling have been proposed, none is established in terms of signaling proteins and inputs and outputs. One exception is the SOS pathway that emerged recently as a result of genetic, molecular, and biochemical analysis (117). An outline of this pathway is shown in Figure 2. Salt stress elicits a cytosolic calcium signal (42). How the calcium signal is different from that triggered by drought, cold, or other stimuli remains a mystery. A myristoylated calcium-binding protein encoded by SOS3 presumably senses the salt-elicited calcium signal and translates it to downstream responses (32, 55). SOS3 interacts with and activates SOS2, a serine/threonine protein kinase (23, 53). SOS2 and SOS3 regulate the expression level of SOS1, a salt tolerance effector gene encoding a plasma membrane Na+/H+ antiporter (92). More importantly, SOS2 and SOS3 are required for the activation of SOS1 transport activity (78). SOS1 by itself can slightly increase the salt tolerance of a yeast mutant strain lacking all endogenous Na+-ATPases and Na+/H+ antiporters (93). Coexpression of SOS3 and SOS2, together with SOS1, can dramatically enhance the salt tolerance of the yeast mutant (J. M. Pardo & J-K. Zhu, unpublished information). Expression of a constitutively activated SOS2 mutant could also increase the salt tolerance activity of SOS1 in the yeast mutant, implying that SOS2 kinase activity is sufficient for SOS1 activation. In a complementary study, Qiu et al. (78) showed that constitutively active SOS2 kinase could enhance a Na+/H+ exchange activity in purified plasma membrane vesicles from wild-type but not sos1-1 mutant (107) plants. In sos2-2 and sos3-1 mutants (121), the plasma membrane Na+/H+ exchange activity is much lower but can be recovered to near wild-type levels by addition of activated SOS2 in vitro to the membrane vesicle preparations (78).
Figure 2.
Regulation of ion (e.g., Na+ and K+) homeostasis by the SOS pathway. High Na+ stress initiates a calcium signal that activates the SOS3-SOS2 protein kinase complex, which then stimulates the Na+/H+ exchange activity of SOS1 and regulates transcriptionally and posttranscriptionally the expression of some genes. SOS3-SOS2 may also stimulate or suppress the activities of other transporters involved in ion homeostasis under salt stress, such as vacuolar H+-ATPases and pyrophosphatases (PPase), vacuolar Na+/H+ exchanger (NHX), and plasma membrane K+ and Na+ transporters.
SOS3 belongs to a novel subfamily of EF-hand-type calcium-binding proteins (21). These proteins share high sequence identities with the B-subunit of calcineurin (type 2B protein phosphatase) and with animal neuronal calcium sensors (21, 55). They are predicted to have three EF-hands and bind to calcium with low affinity compared to calmodulin or caltractin (32; Y. Guo & J-K. Zhu, unpublished information). Only some members of the protein family contain an N-terminal myristoylation motif. SOS3 is myristoylated, which may help target SOS3 and its interacting proteins (e.g., SOS2) to membranes where their target transporters are located (e.g., SOS1). Nevertheless, not all of the SOS3 protein in plant cells is associated with membranes (32), consistent with the presence of nonmembrane-bound protein targets of the SOS3-SOS2 complex. Besides a role in salt tolerance through an interaction with SOS2, SOS3 may also interact with other proteins including SOS2-like proteins (i.e., PKS) (21) to mediate osmotic stress induction of ABA biosynthesis (L. Xiong & J-K. Zhu, unpublished information). SOS3-like proteins (SCaBPs) interact specifically with certain PKS proteins, forming distinct protein kinase complexes that likely mediate calcium signaling in response to other stimuli (21).
SOS2 also represents a novel family of proteins (PKS) so far only found in plants (21). SOS2 contains an SNF1-like catalytic domain and a unique regulatory domain that interacts with SOS3 (23, 53). The regulatory and catalytic domains of SOS2 interact to keep the kinase inactive in substrate phosphorylation, presumably by preventing substrate access to the catalytic site (21). SOS3 binding to the regulatory domain appears to disrupt the intramolecular interaction of SOS2, thus opening up the catalytic site. Serial deletion analysis identified a 21 amino acid sequence (i.e., FISL motif) in the SOS2 regulatory domain, which is necessary and sufficient for SOS3 binding. Deletion of the entire SOS2 regulatory domain (21) or simply of the FISL motif (Y. Guo & J-K. Zhu, unpublished information) results in constitutive activation of the kinase. Expression of the constitutively active SOS2 under the CaMV 35S promoter in sos2 or sos3 mutant plants can rescue the salt sensitive phenotype in the mutant shoot but, curiously, not in the root (Y. Guo & J-K. Zhu, unpublished information), suggesting that SOS2 kinase activity is sufficient for the SOS pathway in salt tolerance at least in the shoot.
The plasma membrane Na+/H+ antiporter SOS1 has a very long tail that is predicted to be on the cytoplasmic side (92). Membrane transporters with long cytoplasmic tails have been proposed to function as sensors of the solutes they transport. For example, evidence suggests that the glucose transporters Snf3 and Rgt2 in yeast can serve as glucose sensors and regulate gene expression in response to glucose starvation (75). The possibility of SOS1 being both a transporter and a sensor cannot be dismissed. If it is a Na+ sensor, it may control SOS2 activation in plants, forming a regulatory loop because an activated SOS2 would stimulate its capacity for Na+ efflux. Future determination of SOS2 kinase activity in response to salt treatment in wild-type and sos1 mutants may prove or disprove this speculation. Besides being regulated by SOS2, SOS1 activity may also be regulated by SOS4. SOS4 catalyzes the formation of pyridoxal-5-phosphate, a cofactor that may serve as a ligand for SOS1 because the latter contains a putative binding sequence for this cofactor (94; H. Shi & J-K. Zhu, unpublished information).
It will not be surprising if SOS3 and SOS2 regulate Na+ influx as well as vacuolar compartmentation systems because these systems are also vital for salt tolerance. Mutations in the AtHKT1 gene suppressed sos3 mutant phenotypes (81). AtHKT1 mediates Na+ influx in oocytes and yeast (103). Ion content analysis in the sos3hkt1 mutant supports that AtHKT1 mediates Na+ influx in planta (81). It is possible that SOS3 and SOS2 may downregulate AtHKT1 activity under salt stress. Nonselective cation channels contribute to Na+ influx (2, 101). Cyclic nucleotide–gated ion channels (CNGCs) are a group of nonselective cation channels that are inhibited by calcium and calmodulin. When expressed in heterologous systems (50), CNGC2 was activated by cyclic nucleotides and but did not show Na+ permeability. However, patch clamping of Arabidopsis root cells showed that nonselective cation channels were inhibited by cAMP or cGMP in vivo (59). Arabidopsis seedling growth on 100 mM NaCl was improved by inclusion of cGMP and to a lesser extent by cAMP. Consistent with an active role of cyclic nucleotides in salt tolerance, cGMP-treated plants had less Na+ accumulation (59). Some CNGCs may also mediate calcium influx and thus are potentially upstream of SOS3 if they can generate cytosolic calcium transients under salt stress.
PROTEIN KINASE PATHWAYS FOR OSMOTIC STRESS SIGNALING
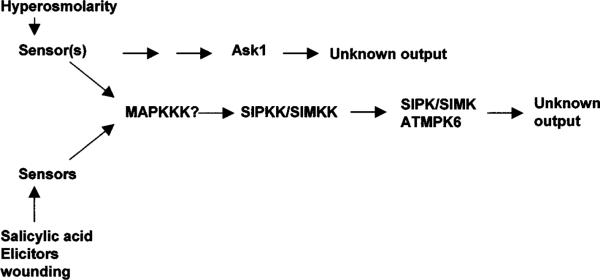
Protein phosphorylation is such a central theme in cell signaling that its involvement in osmotic stress adaptation was predicted a while ago. Only recently, though, has direct experimental evidence been reported (118). In fact, several plant protein kinases have now been found to be activated by osmotic stress. Because of the well-known osmosensing pathway in yeast (22), much attention has been directed toward identifying a homologous pathway in plants. The yeast osmoregulatory pathways begin with either an Src-homology 3 (SH3) domain-containing membrane protein or a two-component histidine kinase, which then activates a MAP kinase cascade and leads to increased osmolyte synthesis and accumulation (22). Although plants accumulate compatible osmolytes for osmotic adjustment, whether they use similar membrane sensors and MAP kinase cascades to regulate osmolyte synthesis is unclear. Recent studies using in-gel kinase assays identified several protein kinases that are activated by osmotic stress in plants, but whether any of the pathways is directly activated by osmotic stress or by a derived stress needs to be addressed (Figure 3). In addition, a central question remains as to the outputs of the pathways, i.e., whether they regulate osmolyte biosynthesis or other stress responses.
Figure 3.
Activation of protein kinases by hyperosmotic stress. The MAP kinase cascade shown is also activated by other stresses. Currently, the functional significance of the kinase activation is unclear (hence the “unknown output”). SIPK, SIMK, and ATMPK6 are homologous MAP kinases from tobacco, alfalfa, and Arabidopsis, respectively.
Plants have several MAP kinases that are activated by hyperosmotic stress (Figure 3). In alfalfa cells, a 46-kD MAP kinase named SIMK became activated in response to moderate hyperosmotic stress (67). It is interesting to note that at severe hyperosmotic stress (>750 mM NaCl) SIMK was no longer activated. Instead, a smaller kinase became activated, suggesting that the two kinases function at different stress levels. In tobacco cells, an SIMK-like MAP kinase named SIPK (salicylic acid–induced protein kinase) was activated by hyperosmotic stress (64). In addition to hyperosmotic stress, hypoosmotic stress, salicylic acid, or fungal elicitors also could activate the tobacco MAP kinase (13). An Arabidopsis protein crossreacting with antibodies against the tobacco MAP kinase was found to be activated by hyperosmotic stress (29). A MAP kinase kinase that interacts with and activates the alfalfa SIMK was reported (41). A tobacco MAP kinase kinase that interacts with SIPK was also found (57). The enormous complexity of MAP kinases can be seen in a study that showed that at least three MAP kinases in Arabidopsis were enzymatically activated by salt as well as by cold, wounding, and other environmental signals (30). The picture is equally complicated in other plants. For example, other MAP kinases, e.g., WIPK in tobacco and SAMK in alfalfa, are activated by cold, drought, wounding, and biotic signals (37, 89).
In cultured tobacco cells, a 42-kD protein kinase is activated rapidly in response to hyperosmotic stress treatment (64). Partial peptide sequences from this kinase suggest that it is an ortholog of Arabidopsis ASK1, a SNF1-related kinase without a known function. In Arabidopsis seedlings, a 40-kD kinase was also identified as being rapidly activated by salt or sorbitol stress in a calcium- and ABA-independent manner (29). Although the amino acid sequence of this 40-kD Arabidopsis protein is not known, it is probably the homolog of the 42-kD kinase of tobacco. Osmotic stress activation of the Arabidopsis 40-kD protein kinase was independent of the SOS pathway because it was not affected by the sos3 mutation. As these 40–42-kD plant kinases are clearly not of the MAP kinase type, their activation may represent a novel osmotic stress signaling pathway in plants (Figure 3).
Because osmotic stress elicits calcium signaling (42), calcium-dependent protein kinases are prime candidates that link the calcium signal to downstream responses. In a maize protoplast transient expression system, a constitutively active CDPK (calcium-dependent protein kinase) mutant activated the expression of a reporter gene that was normally responsive to osmotic stress, cold, or ABA (91). A dominant negative form of the CDPK was able to block stress or ABA induction of the reporter gene. If protein kinase specificity is maintained in the protoplast transient assay system, then the finding would be very significant in that it links calcium signaling with gene induction by osmotic stress.
Transcript levels for a number of protein kinases including a two-component histidine kinase, MAPKKK, MAPKK, and MAPK increase in response to osmotic and other stress treatments (65). It is unclear whether the protein or, more importantly, the activity levels of these kinases change upon osmotic stress treatment. For these and the osmotic stress–activated kinases discussed above, it is vital to identify the input and output of the kinases and of the pathways. The input signal could be osmotic stress (e.g., turgor changes) or derived from osmotic stress injury. The output could be osmolyte accumulation that helps establish osmotic homeostasis, stress damage protection, or repair mechanisms (e.g., induction of LEA/dehydrin-type stress genes).
OSMOTIC STRESS–ACTIVATED PHOSPHOLIPID SIGNALING
Membrane phospholipids constitute a dynamic system that generates a multitude of signal molecules (e.g., IP3, DAG, PA, etc.) in addition to serving important structural roles during stress responses. The potential of phospholipid-based signaling in plants is still underexplored and cannot be overappreciated (68). However, it must also be pointed out that the phospholipid system, like the reactive oxygen species that is associated with it (84), is a double-edged sword: As signaling molecules at low levels, the phospholipid messengers may activate downstream adaptive responses, whereas at high levels, phospholipid-generated products may reflect stress damage or may be damaging.
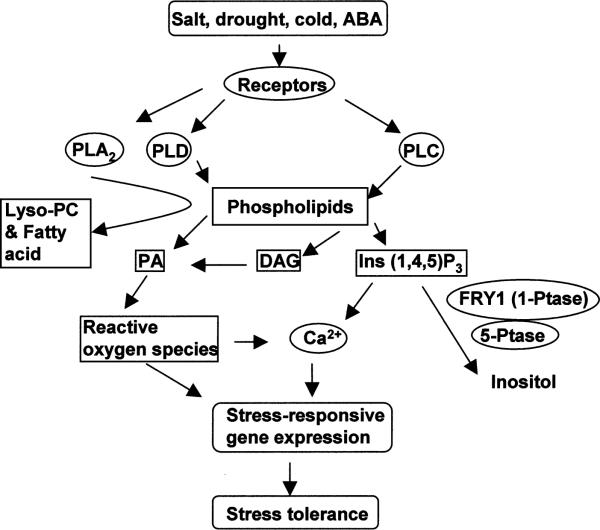
Phospholipid signaling systems are typically grouped according to the phospholipases that catalyze the formation of lipid and other messengers (Figure 4). There are also novel pathways involving the formation of lipid messengers that are not the direct products of phospholipases, such as diacylglycerol pyrophosphate (DGPP) and phosphatidylinositol 3,5-bisphosphate [PI(3,5)P2] (68). The phospholipase C (PLC) pathway has been the best characterized, particularly in nonplant systems. PLC catalyzes the hydrolysis of phosphotidylinositol 4,5-bisphosphate (PIP2), generating the second messengers inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG). IP3 releases Ca2+ from internal stores, whereas DAG activates protein kinase C. Several studies have shown that in various plant systems IP3 levels rapidly increase in response to hyperosmotic stress (10, 12, 25, 98). IP3 levels also increased upon treatment with exogenous ABA in Vicia faba guard cell protoplasts (48) and in Arabidopsis seedlings (112). The IP3 precursor, PIP2, is synthesized via phosphatidylinositol 4-phosphate 5-kinase. An Arabidopsis gene encoding this enzyme, PIP5K, is induced by osmotic stress and ABA (63). Similarly, an Arabidopsis PLC gene, AtPLC1, is also induced by salt and drought stress (26).
Figure 4.
Phospholipid signaling under salt stress, drought, cold, or ABA. Osmotic stress, cold, and ABA activate several types of phospholipases that cleave phospholipids to generate lipid messengers (e.g., PA, DAG, and IP3), which regulate stress tolerance partly through modulation of stress-responsive gene expression. FRY1 (a 1-phosphatase) and 5-phosphatase-mediated IP3 degradation attenuates the stress gene regulation by helping to control cellular IP3 levels. PLC, phospholipase C; PLD, phospholipase D; PLA2, phospholipase A2; PA, phosphatidic acid; DAG, diacyglycerol.
In guard cells, caged IP3 induced Ca2+ increase in the cytoplasm and triggered stomatal closure (83). Exogenous IP3 releases Ca2+ from isolated vacuoles or tonoplast vesicles (86). Microinjection as well as pharmacological experiments suggested that increases in cytoplasmic Ca2+ could lead to the expression of osmotic stress-responsive genes (108). Recently, a connection between phosphoinositides and stress gene expression has also been demonstrated genetically. In the Arabidopsis fry1 mutant, superinduction of ABA- and stress-responsive gene transcription correlated with elevated IP3 accumulation (112). In transgenic plants expressing an antisense PLC gene or overexpressing an Ins(1,4,5)P3 5-phosphatase gene, IP3 levels decreased, and the plants were less sensitive to osmotic stress or ABA in germination and in stress gene induction (82). Like IP3, other inositol phosphates such as IP6 and I(1,3,4)P3 may also function in releasing Ca2+ from internal stores (48, 49). Plants do not seem to have PKC genes. Thus, DAG signaling may be indirect as it can be rapidly phosphorylated to phosphatidic acid (PA), which is an important signaling molecule (66).
The cellular levels of phosphoinositide messengers are a result of both synthesis and degradation. Hence the role of phosphoinositide turnover should not be overlooked (Figure 4). Xiong et al. (112) showed that, in Arabidopsis, loss-of-function mutations in the FRY1 gene encoding an inositol polyphosphate-1-phosphatase result in enhanced osmotic- and ABA-induction of gene transcription. In animal cells, there are two major routes for IP3 breakdown. These are the 5-phosphatase pathway and the 3-kinase pathway, resulting in the accumulation of inositol 1, 4-bisphosphate [Ins(1,4)P2] and inositol 1,3,4,5-tetraphospahte [Ins(1,3,4,5)P4] intermediates, respectively (5, 60, 90). Ins(1,3,4,5)P4 can be further dephosphorylated by 5-phosphatases to generate inositol 1,3,4-trisphosphate [Ins(1,3,4)P3]. Animal inositol polyphosphate 1-phosphatase (IPP) hydrolyzes Ins(1,4)P2 and Ins(1,3,4)P3, at the 1-position (60). FRY1, also known as SAL1, was able to hydrolyze both of these two inositol polyphosphates (79). Although animal IPP isoforms can hydrolyze IP3 directly in certain cell types, in many other cells, the 1-phosphatase does not have this ability to directly hydrolyze IP3. Recombinant FRY1 protein was also active in directly hydrolyzing IP3 (112). Even the 1-phosphatase activity of FRY1 toward Ins(1,4)P2 and Ins(1,3,4)P3 may affect the catabolism of IP3 as the accumulation of these intermediates would slow-down IP3 degradation.
It is interesting that, despite increased IP3 levels and enhanced stress gene expression in fry1, the mutant plants were more susceptible to damage by salt, drought, or freezing stress (112). This raises the possibility that FRY1 is somehow directly involved in the homeostasis pathway of stress responses, and the fry1 mutations disrupt cellular homeostasis and thereby lead to increased stress injury (Figure 1). Perhaps the increased stress gene expression is a compensatory mechanism to limit or repair stress injury. Similar compensatory increases in detoxification responses occur in the sos1 mutant where high levels of proline were found (54). Although the increased stress gene expression or proline level did not correct the stress sensitive phenotypes of the respective mutants, it may be speculated that without these compensatory responses, the mutants would be even more susceptible to stress damage. In this regard, FRY1 represents an interesting point of crosstalk between the stress homeostasis and detoxification pathways. In other words, a functional FRY1 attenuates the detoxification response.
Phospholipase D cleaves membrane phospholipids to produce phosphatidic acid (PA) and free head groups. PA is a second messenger in animal cells by activating targets such as PLC and PKC (16). Osmotic stress activates PLD activity in suspension cells of Chlamydomonas, tomato, and alfalfa (69). In the resurrection plant Craterostigma plantagineum and Arabidopsis, PLD is rapidly activated by dehydration stress (18, 39). Two PLD genes were cloned from the resurrection plant; one was constitutive and the other was induced by dehydration or ABA treatment (18). When drought stress–induced PLD activity was compared between drought-resistant and -sensitive cultivars of cowpea, it was found that the activity was higher in the drought-sensitive cultivar (15). This suggests that PLD activation reflects lipolitic membrane disintegration during stress injuries. Consistent with this view, blocking PLD activity resulted in reduced stress injury and improved freezing tolerance (X-M. Wang, personal communication). In this context, it is interesting that the PLD product, PA, has evolved a signaling role to perhaps mitigate stress injury. In guard cells, PLD activity increased shortly after ABA treatment, and the application of PA mimics the effect of ABA in inducing stomatal closure (35). Sphingosine-1-phosphate (S1P), which shares the same head group with PA, has also been implicated in drought stress signaling (73). S1P concentrations increased upon drought stress, and exogenous application of S1P induced Ca2+ oscillations and stomatal closure.
In addition to PLC- and PLD-based lipid signaling, there are other lipid metabolizing enzymes that also respond to osmotic stress. PLA2 cleaves phospholipids at the sn-2 position to generate lyso-phospholipids and free fatty acids. Hyperosmotic stress stimulates PLA2 activity in algae (14, 62). The role of PLA2 in osmotic stress adaptation and whether it is activated in higher plants is currently unclear. In yeast, phosphatidylinositol 3-phosphate is converted to PI(3,5)P2 by the Fab1p enzyme when subjected to severe hyperosmotic stress (11). PI(3,5)P2 levels also increase rapidly and transiently in several plants (62). This novel lipid messenger may function in osmoregulation by stimulating tonoplast H+-ATPase activity (68).
ABA AND OSMOTIC STRESS SIGNALING
Role of ABA in Water Stress Tolerance
Although ABA has broad functions in plant growth and development, its main function is to regulate plant water balance and osmotic stress tolerance. This point is best illustrated by plant mutants that cannot produce ABA. Several ABA-deficient mutants have been reported for Arabidopsis, namely aba1, aba2, and aba3 (43). There are also ABA-deficient mutants for tobacco, tomato, and maize (52). Without water or temperature stress, ABA-deficient mutants grow and develop relatively normally (43). The mutants, such as the Arabidopsis aba1, aba2, and aba3, have slightly smaller statures, which may be caused by unavoidable stress even under the best growth conditions. Additionally, the smaller stature of the aba mutants may also be due to ABA regulation of the cell cycle and other cellular activities. Under drought stress, ABA-deficient mutants readily wilt and die if the stress persists. Under salt stress, ABA-deficient mutants also perform poorly (110). The role of ABA in drought and salt stress is at least twofold: water balance and cellular dehydration tolerance. Whereas the role in water balance is mainly through guard cell regulation, the latter role has to do with induction of genes that encode dehydration tolerance proteins in nearly all cells. In addition, ABA is required for freezing tolerance, which also involves the induction of dehydration tolerance genes (58, 110).
Salt and Drought Stress Regulation of ABA Biosynthesis and Degradation
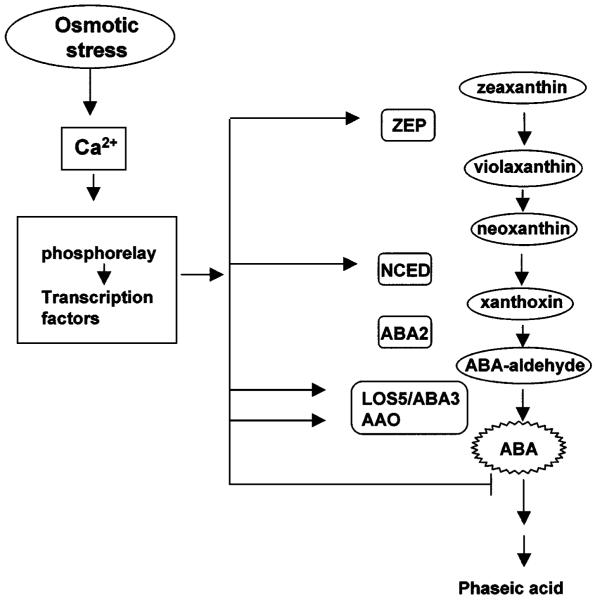
Osmotic stress induction of ABA accumulation is a well-known fact. Recently, some of the underlying molecular mechanisms became clear. Osmotic stress–induced ABA accumulation is a result of both activation of synthesis and inhibition of degradation. Several ABA biosynthesis genes have now been cloned (Figure 5). Zeathanxin epoxidase (known as ABA2 in tobacco and ABA1 in Arabidopsis) catalyzes the epoxidation of zeaxanthin and antheraxanthin to violaxanthin (61). The 9-cis-epoxycarotenoid dioxygenase (NCED) gene was first cloned from the maize vp14 mutant (99). ABA aldehyde oxidase catalyzes the last step. ABA3, also known as LOS5, encodes a sulfurylase that generates the active form of the molybdenum cofactor required by ABA aldehyde oxidase (110).
Figure 5.
ABA metabolism is regulated by osmotic stress at multiple steps. The ABA biosynthesis genes ZEP, NCED, LOS5/ABA3, and AAO are upregulated by salt and drought stresses. ABA degradation is also important in controlling cellular ABA content, and biochemical evidence suggests osmotic stress inhibition of the first step of catabolism.
With the cloning of these biosynthesis genes, it has been possible to determine which of them may be activated by osmotic stress. Biochemical studies suggested that the rate-limiting step is the reaction catalyzed by NCED (43). Indeed, when VP14 and its homologous genes became available, their expression was seen to be upregulated by drought stress (34, 77, 100). It is surprising, however, that other ABA biosynthesis genes are also upregulated by osmotic stress. This is true for the Arabidopsis ABA1 (52), for AAO3 (88), and also for ABA3 (110). Admittedly, the protein amount or activity has not been shown to increase in response to osmotic stress for all these genes. Nevertheless, it is evident that ABA biosynthesis is subjected to osmotic stress regulation at multiple steps (Figure 5). To date, genes responsible for ABA degradation have not been identified. Biochemical studies suggest that a cytochrome P450 monoxygenase catalyzes the first step in the oxidative degradation of ABA (45). It should not be long before this gene, which is expected to be induced by ABA but repressed by osmotic stress, is identified.
Nothing is known about the signaling between osmotic stress perception and the induction of ABA biosynthesis genes. Presumably, it involves calcium signaling and protein phosphorylation cascades.
Stress- and ABA-Regulated Gene Expression
The expression of numerous plant genes has been reported to be regulated by salt and/or drought stress (6, 95, 120). A substantial set of these genes is also responsive to ABA or cold stress. High-throughput technologies using DNA microarrays or chips are quickly replacing traditional differential screening methods in identifying new stress-regulated genes (40). Nonetheless, the reliability of microarray data is sometimes still questionable, and many resort to RNA blot analysis for confirmation. Considering the complexity of salt and drought stress responses, it is not surprising that the limited amount of DNA microarray data suggest that a substantial proportion of the genome is subjected to regulation by these stresses. For example, even in the unicellular yeast Saccharomyces cerevisiae, salt stress affected the expression of ~8% of the genes in the entire genome (76, 80, 114).
One way to make sense of the large number of stress-responsive genes is to group them functionally. For salt stress, many of the induced genes function in ionic homeostasis; these include, e.g., plasma membrane Na+/H+ antiporters for Na+ extrusion (92), vacuolar Na+/H+ antiporters for Na+ compartmentation in the vacuole (3), and high-affinity K+ transporters for K+ acquisition. By increasing the ion concentration in the vacuole, the vacuolar Na+/H+ antiporters also function in osmotic homeostasis. Other salt- or drought-induced genes for osmotic homeostasis include, e.g., those coding for aquaporins and enzymes in osmolyte biosynthesis (6, 95, 120). Besides a potential, but still unproven, role of organic compatible osmolytes in osmotic adjustment, these osmolytes also seem to function in detoxification or damage prevention or repair (119). In fact, the majority of salt- and drought-induced genes appear to function in damage limitation or repair (119). These include the large number of osmolyte biosynthesis genes, LEA/dehydrin-type genes, detoxification enzymes, chaperones, proteases, and ubiquitination-related enzymes. Many of the LEA/dehydrin-type genes have other names that pertain to how they were initially identified; in Arabidopsis, some of them are designated as RD/COR/KIN/LTIs (118).
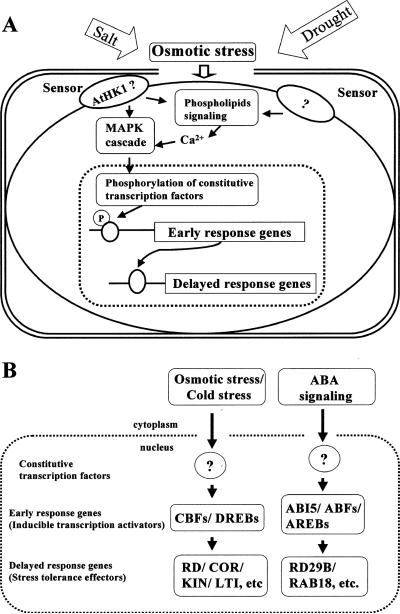
Evidence indicates that salt- or drought-responsive genes are under complex regulation. To study their regulation, it may be of value to consider the stress-responsive genes as either “early-response genes” or “delayed-response genes” (Figure 6). Early-response genes are induced very quickly (within minutes) and often transiently. Their induction does not require new protein synthesis because all signaling components are already in place. In contrast, delayed-response genes, which constitute the vast majority of the stress-responsive genes, are activated by stress more slowly (within hours), and their expression is often sustained. The early-response genes typically encode transcription factors that activate downstream delayed-response genes (Figure 6).
Figure 6.
Model showing osmotic stress regulation of early-response and delayed-response genes. (A) Model integrating stress sensing, activation of phospholipid signaling and MAP kinase cascade, and transcription cascade leading to the expression of delayed-response genes. (B) Examples of early-response genes encoding inducible transcription activators and their downstream delayed-response genes encoding stress tolerance effector proteins. Question marks denote unknown transcription factors that activate the early-response genes.
Several examples of early-response genes in salt, drought, cold, and ABA regulation have emerged. They include, e.g., the CBF/DREB gene family, RD22BP, AtMyb, and ABF/ABI5/AREB (Table 1). These genes are all rapidly induced by either ABA or one or more of the stress cues. A major research emphasis should be to define the cis-regulatory promoter elements in these genes that confer stress inducibility and to identify transcription factors that bind these elements and activate the early-response genes. The upstream transcription factors are typically constitutively expressed and are regulated by stress at the posttranslational level, i.e., by phosphorylation changes (Figure 6).
TABLE 1.
cis-acting promoter elements on delayed-stress response genes and transcription factors (encoded by early-response genes) that bind to them
| cis element | Gene | Transcription factor name | Transcription factor type | Transcription factor expression is induced by | References | ||
|---|---|---|---|---|---|---|---|
| DRE | TACCGACAT | rd29A | DREB2A | AP2 | Dehydration, ABA | Arabidopsis | (56, 71) |
| DRE | rd29A | DREB2B | AP2 | Dehydration, salt | Arabidopsis | (56, 71) | |
| DRE | rd29A | CBF1/DREB1B | AP2 | Cold | Arabidopsis | (19, 96, 97) | |
| DRE | rd29A | CBF2/DREB1C | AP2 | Cold | Arabidopsis | (19, 96, 97) | |
| DRE | rd29A | CBF3/DREB1A | AP2 | Cold | Arabidopsis | (19, 96, 97) | |
| ABRE | CACGTGGC | Em | EmBPl | bZIP | Wheat | (20) | |
| ABRE | CCACGTGG | TAF-1 | bzip | Tobacco | (74) | ||
| ABRE | OSBZ8 | bZIP | ABA | Rice | (70) | ||
| ABRE | osZIP-1a | bZIP | Rice | (72) | |||
| ABRE | (T/G/C)ACGT(G/T)GC | Osem | TRAB1 | bZIP | ABA | Rice | (27) |
| ABRE | rd29B | AREB1 | bZIP | Dehydration, salt | Arabidopsis | (9, 17, 102) | |
| GNGGTG/GTGGNG | MsPRP2 | Alfin1 | zink-finger | Alfalfa | (4) | ||
| MYCRS | CANNTG | rd22 | RD22BP1 | myc | Dehydration, ABA | Arabidopsis | (1) |
| MYBRS | NyAACPyPu | rd22 | AyMyb2 | myb | Dehydration, ABA | Arabidopsis | (104) |
Major achievements in stress molecular biology in the last decade have been to define the cis-regulatory elements in the delayed-response genes and to clone the early-response genes coding for the element-binding proteins. Table 1 lists some of the cis-elements and their binding proteins known to date for salt, drought, cold, or ABA responses.
The transcriptional cascades in stress gene regulation have provided excellent opportunities for producing stress-hardy plants by regulon engineering. For example, ectopic expression of CBF1 or CBF3 leads to constitutive expression of downstream delayed-response genes such as RD29A, COR15, and KIN1 and consequently to enhanced stress tolerance (36, 38). The use of a stress-activated promoter to drive the expression of the transcription factors avoids the pitfalls associated with constitutive expression and appears to be advantageous (38).
ABA-Dependent and ABA-Independent Signaling
Because salt and drought stress enhance ABA accumulation in plants and exogenous application of ABA can have similar effects as osmotic stress, such as in gene induction, it is reasonable to hypothesize that ABA mediates osmotic stress responses. Whether all or most of the osmotic stress responses are dependent on ABA has been of great interest. This question has often been addressed in the context of stress gene regulation, partly made possible by the availability of ABA-deficient and ABA-insensitive mutants. In particular, the Arabidopsis aba1, aba2, and abi1 and abi2 mutants have been extensively used for this type of study (43). One caveat of these studies is that none of the mutants are completely deficient or insensitive to ABA, often making the interpretation of ABA independence equivocal. Nevertheless, a number of studies showed that some osmotic stress responsive genes are induced completely independent of ABA, some are fully dependent on ABA, and others are only partially ABA dependent (95).
The RD29A gene has served as an excellent paradigm of ABA-dependent and -independent gene regulation. Early studies showed that osmotic stress induction of RD29A transcript accumulation is only partially blocked by aba1 or abi1 mutations, thus suggesting both ABA-dependent and -independent regulation (115). The ABA-independent regulation received a strong boost from a landmark paper published in 1994 (116), which reported the identification of the DRE (dehydration responsive element) sequence in the RD29A promoter as sufficient and necessary for osmotic stress induction. ABA cannot activate the DRE element. Whether ABA is still necessary for osmotic stress activation of DRE has not been investigated. This question could be addressed by expressing a reporter gene under a synthetic DRE-containing promoter in ABA-deficient and -insensitive mutants.
There is evidence that although ABA does not activate the DRE element, it may be required for full activation of DRE by osmotic stress. In the los5/aba3 mutant that is deficient in ABA synthesis owing to a defective molybdenum cofactor, high salt or PEG induction of RD29A and other DRE-type genes is virtually abolished (110). It is still curious why the aba3 mutation has a stronger effect than aba1 or aba2 in blocking the osmotic stress induction. LOS5/ABA3 encodes a molybdenum cofactor sulfurylase that catalyzes the synthesis of active MoCo factor required for ABA aldehyde oxidase in the last step of ABA biosynthesis. MoCo is also required for several other enzymes such as xanthine dehydrogenases (110). These latter enzymes do not appear to have a role in osmotic stress responses. Therefore, the more severe osmotic stress phenotypes of aba3 are probably a consequence of more severe defects in ABA synthesis. Xiong et al. (110) have proposed that activation of DRE by DREB2A and related transcription factors may require ABA-dependent factor(s). Consistent with this, full activation of RD29A transcription depends upon the synergy between the DRE and ABRE elements (K. Shinozaki, personal communication).
The role of ABA in cold responses is still unclear. Only a few years ago, ABA was thought to have a major role in cold responses. Lang et al. (46) found transiently increased ABA accumulation in response to chilling treatment. However, other studies do not seem to find ABA accumulation under cold stress. It is clear that there is no dramatic ABA synthesis in the cold owing to the general slowdown effect of cold on cellular metabolism. Several studies found that exogenous ABA application increased the freezing tolerance of plants (8). Furthermore, cold and ABA induce a common set of genes. However, the cold stress induction appears to be completely independent of ABA. Mutations that enhance ABA induction of the RD29A-LUC transgene also increased osmotic stress but not cold induction (109). Nevertheless, the involvement of ABA in cold acclimation and cold-responsive gene expression cannot be ruled out. Besides blocking osmotic stress induction of genes and osmotic stress tolerance, the los5/aba3 mutations also substantially reduce cold-responsive gene expression and freezing tolerance (110).
Although specific branches and components exist (47), the signaling pathways for salt, drought, cold, and ABA interact and even converge at multiple steps (109, 111). This was suggested by a comprehensive mutational analysis in which Arabidopsis single-gene mutations were found to affect responses to all or combinations of these signals (33). A nice example of the pathway convergence is provided by the fry1 (fiery1) mutation. The mutation increases the amplitude and sensitivity of stress gene induction not only by ABA, but also by salt, drought, and cold stresses (112). An analysis of double mutants between fry1 and aba1 or abi1 indicated that the cold or osmotic stress hypersensitivity in the mutant is not dependent on ABA (L. Xiong & J-K. Zhu, unpublished information). FRY1 encodes an inositol polyphosphate 1-phosphatase that is required for IP3 turnover. In response to ABA, wild-type plants accumulated IP3 transiently. The IP3 accumulation in fry1 mutants in response to ABA was more sustained and reached higher levels. It will not be surprising if osmotic or cold stress also leads to more IP3 accumulation in fry1. The results are consistent with IP3 being a second messenger that mediates not only ABA but also salt, drought, or cold stress regulation of gene expression. Another important implication of the study is that ABA and stress signaling is desensitized by IP3 turnover.
As discussed above, it is very intriguing that fry1 mutant plants are less tolerant to salt, drought, or freezing stress despite enhanced expression of stress genes in the mutant (112). This observation may be explained by one or more of the following models: The first model is that too much IP3 may compromise stress tolerance. For example, uncontrolled IP3 accumulation could lead to unbalanced calcium signaling, which certainly would impact stress tolerance. This model can be tested by finding and analyzing other mutants with enhanced IP3 accumulation due either to increased production or decreased degradation. Another model is that the FRY1 inositol polyphosphatase is critical for stress tolerance. Loss-of-function mutations in IP3 biosynthesis genes such as phospholipase C are expected to suppress the IP3 overaccumulation phenotype of fry1. It would be consistent with this model if the hypothetical suppressor mutations did not suppress the stress-tolerance defect of fry1. An interesting implication of the second model is that more stress damage leads to more IP3 and thereby more RD/COR/KIN-type stress gene expression, which would support that notion of a detoxification function of RD/COR/KIN gene products.
FUTURE PROSPECTIVES
Salt and drought stress signaling has largely remained a mystery until recently. Now the molecular identities of some signaling elements have been identified. But we are still far from having a clear picture. The foremost difficulty in putting together the puzzle is not having enough pieces. Therefore, the challenge in the near future remains to identify more signaling elements. Once more components are known, signaling specificities and crosstalks can be properly addressed. Changes in gene expression or even protein amount or activity in response to water stress are not sufficient to establish whether an element is part of salt or drought signaling pathways. Any signaling component has to be established by functional necessity and functional sufficiency when possible. That is to say, plant phenotypes, be they molecular, biochemical, or physiological, are required to establish that a particular component functions in water stress signaling. In this regard, few genes believed to be involved in salt or drought signaling meet this criterion yet.
The most prominent missing elements in salt, drought, ABA, or cold signaling are the sensors or receptors. An Arabidopsis histidine kinase, AtHK1, is a candidate osmosensor because it could complement a yeast osmosensing mutant (105). As with a number of other potential regulatory genes, AtHK1 transcript level is upregulated by osmotic stress. In addition, expression of a receptor-like kinase gene was induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopsis (28). However, the functional significance, if any, of these transcript upregulations in osmotic stress responses is obscure.
Through more widespread application of forward and reverse genetic analysis in model plants and with the growing power of genomics and proteomics tools, progress in understanding water stress signaling will certainly accelerate. With a better understanding come more effective ways to improve plant salt or drought hardiness. The first phase of genetic engineering of stress hardiness has been to simply express one or several tolerance effector genes under constitutive or stress-inducible promoters. The second phase is to improve stress tolerance through engineering more effective signaling. Part of this has been achieved by overexpressing early-response transcription activators to turn on many downstream effector genes (36, 56). Components upstream of transcription factors can also be manipulated to improve stress tolerance (44). The future promises to see a much clearer picture of salt and drought signal transduction pathways and more examples of genetic improvement of water stress tolerance by fine-tuning plant sensing and signaling systems.
ACKNOWLEDGMENTS
I thank Liming Xiong, Becky Stevenson, Masaru Ohta, and Satoshi Iuchi for assistance in the preparation of this manuscript. Research in my laboratory is supported by grants from the U.S. National Institutes of Health, the U.S. National Science Foundation, and the U.S. Department of Agriculture.
LITERATURE CITED
- 1.Abel H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–68. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amtmann A, Sanders D. Mechanisms of Na+ uptake by plant cells. Adv. Bot. Res. 1999;29:75–112. [Google Scholar]
- 3.Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+-antiport in Arabidopsis. Science. 1999;285:1256–58. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- 4.Bastola DR, Pethe VV, Winicov I. Alfin1, a novel zinc-finger protein in alfalfa roots that binds to promoter elements in the salt-inducible MsPRP2 gene. Plant Mol. Biol. 1998;38:1123–35. doi: 10.1023/a:1006081926699. [DOI] [PubMed] [Google Scholar]
- 5.Berridge MJ, Irvine RF. Inositol phosphate and cell signaling. Nature. 1989;341:197–205. doi: 10.1038/341197a0. [DOI] [PubMed] [Google Scholar]
- 5a.Boyer JS. Plant productivity and environment. Science. 1982;218:443–48. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- 6.Bray EA. Molecular responses to water deficit. Plant Physiol. 1993;103:1035–40. doi: 10.1104/pp.103.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burssens S, Himanen K, van de Cotte B, Beeckman T, van Montagu M. Expression of cell cycle regulatory genes and morphological alterations in response to salt stress in Arabidopsis thaliana. Planta. 2000;211:632–40. doi: 10.1007/s004250000334. [DOI] [PubMed] [Google Scholar]
- 8.Chen HH, Li PH, Brenner ML. Involvement of abscisic acid in potato cold acclimation. Plant Physiol. 1983;71:362–65. doi: 10.1104/pp.71.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi HI, Hong JH, Ha JO, Kang JY, Kim SY. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000;275:1723–30. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- 10.DeWald DB, Torabinejad J, Jones CA, Shope JC, Cangelosi AR, et al. Rapid accumulation of phosphatidylinositol 4,5-bisphosphate and inositol 1,4, 5-trisphosphate correlates with calcium mobilization in salt-stressed Arabidopsis. Plant Physiol. 2001;126:759–69. doi: 10.1104/pp.126.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390:187–92. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- 12.Drobak BK, Watkins PA. Inositol (1,4,5)trisphosphate production in plant cells: an early response to salinity and hyperosmotic stress. FEBS Lett. 2000;481:240–44. doi: 10.1016/s0014-5793(00)01941-4. [DOI] [PubMed] [Google Scholar]
- 13.Droillard MJ, Thibivilliers S, Cazale AC, Barbier-Brygoo H, Lauriere C. Protein kinases induced by osmotic stresses and elicitor molecules in tobacco cell suspensions: two crossroad MAP kinases and one osmoregulation-specific protein kinase. FEBS Lett. 2000;474:217–22. doi: 10.1016/s0014-5793(00)01611-2. [DOI] [PubMed] [Google Scholar]
- 14.Einspahr KJ, Maeda M, Thompson GA., Jr. Concurrent changes in Dunaliella salina ultrastructure and membrane phospholipid metabolism after hyperosmotic shock. J. Cell Biol. 1988;107:529–38. doi: 10.1083/jcb.107.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El Maarouf H, Zuily-Fodil Y, Gareil M, d'Arcy-Lameta A, Pham-Thi AT. Enzymatic activity and gene expression under water stress of phospholipase D in two culitivars of Vigna unguiculata L. Walp. differing in drought tolerance. Plant Mol. Biol. 1999;39:1257–65. doi: 10.1023/a:1006165919928. [DOI] [PubMed] [Google Scholar]
- 16.English D. Phosphatidic acid: a lipid messenger involved in intracellular and extracellular signaling. Cell. Signal. 1996;8:341–47. doi: 10.1016/0898-6568(95)00076-3. [DOI] [PubMed] [Google Scholar]
- 16a.Epstein E, Norlyn JD, Rush DW, Kingsbury RW, Kelly DB, et al. Saline culture of crops: a genetic approach. Science. 1980;210:399–404. doi: 10.1126/science.210.4468.399. [DOI] [PubMed] [Google Scholar]
- 17.Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcriptional factor. Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank W, Munnik T, Kerkmann K, Salamini F, Bartels D. Water deficit triggers phospholipase D activity in the resurrection plant Craterostigma plantagineum. Plant Cell. 2000;12:111–23. doi: 10.1105/tpc.12.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16:433–42. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- 20.Guiltinan MJ, Marcotte WR, Quatrano RS. A plant leucine zipper protein that recognizes an abscisic acid response element. Science. 1990;250:267–71. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- 21.Guo Y, Halfter U, Ishitani M, Zhu JK. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell. 2001;13:1383–400. doi: 10.1105/tpc.13.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gustin MC, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1998;62:1264–300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halfter U, Ishitani M, Zhu JK. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA. 2000;97:3730–34. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:463–99. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- 25.Heilmann I, Perera IY, Gross W, Boss WF. Changes in phosphoinositide metabolism with days in culture affect signal transduction pathways in Galdieria suphuraria. Plant Physiol. 1999;119:1331–39. doi: 10.1104/pp.119.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirayama T, Ohto C, Mizoguchi T, Shinozaki K. A gene encoding a phosphatidylinositol-specific phospholipase C is induced by dehydration and salt stress in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 1995;92:3903–7. doi: 10.1073/pnas.92.9.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hobo T, Kowyama Y, Hattori T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA. 1999;96:15348–53. doi: 10.1073/pnas.96.26.15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong SW, Jon JH, Kwak JM, Nam HG. Identification of a receptor-like protein kinase gene rapidly induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopsis thaliana. Plant Physiol. 1997;113:1203–12. doi: 10.1104/pp.113.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoyos ME, Zhang S. Calcium-independent activation of salicylic acid–induced protein kinase and a 40-kilo-dalton protein kinase by hyperosmotic stress. Plant Physiol. 2000;122:1355–63. doi: 10.1104/pp.122.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K. Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 2000;24:655–65. doi: 10.1046/j.1365-313x.2000.00913.x. [DOI] [PubMed] [Google Scholar]
- 31.Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- 32.Ishitani M, Liu J, Halfter U, Kim CS, Wei M, Zhu JK. SOS3 function in plant salt tolerance requires myristoylation and calcium-binding. Plant Cell. 2000;12:1667–77. doi: 10.1105/tpc.12.9.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishitani M, Xiong L, Stevenson B, Zhu JK. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis thaliana: interactions and convergence of abscisic acid–dependent and abscisic acid–independent pathways. Plant Cell. 1997;9:1935–49. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, et al. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001;27:325–33. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- 35.Jacob T, Ritchie S, Assmann SM, Gilroy S. Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. USA. 1999;96:12192–97. doi: 10.1073/pnas.96.21.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–6. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- 37.Jonak C, Kiegerl S, Ligterink W, Barker PJ, Huskisson NS, Hirt H. Stress signaling in plants: a mitogen-activated protein kinase pathway is activated by cold and drought. Proc. Natl. Acad. Sci. USA. 1996;93:11274–79. doi: 10.1073/pnas.93.20.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999;17:287–91. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- 39.Katagiri T, Takahashi S, Shinozaki K. Involvement of a novel Arabidopsis phospholipase D, AtPLD delta, in dehydration-inducible accumulation of phosphatidic acid in stress signaling. Plant J. 2001;26:595–605. doi: 10.1046/j.1365-313x.2001.01060.x. [DOI] [PubMed] [Google Scholar]
- 40.Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, et al. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell. 2001;13:889–906. doi: 10.1105/tpc.13.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiegerl S, Cardinale F, Siligan C, Gross A, Baudouin E, et al. SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell. 2000;12:2247–58. doi: 10.1105/tpc.12.11.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knight H, Trewavas AJ, Knight MR. Calcium signaling in Arabidopsis thaliana responding to drought and salinity. Plant J. 1997;12:1067–78. doi: 10.1046/j.1365-313x.1997.12051067.x. [DOI] [PubMed] [Google Scholar]
- 43.Koornneef M, Léon-Kloosterziel KM, Schwartz SH, Zeevaart JAD. The genetic and molecular dissection of abscisic acid biosynthesis and signal transduction in Arabidopsis. Plant Physiol. Biochem. 1998;36:83–89. [Google Scholar]
- 44.Kovtun Y, Chiu WL, Tena G, Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA. 2000;97:2940–45. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krochko JE, Abrams GD, Loewen MK, Abrams SR, Culter AJ. (+)-abscisic acid 8′-hydroxylase is a cytochrome P450 monooxygenase. Plant Physiol. 1998;118:849–60. doi: 10.1104/pp.118.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lang V, Mantyla E, Welin B, Sundberg B, Palva ET. Alterations in water status, endogenous abscisic acid content, and expression of rab18 gene during the development of freezing tolerance in Arabidopsis thaliana. Plant Physiol. 1994;104:1341–49. doi: 10.1104/pp.104.4.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING-finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Gene Dev. 2001;15:912–24. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee Y, Choi YB, Suh J, Lee J, Assmann SM, et al. Abscisic acid–induced phosphoinositide turnover in guard cell protoplasts of Vicia faba. Plant Physiol. 1996;110:987–96. doi: 10.1104/pp.110.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lemtiri-Chlieh F, MacRobbie EAC, Brearley CA. Inositol hexakisphosphate is a physiological signal regulating the K+-inward rectifying conductance in guard cells. Proc. Natl. Acad. Sci. USA. 2000;97:8687–92. doi: 10.1073/pnas.140217497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leng Q, Mercier RW, Yao W, Berkowitz GA. Cloning and first functional chracterization of a plant cyclic nucleotide–gated cation channel. Plant Physiol. 1999;121:753–61. doi: 10.1104/pp.121.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leung J, Giraudat J. Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- 52.Liotenberg S, North H, Marion-Poll A. Molecular biology and regulation of abscisic acid biosynthesis in plants. Plant Physiol. Biochem. 1999;37:341–50. [Google Scholar]
- 53.Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA. 2000;97:3735–40. doi: 10.1073/pnas.060034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J, Zhu JK. Proline accumulation and salt-stress-induced gene expression in a salt-hypersensitive mutant of Arabidopsis. Plant Physiol. 1997;114:591–96. doi: 10.1104/pp.114.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu J, Zhu JK. A calcium sensor homolog required for plant salt tolerance. Science. 1998;280:1943–45. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- 56.Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA-binding domain separate two cellular signal transduction pathways in drought- and low temperature–responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y, Zhang S, Klessig DF. Molecular cloning and characterization of a tobacco MAP kinase kinase that interacts with SIPK. Mol. Plant Microbe Interact. 2000;13:118–24. doi: 10.1094/MPMI.2000.13.1.118. [DOI] [PubMed] [Google Scholar]
- 58.Llorente F, Oliveros JC, Martinez-Zapater JM, Salinas J. A freezing-sensitive mutant of Arabidopsis, frs1, is a new aba3 allele. Planta. 2000;211:648–55. doi: 10.1007/s004250000340. [DOI] [PubMed] [Google Scholar]
- 59.Maathuis F, Sanders D. Sodium up-take in Arabidopsis roots is regulated by cyclic nucleotides. Plant Physiol. 2001 In press. [PMC free article] [PubMed] [Google Scholar]
- 60.Majerus PW. Inositol phosphate biochemistry. Annu. Rev. Biochem. 1992;61:225–50. doi: 10.1146/annurev.bi.61.070192.001301. [DOI] [PubMed] [Google Scholar]
- 61.Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, et al. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 1996;15:2331–42. [PMC free article] [PubMed] [Google Scholar]
- 62.Meijer HJG, Divecha N, van den Ende H, Musgrave A, Munnik T. Hyperosmotic stress induces rapid synthesis of phosphatidyl-D-inositol 3,5-bis-phosphate in plant cells. Planta. 1999;208:294–98. [Google Scholar]
- 63.Mikami K, Katagiri T, Luchi S, Yamaguchi-Shinozaki K, Shinozaki K. A gene encoding phosphatidylinositol 4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J. 1998;15:563–68. doi: 10.1046/j.1365-313x.1998.00227.x. [DOI] [PubMed] [Google Scholar]
- 64.Mikolajczyk M, Olubunmi SA, Muszynska G, Klessig DF, Dobrowolska G. Osmotic stress induces rapid activation of a salicylic acid–induced protein kinase and a homolog of protein kinase ASK1 in tobacco cells. Plant Cell. 2000;12:165–78. [PMC free article] [PubMed] [Google Scholar]
- 65.Mizoguchi T, Ichimura K, Yoshida R, Shinozaki K. MAP kinase cascades in Arabidopsis: their roles in stress and hormone responses. Results Probl. Cell Differ. 2000;27:29–38. doi: 10.1007/978-3-540-49166-8_3. [DOI] [PubMed] [Google Scholar]
- 66.Munnik T, Irvine RF, Musgrave A. Phospholipid signaling in plants. Biochim. Biophys. Acta. 1998;1389:222–72. doi: 10.1016/s0005-2760(97)00158-6. [DOI] [PubMed] [Google Scholar]
- 67.Munnik T, Ligterink W, Meskiene I, Calderini O, Beyerly J, et al. Distinct osmosensing protein kinase pathways are involved in signalling moderate and severe hyperosmotic stress. Plant J. 1999;20:381–88. doi: 10.1046/j.1365-313x.1999.00610.x. [DOI] [PubMed] [Google Scholar]
- 68.Munnik T, Meijer HJG. Osmotic stress activates distinct lipid and MAPK signaling pathways in plants. FEBS Lett. 2001;498:172–78. doi: 10.1016/s0014-5793(01)02492-9. [DOI] [PubMed] [Google Scholar]
- 69.Munnik T, Meijer HJG, ter Riet B, Frank W, Bartels D, Musgrave A. Hyper-osmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol pyrophosphate. Plant J. 2000;22:147–54. doi: 10.1046/j.1365-313x.2000.00725.x. [DOI] [PubMed] [Google Scholar]
- 70.Nakagawa H, Ohmiya K, Hattori T. A rice bZIP protein, designated OSBZ8, is rapidly induced by abscisic acid. Plant J. 1996;9:217–27. doi: 10.1046/j.1365-313x.1996.09020217.x. [DOI] [PubMed] [Google Scholar]
- 71.Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S, et al. Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol. Biol. 2000;42:657–65. doi: 10.1023/a:1006321900483. [DOI] [PubMed] [Google Scholar]
- 72.Nantel A, Quatrano RS. Characterization of three rice basic/leucine zipper factors, including two inhibitors of EmBP-1 DNA binding activity. J. Biol. Chem. 1996;271:31296–305. doi: 10.1074/jbc.271.49.31296. [DOI] [PubMed] [Google Scholar]
- 73.Ng CK, Carr K, McAinsh MR, Powell B, Hetherington AM. Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature. 2001;410:596–99. doi: 10.1038/35069092. [DOI] [PubMed] [Google Scholar]
- 74.Oeda K, Salinas J, Chua NH. A tobacco bZip transcription activator (TAF-1) binds to a G-box-like motif conserved in plant genes. EMBO J. 1991;10:1793–802. doi: 10.1002/j.1460-2075.1991.tb07704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ozcan S, Dover J, Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:2566–73. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Posas F, Chambers JR, Heyman JA, Hoeffler JP, de Nada E, Arino J. The transcriptional response of yeast to salt stress. J. Biol. Chem. 2000;275:17249–55. doi: 10.1074/jbc.M910016199. [DOI] [PubMed] [Google Scholar]
- 77.Qin X, Zeevaart JAD. The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc. Natl. Acad. Sci. USA. 1999;96:15354–61. doi: 10.1073/pnas.96.26.15354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qiu Q, Guo Y, Dietrich M, Schumaker KS, Zhu JK. Characterization of the plasma membrane Na+/H+ exchanger in Arabidopsis thaliana. Abstr. Int. Workshop Plant Membr. Biol., 12th, Madison, Wis. 2001:235. [Google Scholar]
- 79.Quintero FJ, Garciadeblas B, Rodriguez-Navarro A. The SAL1 gene of Arabidopsis, encoding an enzyme with 3′(2′), 5′-bisphosphate nucleotide and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell. 1996;8:529–37. doi: 10.1105/tpc.8.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rep M, Krantz M, Thevelein JM, Hoh-mann S. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. J. Biol. Chem. 2000;275:8290–300. doi: 10.1074/jbc.275.12.8290. [DOI] [PubMed] [Google Scholar]
- 81.Rus A, Yokoi S, Sharkhuu A, Reddy M, Lee BH, et al. AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc. Natl. Acad. Sci. USA. 2001;98:14150–55. doi: 10.1073/pnas.241501798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sanchez JP, Chua NH. Arabidopsis PLC1 is required for secondary responses to abscisic acid signals. Plant Cell. 2001;13:1143–54. doi: 10.1105/tpc.13.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sanders D, Brownlee C, Harper JF. Communicating with calcium. Plant Cell. 1999;11:691–706. doi: 10.1105/tpc.11.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sang Y, Cui D, Wang X. Phospholipase D and phosphatidic acid–mediated generation of superoxide in Arabidopsis. Plant Physiol. 2001;126:1449–58. doi: 10.1104/pp.126.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D. Guard cell signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:627–58. doi: 10.1146/annurev.arplant.52.1.627. [DOI] [PubMed] [Google Scholar]
- 86.Schumaker KS, Sze H. Inositol 1,4,5-trisphosphate releases Ca2+ from vacuolar membrane vesicles of oat roots. J. Biol. Chem. 1987;262:3944–46. [PubMed] [Google Scholar]
- 87.Schuppler U, He PH, John PCL, Munns R. Effects of water stress on cell division and cell-division-cycle-2-like cell-cycle kinase activity in wheat leaves. Plant Physiol. 1998;117:667–78. doi: 10.1104/pp.117.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Seo M, Peeters AJM, Koiwai H, Oritani T, Marion-Poll A, et al. The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. USA. 2000;97:12908–13. doi: 10.1073/pnas.220426197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Seo S, Okamoto M, Seto H, Ishizuka K, Sano H, Ohashi Y. Tobacco MAP kinase: a possible mediator in wound signal transduction pathways. Science. 1995;270:1988–92. doi: 10.1126/science.270.5244.1988. [DOI] [PubMed] [Google Scholar]
- 90.Shears SB. Metabolism of inositol phosphates. Adv. Second Messenger Phosphoprot. Res. 1992;26:63–92. [PubMed] [Google Scholar]
- 91.Sheen J. Ca2+ dependent protein kinases and stress signal transduction in plants. Science. 1996;274:1900–2. doi: 10.1126/science.274.5294.1900. [DOI] [PubMed] [Google Scholar]
- 92.Shi H, Ishitani M, Kim C, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA. 2000;97:6896–901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shi H, Quintero FJ, Pardo JM, Zhu JK. The putative plasma membrane Na+/H+ antiporter SOS1 controls long distance Na+ transport in plants. Plant Cell. 2002 doi: 10.1105/tpc.010371. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shi H, Xiong L, Stevenson B, Lu T, Zhu J-K. The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell. 2002 doi: 10.1105/tpc.010417. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shinozaki K, Yamaguchi-Shinozaki K. Gene expression and signal transduction in water-stress response. Plant Physiol. 1997;115:327–34. doi: 10.1104/pp.115.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shinwari ZK, Nakashima K, Miura S, Kasuga M, Seki M, et al. An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem. Biophys. Res. Commun. 1998;250:161–70. doi: 10.1006/bbrc.1998.9267. [DOI] [PubMed] [Google Scholar]
- 97.Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA. 1997;94:1035–40. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Takahashi S, Katagiri T, Hirayama T, Yamaguchi-Shinozaki K, Shinozaki K. Hyperosmotic stress induced a rapid and transient increase in inositol 1,4,5-trisphosphate independent of abscisic acid in Arabidopsis cell culture. Plant Cell Physiol. 2001;42:214–22. doi: 10.1093/pcp/pce028. [DOI] [PubMed] [Google Scholar]
- 99.Tan BC, Schwartz SH, Zeevaart JAD, Mc-Carty DR. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA. 1997;94:12235–40. doi: 10.1073/pnas.94.22.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thompson AJ, Jackson AC, Symonds RC, Mulholland BJ, Dadswell AR, et al. Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes overproduction of abscisic acid. Plant J. 2000;23:363–74. doi: 10.1046/j.1365-313x.2000.00789.x. [DOI] [PubMed] [Google Scholar]
- 101.Tyerman SD, Skerrett IM. Root ion channels and salinity. Sci. Hortic. 1999;78:175–235. [Google Scholar]
- 102.Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA. 2000;97:11632–37. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, et al. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 2000;122:1249–60. doi: 10.1104/pp.122.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Urao T, Noji M, Yamaguchi-Shinozaki K, Shinozaki K. A transcriptional activation domain of ATMYB2, a drought-inducible Arabidopsis Myb-related protein. Plant J. 1996;10:1145–48. doi: 10.1046/j.1365-313x.1996.10061145.x. [DOI] [PubMed] [Google Scholar]
- 105.Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki B, et al. A trans-membrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell. 1999;11:1743–54. doi: 10.1105/tpc.11.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang H, Qi Q, Schorr P, Cutler AJ, Crosby WL, Fowke LC. ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 1998;15:501–10. doi: 10.1046/j.1365-313x.1998.00231.x. [DOI] [PubMed] [Google Scholar]
- 107.Wu SJ, Lei D, Zhu JK. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell. 1996;8:617–27. doi: 10.1105/tpc.8.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu Y, Kuzma J, Marechal E, Graeff R, Lee HC, et al. Abscisic acid signaling through cyclic ADP-ribose in plants. Science. 1997;278:2126–30. doi: 10.1126/science.278.5346.2126. [DOI] [PubMed] [Google Scholar]
- 109.Xiong L, Ishitani M, Lee H, Zhu JK. HOS5—a negative regulator of osmotic stress-induced gene expression in Arabidopsis thaliana. Plant J. 1999;19:569–78. doi: 10.1046/j.1365-313x.1999.00558.x. [DOI] [PubMed] [Google Scholar]
- 110.Xiong L, Ishitani M, Lee H, Zhu JK. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold and osmotic stress responsive gene expression. Plant Cell. 2001;13:2063–83. doi: 10.1105/TPC.010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xiong L, Ishitani M, Zhu JK. Interaction of osmotic stress, ABA and low temperature in the regulation of stress gene expression in Arabidopsis thaliana. Plant Physiol. 1999;119:205–11. doi: 10.1104/pp.119.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xiong L, Lee BH, Ishitani M, Lee H, Zhang C, Zhu JK. FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 2001;15:1971–84. doi: 10.1101/gad.891901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xiong L, Zhu JK. Abiotic stress signal transduction in plants: molecular and genetic perspectives. Physiol. Plant. 2001;112:152–66. doi: 10.1034/j.1399-3054.2001.1120202.x. [DOI] [PubMed] [Google Scholar]
- 114.Yale J, Bohnert HJ. Transcript expression in S. cerevisiae at high salinity. J. Biol. Chem. 2001;276:15996–6007. doi: 10.1074/jbc.M008209200. [DOI] [PubMed] [Google Scholar]
- 115.Yamaguchi-Shinozaki K, Shinozaki K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Gen. Genet. 1993;236:331–40. doi: 10.1007/BF00277130. [DOI] [PubMed] [Google Scholar]
- 116.Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–64. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhu JK. Genetic analysis of plant salt tolerance using Arabidopsis thaliana. Plant Physiol. 2000;124:941–48. doi: 10.1104/pp.124.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhu JK. Cell signaling under salt, water and cold stresses. Curr. Opin. Plant Biol. 2001;4:401–6. doi: 10.1016/s1369-5266(00)00192-8. [DOI] [PubMed] [Google Scholar]
- 119.Zhu JK. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/s1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- 120.Zhu JK, Hasegawa PM, Bressan RA. Molecular aspects of osmotic stress in plants. Crit. Rev. Plant Sci. 1996;16:253–77. [Google Scholar]
- 121.Zhu JK, Liu J, Xiong L. Genetic analysis of salt tolerance in Arabidopsis thaliana: evidence of a critical role for potassium nutrition. Plant Cell. 1998;10:1181–92. doi: 10.1105/tpc.10.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]