Summary
Salt Overly Sensitive 1 (SOS1), a plasma membrane Na+/H+ antiporter in Arabidopsis, is a salt tolerance determinant crucial for the maintenance of ion homeostasis in saline stress conditions. SOS1 mRNA is unstable at normal growth conditions, but its stability is substantially increased under salt stress and other ionic and dehydration stresses. In addition, H2O2 treatment increases the stability of SOS1 mRNA. SOS1 mRNA is inherently unstable and rapidly degraded with a half-life of approximately 10 min. Rapid decay of SOS1 mRNA requires new protein synthesis. Stress-induced SOS1 mRNA stability is mediated by reactive oxygen species (ROS). NADPH oxidase is also involved in the upregulation of SOS1 mRNA stability, presumably through the control of extracellular ROS production. The cis-element required for SOS1 mRNA instability resides in the 500-bp region within the 2.2 kb at the 3′ end of the SOS1 mRNA. Furthermore, mutations in the SOS1 gene render sos1 mutants more tolerant to paraquat, a non-selective herbicide causing oxidative stress, indicating that SOS1 plays negative roles in tolerance of oxidative stress. A hypothetical model for the signaling pathway involving SOS1-mediated pH changes, NADPH oxidase activation, apoplastic ROS production and downstream signaling transduction is proposed, and the biological significance of ROS-mediated induction of SOS1 mRNA stability is discussed.
Keywords: mRNA stability, salt stress, ROS, SOS1, pH regulation, signal transduction
Introduction
Plants cope with environmental changes by activating signal transduction cascades that control and coordinate the physiological and biochemical responses necessary for adaptation (Hasegawa et al., 2000; Zhu, 2002). A number of signaling pathways have been implicated in abiotic stress responses in plants (Yamaguchi-Shinozaki and Shinozaki, 2006). For example, the MAP kinase signaling pathway is involved in oxidative stress and other abiotic stress responses (Nakagami et al., 2005). Calcium signaling is incorporated in different signaling pathways during abiotic stress response, e.g. to heat, cold, drought, and salt (Sanders et al., 2002). The recently identified salt overly sensitive (SOS) signaling pathway is crucial for salt tolerance in plants (Shi et al., 2005). The SOS pathway includes three key components: SOS3, a Ca2+ sensor (Liu and Zhu, 1998); SOS2, a serine/threonine protein kinase (Liu et al., 2000); and SOS1, a plasma membrane Na+/H+ antiporter (Shi et al., 2000). The plasma membrane-localized SOS1 contains a large cytosolic C-terminus that could interact with different molecules. Therefore, SOS1 might also serve as a signaling component to sense and transduce salt stress signals.
Signal perception and signal transduction mediate changes in gene expression. Gene regulation is controlled at several checkpoints, including transcription, pre-mRNA processing and editing, nuclear export, and mRNA stability and translational efficiency (Fedoroff, 2002; Kuhn and Schroeder, 2003). The control mechanisms operating through post-transcriptional events are now emerging as important signal control nodes in eukaryotes (Maniatis and Reed, 2002; Reed, 2003; Tourrière et al., 2002). As an important checkpoint, mRNA decay is a highly regulated process and plays key roles in determining the expression pattern of many genes (Wilusz and Wilusz, 2004). Differential RNA turnover contributes substantially to the control of gene expression during development and in response to chemical and environmental stimuli (Guhaniyogi and Brewer, 2001; Shim and Karin, 2002; Tourrière et al., 2002). In mammalian cells, mRNA decay machinery and signal transduction pathways that transduce environmental stimuli into changes in mRNA stability are now being identified and characterized. Ca2+-dependent signaling and involvement of c-Jun amino-terminal kinase (JNK), PKC, MAP kinase and calcineurin pathways are implicated (Shim and Karin, 2002). The stability of mRNA is dependent on interactions between structural elements that exist in the 5′-untranslated region (UTR), coding, and 3′-UTR sequences (cis-acting elements) and RNA-binding proteins (trans-acting factors). Some cis-element and RNA-binding protein interactions are ubiquitous, e.g. 5′-cap structure and cap-binding proteins and poly(A) and poly(A)-binding proteins(PABPs), while others are mRNA-specific and mediate differential message decay. Studies of AUF1/hnRNPD, HuR, TIA-1 or tristetraprolin trans-acting proteins indicate that interaction with the cis-element may enhance the stability or decay of RNA (Wilusz et al., 2001). In plants, although a number of putative mRNA cis-elements have been identified (Gutierrez et al., 1999; Johnson et al., 2000), there is little experimental evidence revealing trans-acting factors that bind to cis-elements and regulate mRNA stability, despite annotation that indicates the presence of more than 200 putative RNA-binding proteins in Arabidopsis (Fedoroff, 2002; Kastenmayer and Green, 2000). Notable exceptions are the RNA-binding complex that mediates the light-regulated translation of chloroplast mRNA (Rochaix, 2001) and the single-stranded RNA-binding protein AKIP1 that is activated by ABA-activated protein kinase (AAPK) to interact with dehydrin mRNA (Li et al., 2002). Besides AKIP, recent reports have implicated other RNA-binding proteins in stress and ABA signaling, including ABH1 (Hugouvieux et al., 2001), SAD1 (Xiong et al., 2001), HYL1 (Lu et al., 2002) and CPL1/FRY2 (Koiwa et al., 2002; Xiong et al., 2002).
We previously found that SOS1 mRNA is unstable under normal growth conditions, and that its stability is substantially increased upon salt stress treatment (Shi et al., 2003a). This finding suggests that, besides transcriptional regulation, post-transcriptional control of the mRNA stability of important salt determinant genes may also play a crucial role in the response of plants to salt stress. In this study, we attempted to initiate the identification of the cis-element in SOS1 mRNA responsible for the stability-regulation and signaling pathways mediating salt-induced mRNA stability.
Results
SOS1 mRNA stability is increased in response to Na+ and other abiotic stress treatments
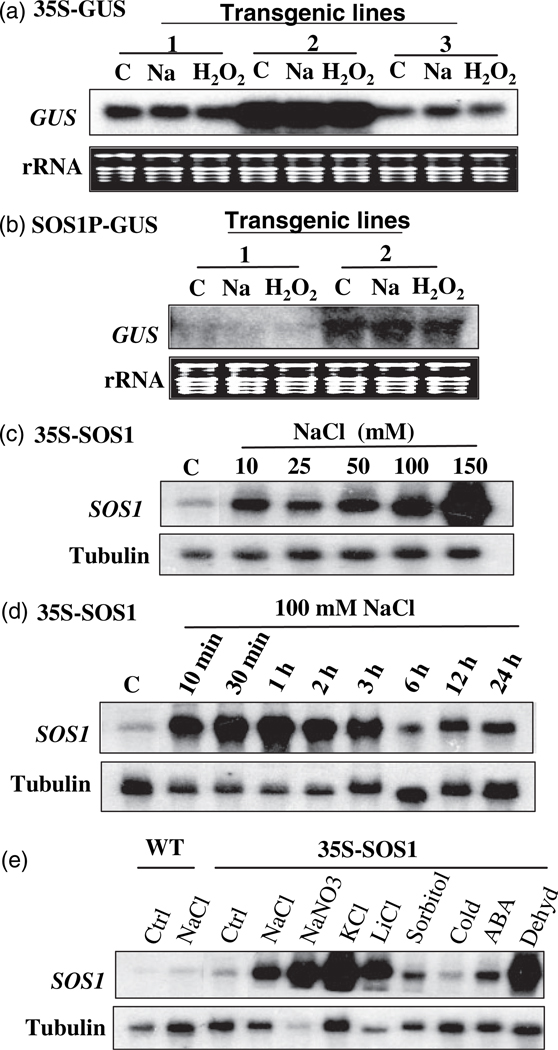
Our previous studies suggested that SOS1 mRNA might be inherently unstable, but that its stability is elevated in response to salt stress (Shi et al., 2003a). Whether the increase in SOS1 transcript level in the 35S:SOS1 transgenic plants upon NaCl treatment is due to increased stability of SOS1 mRNA was further determined in this study. As shown in Figure 1a, the expression of the GUS gene under the control of the 35S promoter was not affected by NaCl treatment, although the expression levels of the GUS gene in three independent lines are apparently different. The expression of other reporter genes, including GFP and aequorin, under the control of the 35S promoter was also tested, and no induction of gene expression was observed after NaCl treatment (data not shown). Therefore, the 35S promoter is indeed not a Na+-inducible promoter, and change in SOS1 transcript level in response to NaCl treatment in the 35S:SOS1 transgenic plants is attributed to mRNA stability. Moreover, NaCl treatment did not induce GUS gene expression under the control of the SOS1 promoter in the transgenic plants (Figure 1b), indicating that the SOS1 promoter is not NaCl-inducible and that the previously observed elevation of transcript level of the native SOS1 gene upon NaCl treatment (Shi et al., 2000) is due to enhanced mRNA stability. Thus, salt-induced SOS1 mRNA stability is an intrinsic mechanism and SOS1 expression in transgenic plants harboring 35S:SOS1 resembles an endogenous situation. The 35S:SOS1 transgenic plants were used to investigate the stability of SOS1 mRNA because SOS1 mRNA can accumulate to high levels under stress conditions and is easily monitored in these transgenic plants.
Figure 1. Stability of SOS1 mRNA is regulated by abiotic stresses.
(a) Northern blot detecting the transcripts of the GUS gene under the control of the 35S promoter. Three independent transgenic lines were used. C, control; Na, 150 mm NaCl treatment for 5 h; H2O2, 10 mm H2O2 treatment for 1 h.
(b) Northern blot detecting the transcripts of the GUS gene under the control of the SOS1 promoter. Two independent transgenic lines were used. C, control; Na, 150 mm NaCl treatment for 5 h; H2O2, 10 mm H2O2 treatment for 1 h.
(c) Induction of stability of SOS1 mRNA by NaCl at different concentrations in 35S:SOS1 transgenic plants.
(d) Time course of induction of stability of SOS1 mRNA by NaCl in 35S:SOS1 transgenic plants.
(e) Northern blotting of SOS1 transcripts in wild-type and 35S:SOS1 transgenic plants after different treatments. Salt treatments were done by treating the seedlings with 150 mm salt in 1/2 MS liquid medium for 5 h. Sorbitol: 300 mm sorbitol for 5 h; Cold, 0°C for 24 h; ABA, 100 µm ABA for 3 h; Dehyd, dehydration by drying seedlings on a filter paper for 15 min. Tubulin and rRNA were used as loading controls.
The induction of SOS1 transcripts by NaCl is concentration dependent (Figure 1c). Higher concentrations of NaCl resulted in more accumulation of SOS1 transcripts. Enhancement of the stability of SOS1 mRNA is rapid (Figure 1d). The SOS1 transcript level significantly increased after 10 min of NaCl treatment and was maintained at high levels after treatment with NaCl for 3 h. However, the transcript levels of SOS1 dropped after treatment with NaCl for longer period of time. To determine whether the increased SOS1 mRNA stability is specific to Na+ treatment, the effects of ionic stress, osmotic stress, cold, dehydration, and ABA on the stability of SOS1 mRNA were determined. As shown in Figure 1e, SOS1 mRNA levels in the 35S:SOS1 transgenic plants are also highly elevated by the treatment with KCl, LiCl, and dehydration, but only slightly increased by treatment with sorbitol and ABA, and unaffected by cold treatment. A marginal increase in SOS1 transcripts by the treatment with sorbitol at a concentration nearly iso-osmotic to NaCl revealed that osmotic stress plays a trivial role in the induction of SOS1 mRNA stability by salts and dehydration. Enhancement of the stability of SOS1 mRNA by different salts and dehydration treatments suggests that there might be common signal molecules elicited by these stress treatments that mediate regulation of SOS1 mRNA stability.
Stability of SOS1 mRNA is induced by H2O2 treatment
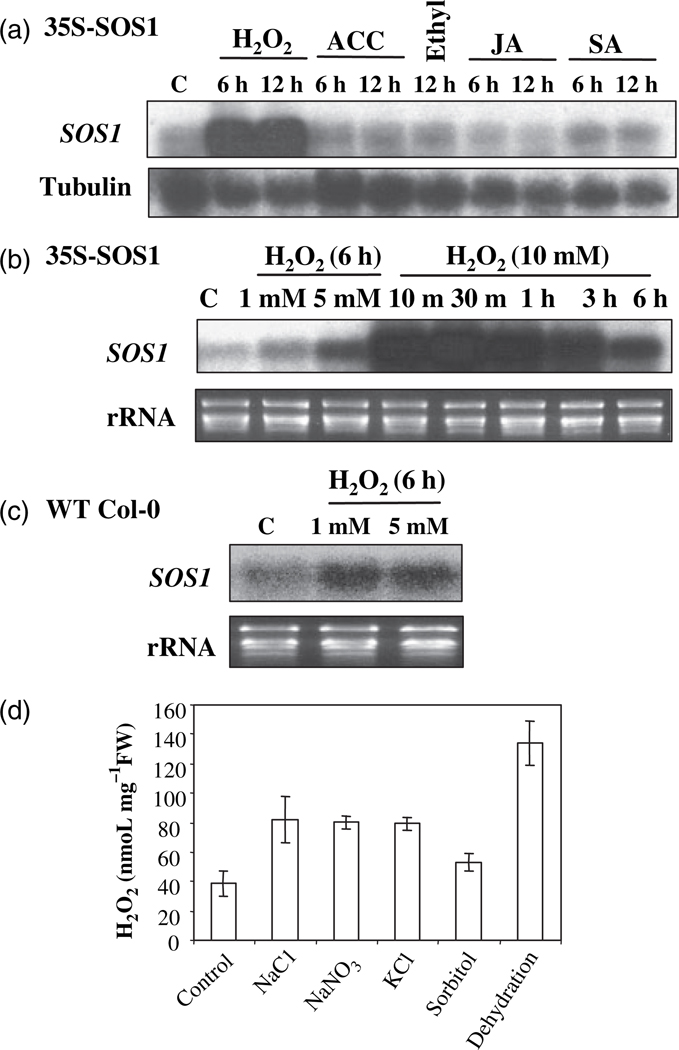
To seek the possible common signal molecules mediating the stress-regulated stability of SOS1 mRNA, the effects of different growth regulators and H2O2 on the stability of SOS1 mRNA were determined. As shown in Figure 2a, 1-aminocyclopropane-1-carboxylic acid (ACC; a precursor of ethylene biosynthesis), ethylene, methyl jasmonate and salicylic acid had no effect on the transcript level of SOS1. However, H2O2 treatment increased the SOS1 transcript level. Increase of SOS1 transcript level by H2O2 treatment is concentration dependent. Higher concentrations of H2O2 (from 1 mm to 10 mm) result in increased accumulation of SOS1 transcripts (Figure 2b). Stability of SOS1 mRNA is rapidly induced by H2O2 treatment. SOS1 transcripts reached peak level at only 10 min with H2O2 treatment, and dropped down at 6 h of H2O2 treatment (Figure 2b). H2O2 treatment also increased the abundance of the native SOS1 transcripts (Figure 2c), but had no effect on GUS gene expression under the control of the SOS1 promoter (Figure 1b). Furthermore, treatment with H2O2 had no effect on GUS gene expression driven by the 35S promoter (Figure 1a). Therefore, H2O2-induced expression of both native and transgene SOS1 is due to increased stability, but not enhanced transcription. Rapid elevation of SOS1 transcripts in response to H2O2 suggests that H2O2 and perhaps other ROS might serve as signaling molecules to mediate the regulation of SOS1 mRNA stability. Supporting this notion is the fact that different abiotic stress treatments enhanced H2O2 production in Arabidopsis seedlings (Figure 2d).
Figure 2. Stability of SOS1 mRNA is enhanced by H2O2 treatment.
(a) SOS1 transcript levels in 35S:SOS1 transgenic plants after treatment with H2O2 or plant growth regulators. H2O2, 10 mm H2O2; ACC, 10 µm 1-aminocyclopropane-1-carboxylic acid (ACC); Ethylene, 100 µl l−1 ethylene gas; JA, 100 µm methyl jasmonate; SA, 0.3 mm salicylic acid.
(b) The H2O2-induced stability of SOS1 mRNA is concentration dependent and the induction is rapid. SOS1 transcripts were detected in the 35S:SOS1 transgenic plants.
(c) The native SOS1 transcript level is also increased by H2O2 treatments.
(d) Salt and dehydration stress treatments promote the production of ROS in Arabidopsis seedlings. The stress treatments are the same as stated in Figure 1e.
ROS production is required for Na+-induced stability of SOS1 mRNA
Two ROS scavengers were used to test the involvement of ROS in salt-induced stability of SOS1 mRNA. Dimethylthiourea (DMTU), a synthetic antioxidant for hydroxyl radical scavenging, and deferoxamine (DF), an iron chelator to prevent hydroxyl radical formation from H2O2, were applied to the seedlings prior to NaCl treatment. As shown in Figure 3a, the induction of SOS1 mRNA stability by NaCl was remarkably reduced by the pre-treatments using these antioxidants, indicating that salt stress-induced stability of SOS1 mRNA is indeed mediated by ROS.
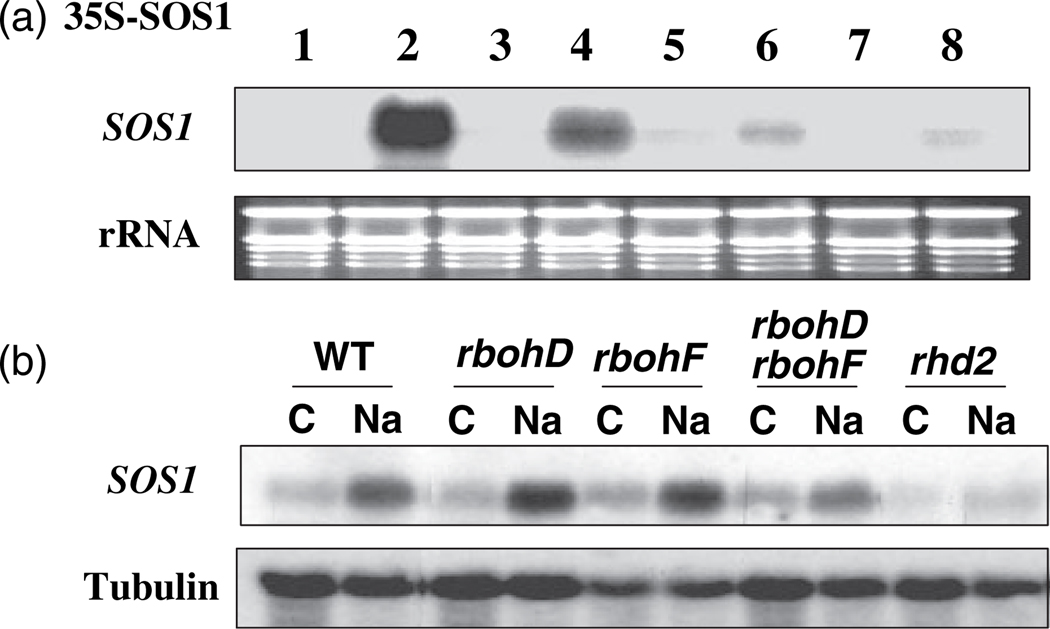
Figure 3. Salt-induced accumulation of SOS1 mRNA is mediated by ROS and dependent on NADPH oxidase activity.
(a) Pharmaceutical study. The SOS1 transcripts were detected in the 35S:SOS1 transgenic plants. The treatments are: 1, Control; 2, 150 mm NaCl for 1 h; 3, 100 µm diphenylene iodonium (DPI) for 3 h; 4, 100 µm DPI for 2 h followed by 150 mm NaCl with DPI for 1 h; 5, 15 mm DMTU for 3 h; 6, 15 mm DMTU for 2 h followed by 150 mm NaCl with DMTU for 1 h; 7, 1 mm DF for 3 h; 8, 1 mm DF for 2 h followed by 150 mm NaCl with DF for 1 h.
(b) Accumulation of native SOS1 transcripts in response to NaCl treatment in wild type and mutants defective in NADPH oxidases. C, control; Na, 200 mm NaCl for 5 h.
Plasma membrane-bound NADPH oxidases have been implicated in ROS production in the apoplastic compartment during pathogen attack and under abiotic stress (Torres and Dangl, 2005). Diphenylene iodonium (DPI) is a potent NADPH oxidase inhibitor and has been used extensively to study the functions of NADPH oxidase. Treatment with DPI alone had little effect on SOS1 mRNA levels (Figure 3a). However, pre-treatment with DPI prior to NaCl treatment significantly attenuated Na+-induced SOS1 mRNA stability (Figure 3a). These results suggest that salt stress may trigger the activation of NADPH oxidases, which results in the production of apoplastic ROS that mediate regulation of the stability of SOS1 mRNA.
The involvement of NADPH oxidases in salt-induced SOS1 mRNA stability was further evaluated by using genetic mutants defective in NADPH oxidases. Mutants of three NADPH oxidases previously shown to produce ROS (Torres and Dangl, 2005) were examined for the accumulation of native SOS1 mRNA. As expected, the SOS1 mRNA level was increased after NaCl treatment in wild-type Arabidopsis plants (Figure 3b). In rbohD and rbohF single mutants and rbohDrbohF double mutants, salt-induced accumulation of SOS1 mRNA was not significantly affected (Figure 3b). However, salt-induced accumulation of SOS1 mRNA was completely abolished in rhd2 mutant plants (Figure 3b), suggesting that Rhd2/RbohC is the major source of production of ROS mediating the control of SOS1 mRNA stability.
Rapid decay of SOS1 mRNA requires new protein synthesis
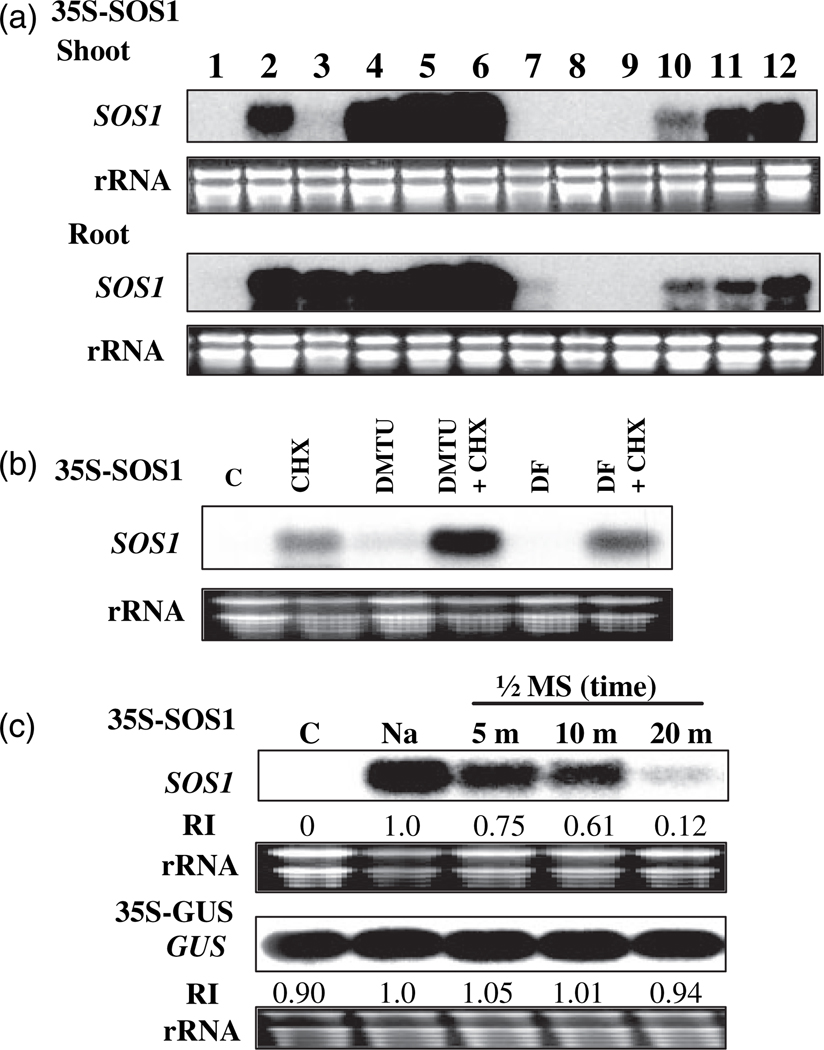
The decay of RNA is executed by protein complexes, including exonucleases, RNA-associated proteins (e.g. PABPs) and sequence-specific RNA-binding proteins. Protein binding on the specific sequence of an mRNA can result in either stabilization or destabilization. Furthermore, mRNA stability can be controlled in a cell- or tissue-specific manner, which results in spatial and temporal gene expression. To determine whether the stability of SOS1 mRNA is different in roots and leaves in response to salt stress, the 35S:SOS1 seedlings were treated with NaCl and roots and shoots were collected separately. Northern hybridization indicated that in both shoot and root SOS1 mRNA is unstable under normal growth conditions, but that its stability is greatly increased after NaCl treatment (Figure 4a, lanes 1 and 2). The involvement of protein biosynthesis in the control of SOS1 mRNA stability was also determined by applying treatments with cycloheximide (CHX), a commonly used translation inhibitor. Treatment with only CHX highly induced accumulation of SOS1 mRNA in both leaves and roots (Figure 4a, lanes 3 and 4). Treatment with CHX for 1 h resulted in high accumulation of SOS1 mRNA in roots, but not in leaves, presumably because CHX did not reach the aerial part of the seedlings. Cycloheximide-induced accumulation of SOS1 mRNA was detected in both roots and leaves after treatment with CHX for 5 h. Cycloheximide treatment and NaCl treatment showed an additive effect on accumulation of SOS1 mRNA (Figure 4a, lanes 5 and 6). The decay of SOS1 mRNA is rapid. Accumulated SOS1 mRNAs were degraded to an undetectable level within 2 h of the seedlings being released from salt treatment (Figure 4a, lane 7). Cycloheximide treatment attenuated the decay of SOS1 mRNA, presumably by inhibition of the synthesis of new protein (Figure 4a, comparing lanes 7, 8, and 9 with 10, 11, and 12). Stabilization of SOS1 mRNA by CHX is time dependent (Figure 4, lanes 10, 11, 12). Pre-treatments with DMTU or DF had no effect on CHX-induced accumulation of SOS1 mRNA (Figure 4b), unlike their effects on NaCl-induced accumulation of SOS1 mRNA (Figure 3a). Therefore, CHX-induced stability of SOS1 mRNA is due to inhibition of protein translation, not ROS production. These results suggest that proteins (perhaps a short-lived RNA-binding protein and other proteins for mRNA decay) are required for the decay of SOS1 mRNA and naked SOS1 mRNAs or SOS1 mRNAs associated with long-lived RNA-binding proteins are more stable.
Figure 4. SOS1 mRNA decay is rapid and requires new protein synthesis Cycloheximide (CHX) was used to inhibit protein translation.
(a) SOS1 mRNA stability in shoot and root is induced by CHX treatments. 1, control; 2. 200 mm NaCl for 5 h; 3. CHX 100 µm for 1 h; 4, CHX 100 µm for 5 h; 5, CHX 100 µm for 1 h followed by 200 mm NaCl for 5 h; 6, CHX 100 µm for 1 h followed by 200 mm NaCl with 100 µm CHX for 5 h; 7, 200 mm NaCl for 5 h followed by 1/2 MS salt solution for 2 h; 8, 200 mm NaCl for 5 h followed by 1/2 MS salt solution for 12 h; 9, 200 mm NaCl for 5 h followed by 1/2 MS salt solution for 24 h; 10, 200 mm NaCl for 5 h followed by 100 µm CHX for 2 h; 11, 200 mm NaCl for 5 h followed by 100 µm CHX for 12 h; 12, 200 mm NaCl for 5 h followed by 100 µm CHX for 24 h.
(b) Cycloheximide-induced stability of SOS1 mRNA is not due to elicited production of reactive oxygen species. C, control; CHX, CHX 100 µm for 1 h; DMTU, 15 mm DMTU for 3 h; DMTU+CHX, 15 mm DMTU for 2 h followed by 100 µm CHX with DMTU for 1 h; DF, 1 mm DF for 3 h; DF+CHX, 1 mm DF for 2 h followed by 100 µm CHX with DF for 1 h.
(c) Northern blotting showing rapid decay of SOS1 mRNA. Ten-day-old 35S:SOS1 and 35S:GUS transgenic seedlings were treated with 200 mm NaCl for 5 h (indicated as Na) and then transferred to 1/2 MS liquid medium. Seedlings were harvested at indicated time points for RNA isolation. C, control; RI, relative signal intensity. rRNA was used as a loading control.
Rapid decay of SOS1 mRNA was determined by measuring the decay rate of SOS1 and GUS transcripts. The 35S:SOS1 transgenic seedlings were treated with NaCl and then transferred into 1/2 MS medium without NaCl. As shown in Figure 4c, the transcript level of SOS1 was reduced to 75% within 5 min of the seedlings being released from NaCl stress. The transcript levels of SOS1 were only 61% and 12% at 10 and 20 min, respectively, after seedlings were released from salt stress. Thus, the half-life of SOS1 mRNA is approximately 10 min. In contrast, the transcript levels of the GUS gene were relatively constant with or without NaCl treatment and after release from salt stress (Figure 4c). Therefore, SOS1 mRNA is inherently unstable under normal growth conditions.
An approximate 500-bp region in SOS1 mRNA is required for its instability
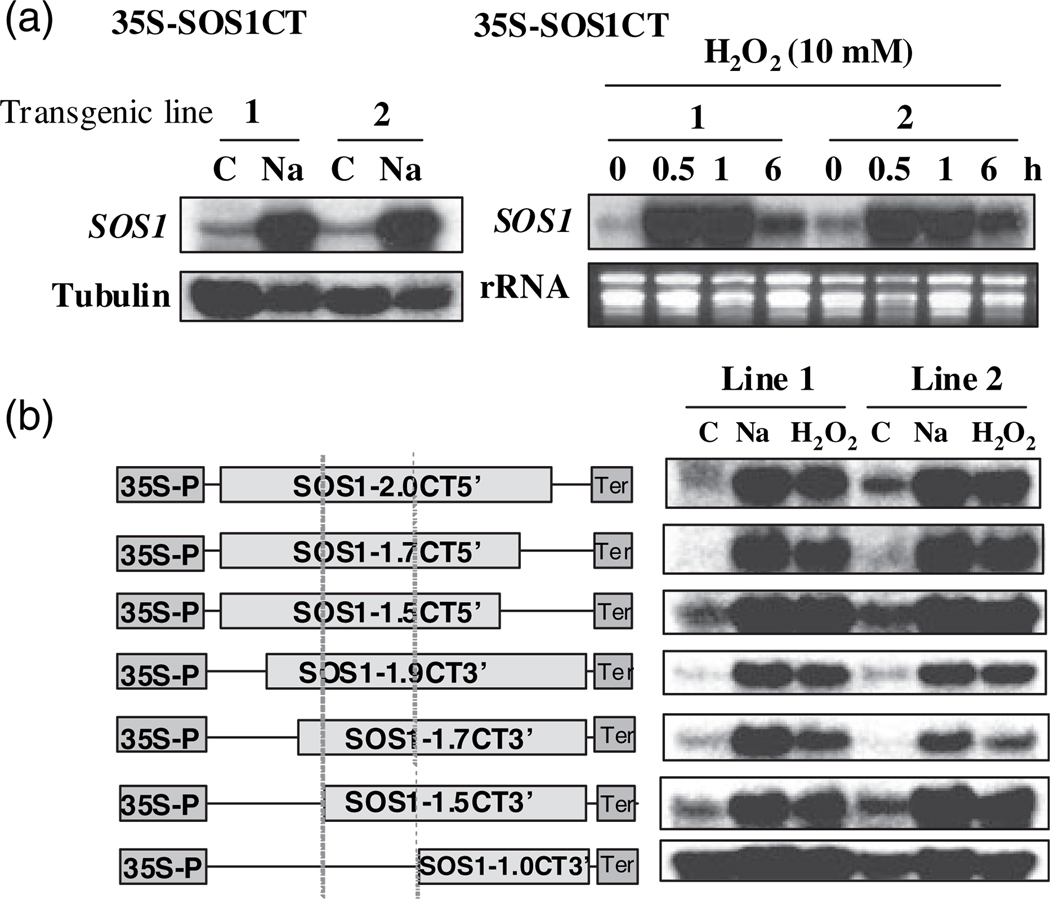
The decay rates of specific mRNAs are determined by sequence elements that are generally located within the 3′-UTR, but have also been observed within the open reading frame (ORF), intron, or 5′-UTR. Since the expression cassette of 35S:SOS1 contained only the coding region of SOS1 cDNA, instability/stability of mRNA is not attributed to decay processes involving cis-elements in UTRs or introns or to pre-mRNA processing. Nonsense-mediated decay (NMD) and non-stop decay (NSD) mechanisms are excluded as well, since the sequence analysis of the cDNA of the coding region revealed the presence of only the appropriate stop codon, and overexpression of the cDNA can complement the sos1-1 NaCl-sensitive phenotype (Shi et al., 2000). Therefore, instability of SOS1 mRNA and salt-induced stability is controlled by transcript-specific cis-elements in the SOS1 coding region. To map the region that is responsible for instability and salt-induced stability of SOS1 mRNA, SOS1 cDNA encoding only the entire C-terminus of the protein (amino acids 447–1146) or SOS1 cDNA encoding only the transmembrane portion (amino acids 1–547) were expressed in Arabidopsis under the control of the 35S promoter. Northern analysis indicates that mRNA derived from the DNA sequence encoding the SOS1 C-terminus are unstable without stress treatments but are stabilized by both NaCl and H2O2 (Figure 5a). Thus, the region responsible for instability and salt-induced stability of SOS1 mRNA resides in the mRNA region encoding the C-terminal cytosolic portion of SOS1. To further dissect the sequence motif, a series of deletions were made (Figure 5b). These constructs were transformed into Arabidopsis and T2 transgenic plants were obtained. Four independent lines of transgenic plants harboring each construct were subjected to both NaCl and H2O2 treatments. Northern analysis revealed that a cDNA fragment of about 500 nucleotides (nt) within the region encoding the SOS1 cytosolic C-terminus is required for instability of SOS1 mRNA (Figure 5b). Deletion of this 500-nt region resulted in increased stability of the mRNA under normal growth conditions (Figure 4b, construct SOS1-1.0CT3′). The expression levels in the transgenic plants harboring SOS1-1.0CT3′ treated with NaCl or H2O2 are similar to those transgenic plants harboring constructs containing the approximate 500-nt region (Figure 5b). Therefore, the approximate 500-nt region confers SOS1 mRNA instability under normal growth conditions rather than conferring induced stability by NaCl and H2O2. Analysis of the RNA sequence in this approximate 500-nt region by using several programs predicting RNA secondary structures revealed a putative secondary structure containing stem–loop structures that are characteristic of those that interact with RNA-binding proteins (Supplementary Figure S1). Interestingly, a putative small RNA target (DBE# 874) within this region was also identified by searching the small RNA database ASRP (http://asrp.cgrb.oregonstate.edu/db/). Although some mismatches between the small RNA and the target sequence in SOS1 mRNA are present, involvement of small RNA in the degradation of SOS1 mRNA is possible.
Figure 5. Identification of the cis-element required for the regulation of the stability of SOS1 mRNA.
(a) The cis-element is located in the 2.2-kb region encoding the C-terminal cytosolic portion of SOS1 protein. C, control; Na, 200 mm NaCl treatment for 5 h. Tubulin and rRNA were used as loading controls.
(b) The approximate 500-bp sequence within the 2.2 kb is required for instability of SOS1 mRNA. Deletion constructs of the 2.2 kb (left panel) were made and transgenic plants were obtained as described in Experimental procedures. Two independent T2 transgenic lines were used to detect the mRNA level in the transgenic plants (right panel). Vertical dotted lines show the sequence region (approximately 500 bp) required for SOS1 mRNA instability. C, control; Na, 200 mm NaCl treatment for 5 h; H2O2, 10 mm H2O2 treatment for 1 h. rRNA (as a loading control) is not shown.
Mutations in the SOS1 gene render mutants tolerant to oxidative stress
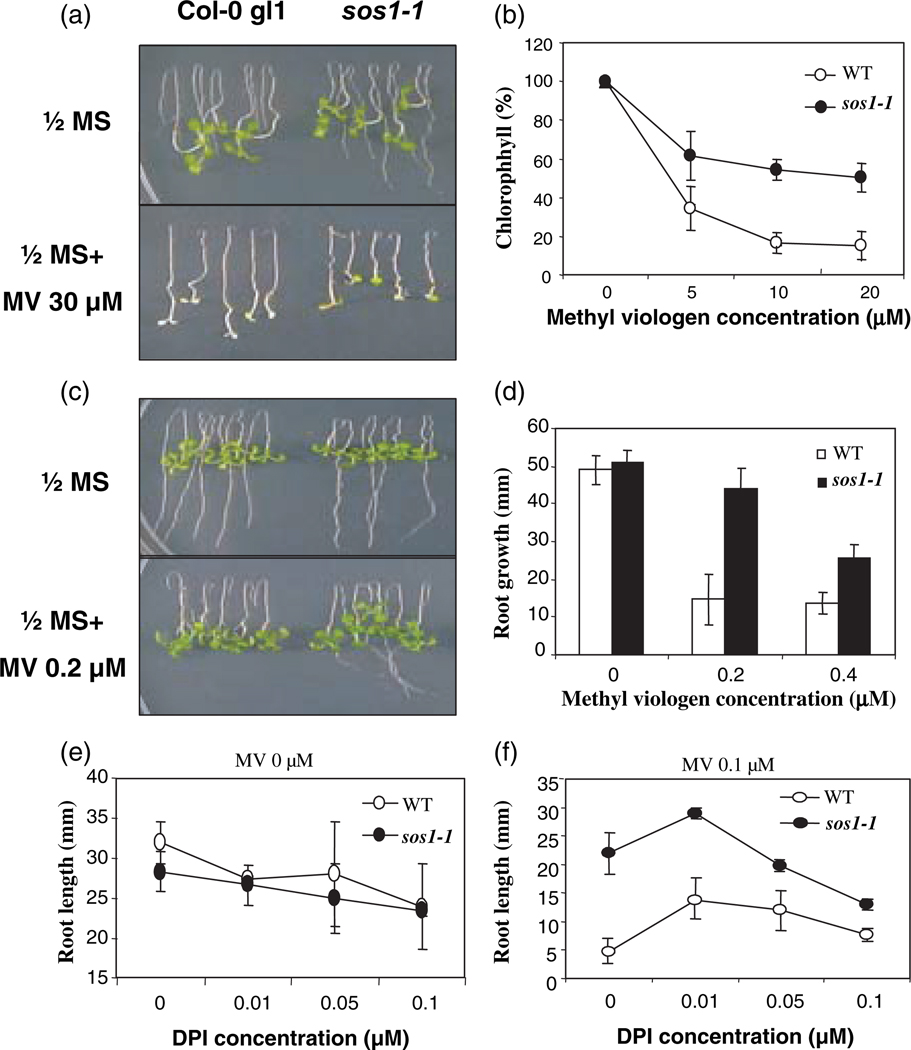
Our finding that salt-induced stability of SOS1 mRNA is mediated by ROS suggests that SOS1 possibly plays a role in the response of the plant to oxidative stress. After testing several ROS-generating compounds, the sos1 mutant seedlings were found to be significantly more tolerant to methyl viologen (MV; also called paraquat, PQ) than wild-type seedlings (Figure 6). Although root growth of both sos1 mutant and wild-type seedlings was completely inhibited at high concentrations of MV, leaves of sos1 mutants remained green while wild-type leaves were totally bleached (Figure 6a). Consistent with its greener color, the chlorophyll content in sos1 mutants was substantially higher than that in wild-type seedlings (Figure 6b). At low concentrations of MV, the root growth of sos1 mutants was much less inhibited compared than the wild-type control (Figure 6c,d). These results suggest that SOS1 functions to render Arabidopsis plants more sensitive to MV. Since our results indicated that the ROS-generating enzyme NADPH oxidase is involved in the control of NaCl-induced stability of SOS1 mRNA (Figure 3), the involvement of this enzyme in MV sensitivity conferred by SOS1 was determined. Low DPI concentrations were used because at these concentrations the root growth of both sos1 mutant and wild-type seedlings was not significantly inhibited (Figure 6e). With 0.1 µm MV only, sos1 root length was fourfold greater that of the wild type. However, addition of 0.1 µm DPI decreased this difference to only 50% (Figure 6f). Interestingly, the MV tolerance of both wild-type and sos1 mutant seedlings was enhanced by the addition of 0.01 µm DPI in the growth medium (Figure 6f). These results indicate that MV sensitivity conferred by SOS1 is dependent on NADPH oxidase activity.
Figure 6. sos1 mutants are more tolerant of methyl viologen (MV).
(a) Root-bending assay of wild-type and sos1 mutant seedlings gown on the 1/2 MS agar medium with or without 30 µm MV.
(b) Measurements of chlorophyll content.
(c) Root-bending assay of wild-type and sos1 mutant seedlings grown on the 1/2 MS agar medium with or without 0.2 µm MV.
(d) Quantification of root growth in response to different concentrations of MV.
(e), (f) Effects of diphenylene iodonium (DPI) on MV tolerance of wild-type and sos1 mutant seedlings. Note that the sensitivity to MV is significantly affected by the presence of low concentrations of DPI (f), at which root growth of both the wild type and sos1 mutants is not significantly affected (e).
Discussion
We have provided evidence that abiotic stress-induced stability of SOS1 mRNA is mediated by ROS. NADPH oxidase activity is at least partially required for the ROS generation needed to stabilize SOS1 transcripts. The role of SOS1 in tolerance of NaCl and oxidative stress is complex, apparently owing to the participation of SOS1 in the biochemistry of Na+ homeostasis and perhaps in signal transduction as well. This is attested to by the fact that sos1 mutants are hypersensitive to NaCl (Shi et al., 2000), but more tolerant of MV (Figure 6; Katiyar-Agarwal et al., 2006).
Reactive oxygen species have long been proposed as signal molecules that regulate various processes such as growth, development, responses to biotic and abiotic environmental stimuli, and programmed cell death (Apel and Hirt, 2004). Several lines of evidence indicate that the plant homolog of NADPH oxidase is involved in the respiratory burst (Rboh) and can initiate, and most likely amplify, ROS production for the purpose of signaling (Foreman et al., 2003; Sagi and Fluhr, 2006; Torres and Dangl, 2005). Plant Rboh proteins contain N-terminal EF-hand calcium-binding motifs and can be activated directly by Ca2+ in vitro (Sagi and Fluhr, 2001). Thus, Ca2+ elevation may be an upstream signaling event that links environmental stimuli and ROS production via NADPH oxidases. Mitogen-activated protein kinase (MAPK) may transduce ROS signals through phosphorylation of target proteins (Pitzschke and Hirt, 2006). The ROS-mediated regulation of mRNA stability can be a rapid signal transduction process, since ROS may mediate signals by affecting targets that are constitutively present in the cells including Ca2+ channels, membrane-bound Rboh proteins, and substrate proteins that are phosphorylated by MAPKs. In fact, the elevation of SOS1 mRNA stability elicited by salt stress and H2O2 treatment is quite rapid. The peak accumulation of SOS1 mRNA appears only about 10 min (or perhaps in an even shorter period of time) after NaCl or H2O2 treatment (Figures 1d and 2b). The stabilization and resulting increase in SOS1 mRNAs could lead to accumulation of SOS1 transporter proteins in the plasma membrane and enhance Na+ exclusion, thus rapidly conferring salt tolerance to plant cells. It has been reported that treatment of rice seedlings with low levels of H2O2 could improve salt and heat tolerance (Uchida et al., 2002). The involvement of MAPKs in signaling control of the stability of SOS1 mRNA is supported by our observation that some MAPK inhibitors can reduce accumulation of SOS1 mRNA under conditions of salt stress (data not shown).
One of the most important functions of the human Na+/H+ exchangers (NHE) is regulation of intracellular pH (Counillon and Pouyssegur, 2000). In plants, vacuolar Na+/H+ antiporters also have been implicated in pH regulation. A mutation in an NHX-like gene of Ipomoea nil (InNHX1) that encodes a vacuolar Na+/H+ antiporter abrogated the capacity of cells to increase vacuolar pH, a requirement for shift of the flower color from reddish-purple to blue (Fukada-Tanaka et al., 2000). The plasma membrane Na+/H+ antiporter SOS1 has also been shown to affect H+ transport even in the absence of salt stress (Shabala et al., 2005). Increased SOS1 activity would lead to apoplastic alkalinization and cytoplasmic acidification. It has been proposed that apoplastic alkalinization might be a general stress response and change in pH could serve as a signal to mediate gene regulation (Felle, 2001; Felle and Hanstein, 2002; Felle et al., 2004, 2005). Thus, SOS1, by promoting apoplastic alkalinization, could play a signaling role in the stress response. Apoplastic alkalinization may also be essential for the activation and/or maintenance of Rboh NADPH oxidase activity and subsequent ROS production. Activation of NADPH oxidases was found to be preceded by apoplastic alkalinization, which is thought to result from elicitor-induced depolarization of the plasma membrane and subsequent K+/H+ exchange, followed by Ca2+ influx/Cl− efflux (Nurnberger and Scheel, 2001; Simon-Plas et al., 1997; Zhao et al., 2005). Moreover, extracellular alkalinization has been shown to be required for the oxidative burst (Bindschedler et al., 2001; Bolwell et al., 1995, 1999). It appears that a change inpHis essential for the initiation of the apoplastic oxidative burst that occurs in response to biotic stress (Bolwell et al., 2002). A recent study has shown that touch stimulation triggers elevation of cytosolic Ca2+, which is required for rapid (within 1 sec) and transient apoplastic alkalinization and cytosolic acidification, as well as production of extracellular ROS in root hairs (Monshausen et al., 2006). In the Arabidopsis mutant rhd2, which lacks the functional NADPH oxidase, touch stimulation can still trigger pH changes but not an increase in ROS production (Monshausen et al., 2006), suggesting that pH changes precede extracellular ROS production. It is conceivable that both biotic and abiotic stresses could activate a common signal transduction pathway that involves changes in pH and the production of extracellular ROS. SOS1 could be one of the signaling components upstream of apoplastic alkalinization.
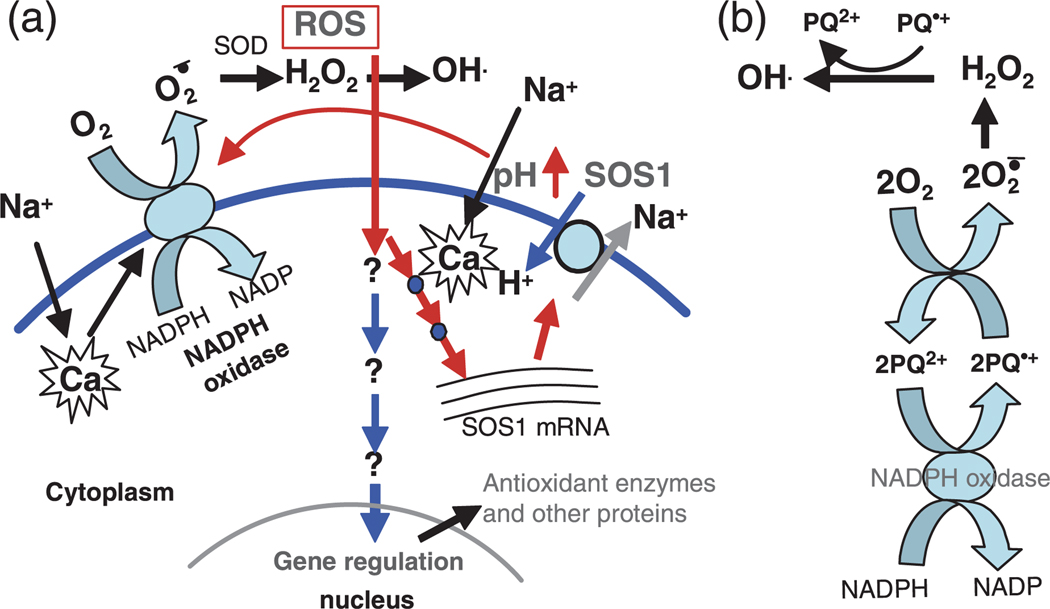
Based on our results and previous studies, we propose the following hypothesis. Salt and other stresses elicit elevation of Ca2+, which activates the SOS signaling pathway. The activation of SOS1 would cause rapid apoplastic alkalinization and cytosolic acidification. This pH change may then activate the plasma membrane-bound NADPH oxidase. Alternatively, salt and other abiotic stresses capable of inducing stability of SOS1 mRNA (such as dehydration) may not employ the SOS signaling pathway, but instead could directly activate NADPH oxidase by inducing a Ca2+ spike specifically localized to or recognized by NADPH oxidase. All plant NADPH oxidases carry a presumably cytosolic 300-amino-acid amino-terminal extension with two EF-hands that bind Ca2+ (Keller et al., 1998). Activation of plasma membrane-bound NADPH oxidase promotes the production of apoplastic superoxide anion (O2−). Superoxide is then catalytically converted by the action of superoxide dismutase (SOD) to hydrogen peroxide (H2O2). In the presence of transition metals such as Fe2+, the extremely reactive hydroxyl radical (OH) can be generated from H2O2. It is likely that SOS1 serves as a fine-tuner of pH, thus maintaining NADPH oxidase activity and ROS production under stress conditions. This notion is supported by our results showing that H2O2 concentrations are higher in SOS1 overexpression transgenic plants than in wild-type plants with or without salt stress (data not shown). Stabilization of SOS1 mRNA may also serve to activate a positive feedback loop. In this feedback loop, SOS1 would be required for activation and maintenance of NADPH oxidase activity. The ROS generated by NADPH oxidase would then stabilize SOS1 mRNA, thereby greatly increasing its activity and subsequently NADPH oxidase activity (Figure 7a). Rapid elevation of the level of SOS1 mRNA upon salt stress and fast degradation after release from stress could exert a fine control on SOS1 protein homeostasis. This could be crucial for maintaining optimal extracellular and intercellular pH during normal growth and after salt stress exposure.
Figure 7. A proposed signaling pathway controlling SOS1 mRNA stability and SOS1-conferred paraquat sensitivity.
(a) Hypothetical model showing the early signaling events and downstream signal transduction regulating the stability of SOS1 mRNA. Under Na+ stress, operation of SOS1 would cause extracellular pH elevation, which could be required for activation and/or maintenance of the plasma membrane-bound NADPH oxidase activity that produces extracellular reactive oxygen species (ROS). Extracellular ROS could serve as signaling molecules to trigger gene regulation. The stability of SOS1 mRNA could be increased through a positive feedback regulation upon salt stress.
(b) Redox cycling of paraquat (PQ) coupling with the plasma membrane NADPH oxidases. The plasma membrane-bound NADPH oxidase could be an enzymatic source of electrons for the formation of free radical paraquat (PQ+) from normal divalent cation paraquat (PQ2+). Rapid reoxidation of PQ+ causes production of ROS through transfer of electrons to molecular oxygen. The PQ redox cycling, which causes enormous extracellular ROS production deleterious to plant cells, could be dependent on SOS1 activity that would be required for activation and/or maintenance of the NADPH oxidase activity. See the Discussion in the text for a detailed explanation.
This model may also explain why mutations in SOS1 confer tolerance to PQ/MV. Paraquat has a redox potential of −446 mV, which is critical to its function as a free-radical generator. In the presence of paraquat, NADPH oxidase could be an enzymatic source of electrons, causing the divalent cation paraquat2+ (PQ2+) to form a free radical paraquat1+ (PQ+). Since this molecule is unstable, it is rapidly reoxidized in the presence of oxygen to yield the original divalent cation. During this oxidation process, electrons are transferred to molecular oxygen to form superoxide anion radicals (O2−). Superoxide radicals are then enzymatically converted to H2O2, which could subsequently produce toxic hydroxyl radicals (Figure 7b). The PQ2+ ion would then be available again for another cycle. Thus this herbicide would function as a catalyst for transferring electrons to oxygen. In this paradigm, formation of free radicals promoted by PQ would require activity of the plasma membrane NADPH oxidase. In fact, NADPH oxidase has been identified as an enzymatic source of electrons that trigger PQ redox cycling and extracellular ROS production in mammalian cells (Bonneh-Barkay et al., 2005; Lee et al., 1990; Wu et al., 2005). According to our model (Figure 7a), SOS1 plays a pivotal role in control of NADPH oxidase activity and apoplastic ROS production. When SOS1 is functional, the activity of NADPH oxidase would be maintained, which enhances PQ redox cycling and extracellular ROS production. Since apoplastic ROS could either be signaling molecules at low concentration or cause cell damage by oxidative stress at high concentration, a large amount of ROS produced through PQ redox cycling would be extremely toxic to plant cells. In contrast, NADPH oxidase activity in the sos1 mutant, according to our model, would be lower than in the wild type due to lack (or reduction) of extracellular alkalinization, which results in less efficient PQ redox cycling and reduced production of extracellular ROS and consequently reduced damage to the plant cells. This is also consistent with our observation that inhibition of NADPH oxidase activity by DPI increased the resistance of Arabidopsis seedlings to PQ (Figure 6f).
Our study on the stability of SOS1 mRNA has revealed the early signaling events involving post-transcriptional control of gene expression. The possible role of SOS1 in regulating apoplastic pH and extracellular ROS production would putatively position SOS1 at a very early signaling step of a signal transduction pathway that is common to several abiotic stresses and is probably initiated by pH alterations. This hypothesis deserves further experimental validation. Moreover, identification of signaling components that mediate stress-induced SOS1 mRNA stability will greatly improve our understanding of stress adaptation.
Experimental procedures
Plant materials and growth conditions
The SOS1 overexpressed transgenic plants were generated as previously described (Shi et al., 2003a). Briefly, SOS1 cDNA containing the complete ORF was subcloned into the vector pIG121-Hm by replacing the GUS gene between the XbaI and SacI sites, resulting in a construct for overexpression of the SOS1 gene under the control of the CaMV 35S promoter. Transgenic Arabidopsis plants harboring this construct were generated and two homologous lines were selected for the study of SOS1 mRNA stability. The vector pIG121-Hm harboring the GUS gene was used to transform Arabidopsis, and transgenic plants were obtained (35S:GUS) and used as controls in the mRNA stability study. Arabidopsis transgenic plants harboring the SOS1 promoter–GUS fusion were obtained as previously described (Shi et al., 2002a). An approximate 2.2 kb sequence of the SOS1 cDNA encoding the C-terminus of the SOS1 protein and an approximate 1.2 kb sequence of the SOS1 cDNA encoding the N-terminal transmembrane domain of SOS1 protein were subcloned into the vector pCAMBIA1207, resulting in transcriptional fusions of the 35S promoter and part of the SOS1 cDNA sequences. Transgenic plants with these constructs, designated as 35S-SOS1CT and 35S-SOS1NT, were obtained, and independent T2 lines were used for the mRNA stability study. The reporter genes EGFP and aequorin were also cloned into the vector pCAMBIA1207, and transgenic plants were generated and used as controls. Unless otherwise stated, Arabidopsis plant seeds were surface sterilized and rinsed with sterile water. The seeds were then suspended in sterile 0.3% (w/v) low-melting-point agarose. After being treated at 4°C for 3 days, the seeds were planted onto 1/2 MS nutrients agar medium (1/2 MS salts, 1.5% sucrose, 0.7% agar, pH 5.7). The plates were placed in a growth chamber at 22°C under a daily cycle of 16 h light and 8 h dark.
Stress treatments and northern hybridization
Ten-day-old seedlings gown on 1/2 MS agar medium were used for different treatments. For salt treatment, the seedlings were transferred onto a Whatman filter paper soaked with NaCl or other salts at different concentrations and treated for different lengths of time as indicated in the figures. For H2O2 treatment, the seedlings were sprayed with H2O2 at different concentrations and kept for different lengths of time as indicated in the figures. Seedlings grown on 1/2 MS agar medium were put in a low-temperature incubator and exposed to 0°C for 24 h as cold treatment. In the ABA treatment, seedlings were sprayed with 100 m ABA solution and kept for 3 h. For the dehydration treatment, seedlings were transferred onto a filter paper and dehydrated for 15 min. Seedlings were sprayed with 10 µm ACC, 100 µm methyl jasmonate, or 300 µm salicylic acid, respectively, and kept for different lengths of time as indicated in Figure 2a. For ethylene treatment, seedlings were placed into a sealed chamber with ethylene gas at a concentration of 100 µl µm for 12 h. For 100 µm DPI, 15 µm DMTU, and 1 µm DF treatments, 10-day-old seedlings were transferred onto a Whatman filter paper soaked with the chemical solution in 1/2 MS liquid medium for 3 h as a control treatment. NaCl was added after 2 h of treatment with these chemicals, and the seedling plants were treated for an additional hour. For CHX treatment, 10-day-old seedling plants were transferred onto a Whatman filter paper soaked with 100 µm CHX for 1 h or 5 h. Combinational treatments by NaCl and CHX are indicated in Figure 4. To determine the mRNA decay rate, the seedling plants were treated with 200 µm NaCl for 5 h and then transferred into 1/2 MS liquid medium without NaCl for 5, 10, or 20 min. Total RNA was extracted from Arabidopsis seedlings according to the method described in Shi and Bressan (2006). Total RNA was separated by electrophoresis and blotted onto a nylon membrane using a vacuum blotter (Bio-Rad model 785 vacuum blotter; http://www.bio-rad.com/). Northern hybridization was carried out at 60°C with 32P-dCTP-labeled specific probes. Blots were washed once in 0.1% SDS and 0.5× SSC for 20 min at RT and twice in 0.1% SDS and 0.2× SSC for 20 min at 50°C. Images were obtained by exposing X-ray films to the blots, and quantification of the bands was carried out by using imagej software.
H2O2 measurement
Quantitative measurement of H2O2 production was performed as described by Shin and Schachtman (2004). Briefly, 10-day-old seedlings were treated with different ionic, osmotic and dehydration stresses as stated in Figures 1e and 2d. The samples were ground in liquid nitrogen, and 200 mg of ground tissues from each sample was placed in an Eppendorf tube and kept frozen. Two hundred microliters of 20 mm sodium phosphate buffer (pH 6.5) was immediately added into the tube and mixed. The extraction was centrifuged at 9500 g for 10 min at 4°C and the supernatant was used for assay. H2O2 concentration was measured using an Amplex red hydrogen peroxide/peroxidase assay kit (Molecular Probes, http://probes.invitrogen.com/). The fluorescence was detected by a fluorospectrometer (Fluorolog FL-1039, Horibagroup; http://www.isainc.com).
Deletion analysis of SOS1 mRNA
The respective deletion constructs (see Figure 5b) were made by inserting the deletion sequences into the cloning site of the vector pCAMBIA1207. The forward primer for SOS1-2.0CT5′, SOS1-1.7CT5′, and SOS1-1.5CT5′ constructs is: 5′-GACTCCATGGGTTCTACGCCTTCTTCGCATGG-3′. The reverse primers for SOS1-2.0CT5′, SOS1-1.7CT5′, and SOS1-1.5CT5′ constructs are: 5′-CAGTGGATCCTCACTGACACGCATGTTTACGG-3′, 5′-CAGTGGATCCTCAGATCTCTTGTT GTTGTTTGGC-3′, and 5′-CAGTGGATCCTCAGCTTTGATTCCCGTTAGAAGG-3′, respectively. The forward primers for SOS1-1.9CT3′, SOS1-1.7CT3′, SOS1-1.5CT3′, and SOS1-1.0CT3′ constructs are 5′-GACTCCATGGTTCATCATCCTCACAATGG-3′, 5′-GACTCCATGGGAGGTCTAAAACCACATGTC-3′, 5′-GACTCCATGGA GAGTAATATTGGTTCC-3′, and 5′-GACTCCATGGGACTCTACGAA GTCCTC-3′, respectively. The reverse primer for SOS1-1.9CT3′, SOS1-1.7CT3′, SOS1-1.5CT3′, and SOS1-1.0CT3′ constructs is: 5′-CAGTGGATCCTCATAGATCGTTCCTGAAAACG-3′.
Determination of sensitivity to methyl viologen/paraquat
A root-bending assay (Shi et al., 2002b, 2003b) was used for root growth measurements in response to MV and DPI. Four-day-old seedlings of wild-type and sos1-1 mutants grown on vertical 1/2 MS agar (1.2%) plates were transferred to 1/2 MS agar (1.2%) medium supplemented with different concentrations of MV and DPI. The plates were placed vertically with seedlings in an upside-down position. The root length was measured at the 6th, 9th, and 15th days after transfer. For measurement of chlorophyll content, seedlings were ground in 90% acetone and then centrifuged at 9500 g for 5 min. The extract was used for chlorophyll measurement. Chlorophyll content was determined by measuring OD at 647, 664, and 750 nm using a spectrophotometer (SmartSpec® Plus, Bio-Rad). The equations for calculation of chlorophylls were Chl a = (11.93*OD664)−(1.93*OD647) and Chl b = (20.36*OD647)−(5.5*OD664) (e.g. OD664 = Abs664−Abs750) (Jeffrey and Humphrey, 1975).
Supplementary Material
Acknowledgements
This work was supported by the US Department of Agriculture National Research Initiative competitive grants 2004-35 100-14 863 to PMH and HS. The rbohD, rbohF, and rbohDrbohF mutants were kindly provided by Dr Jeffery L. Dangl.
Footnotes
Supplementary Material
The following supplementary material is available for this article online:
Figure S1. Predicted secondary structures of the 500-nt region in SOS1 mRNA.
This material is available as part of the online article from http://www.blackwell-synergy.com
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant. Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Bindschedler LV, Minibayeva F, Gardner SL, Gerrish C, Davis DR, Bolwell GP. Early signaling events in the apoplatic oxidative burst in suspension-cultured French bean cells involve cAMP and Ca2+ New Phytol. 2001;151:185–194. doi: 10.1046/j.1469-8137.2001.00170.x. [DOI] [PubMed] [Google Scholar]
- Bolwell GP, Butt VS, Davies DR, Zimmerlin A. The origin of the oxidative burst in plants. Free Radic. Res. 1995;23:517–532. doi: 10.3109/10715769509065273. [DOI] [PubMed] [Google Scholar]
- Bolwell GP, Blee KA, Butt VS, Davies DR, Gardner SL, Gerrish C, Minibayeva F, Rowntree EG, Wojtaszek P. Recent advances in understanding the origin of the apoplastic oxidative burst in plant cells. Free Radic. Res. 1999;31 Suppl.:S137–S145. doi: 10.1080/10715769900301431. [DOI] [PubMed] [Google Scholar]
- Bolwell GP, Bindschedler LV, Blee KA, Butt VS, Davies DR, Gardner SL, Gerrish C, Minibayeva F. The apoplastic oxidative burst in response to biotic stress in plants: a three-component system. J. Exp. Bot. 2002;53:1367–1376. [PubMed] [Google Scholar]
- Bonneh-Barkay D, Reaney SH, Langston WJ, Di Monte DA. Redox cycling of the herbicide paraquat in microglial cultures. Mol. Brain Res. 2005;134:52–56. doi: 10.1016/j.molbrainres.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Counillon L, Pouyssegur J. The expanding family of eucaryotic Na+/H+ exchangers. J. Biol. Chem. 2000;275:1–4. doi: 10.1074/jbc.275.1.1. [DOI] [PubMed] [Google Scholar]
- Fedoroff NV. RNA-binding proteins in plants: the tip of an iceberg? Curr. Opin. Plant Biol. 2002;5:452–459. doi: 10.1016/s1369-5266(02)00280-7. [DOI] [PubMed] [Google Scholar]
- Felle HH. pH: signal and messenger in plant cells. Plant Biol. 2001;3:577–591. [Google Scholar]
- Felle HH, Hanstein S. The apoplastic pH of the substomatal cavity of Vicia faba leaves and its regulation responding to different stress factors. J. Exp. Bot. 2002;53:73–82. [PubMed] [Google Scholar]
- Felle HH, Herrmann A, Hanstein S, Huckelhoven R, Kogel KH. Apoplastic pH signaling in barley leaves attacked by the powdery mildew fungus Blumeria graminis f. sp. hordei. Mol. Plant Microbe Interact. 2004;17:118–123. doi: 10.1094/MPMI.2004.17.1.118. [DOI] [PubMed] [Google Scholar]
- Felle HH, Herrmann A, Huckelhoven R, Kogel KH. Root-to-shoot signaling: apoplastic alkalinization, a general stress response and defence factor in barley (Hordeum vulgare) Protoplasma. 2005;227:17–24. doi: 10.1007/s00709-005-0131-5. [DOI] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JH, et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature. 2003;422:442–446. doi: 10.1038/nature01485. [DOI] [PubMed] [Google Scholar]
- Fukada-Tanaka S, Inagaki Y, Yamaguchi T, Saito N, Iida S. Colour-enhancing protein in blue petals. Nature. 2000;407:581. doi: 10.1038/35036683. [DOI] [PubMed] [Google Scholar]
- Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- Gutierrez RA, MacIntosh GC, Green PJ. Current perspectives on mRNA stability in plants: multiple levels and mechanisms of control. Trends Plant Sci. 1999;4:429–438. doi: 10.1016/s1360-1385(99)01484-3. [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu J-K, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- Hugouvieux V, Kwak JM, Schroeder JI. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell. 2001;106:477–487. doi: 10.1016/s0092-8674(01)00460-3. [DOI] [PubMed] [Google Scholar]
- Jeffrey SW, Humphrey GF. New Spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae, and natural phytoplankton. Biochem. Physiol. Pflanz. 1975;167:191–194. [Google Scholar]
- Johnson MA, Pérez-Amador MA, Lidder P, Green PM. Mutants of Arabidopsis defective in a sequence-specific mRNA degradation pathway. Proc. Natl Acad. Sci. U.S.A. 2000;97:13991–13996. doi: 10.1073/pnas.240354097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenmayer JP, Green PJ. Novel features of the XRN-family in Arabidopsis: evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl Acad. Sci. U.S.A. 2000;97:13985–13990. doi: 10.1073/pnas.97.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Zhu J, Kim K, Agarwal M, Fu X, Huang A, Zhu JK. The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc Natl Acad Sci U.S.A. 2006;103:18816–18821. doi: 10.1073/pnas.0604711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T, Damude HG, Werner D, Doerner P, Dixon RA, Lamb C. A plant homolog of the neutrophil NADPH oxidase gp91phox subunit gene encodes a plasma membrane protein with Ca2+ binding motifs. Plant Cell. 1998;10:255–266. doi: 10.1105/tpc.10.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwa H, Barb AW, Xiong L, et al. C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc. Natl Acad. Sci. U.S.A. 2002;99:10893–10898. doi: 10.1073/pnas.112276199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JM, Schroeder JI. Impacts of altered RNA metabolism on abscisic acid signaling. Curr. Opin. Plant Biol. 2003;6:463–469. doi: 10.1016/s1369-5266(03)00084-0. [DOI] [PubMed] [Google Scholar]
- Lee TC, Lin FM, Ho IC, Liu TY, Wang TC, Chu YI, Chang HY. Paraquat-resistant cell lines derived from Chinese hamster ovary cells. Cell. Biol. Int. Rep. 1990;14:235–246. doi: 10.1016/s0309-1651(05)80006-2. [DOI] [PubMed] [Google Scholar]
- Li J, Kinoshita T, Pandey S, Ng CKY, Gygi SP, Shimazaki KI, Assmann SM. Modulation of an RNA-binding protein by abscisic-acid-activated protein kinase. Nature. 2002;418:793–797. doi: 10.1038/nature00936. [DOI] [PubMed] [Google Scholar]
- Liu J, Zhu JK. A calcium sensor homolog required for plant salt tolerance. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl Acad. Sci. U.S.A. 2000;97:3730–3734. doi: 10.1073/pnas.060034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Han MH, Guevara-Garcia A, Fedoroff NV. Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc. Natl Acad. Sci. U.S.A. 2002;99:15812–15817. doi: 10.1073/pnas.242607499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Monshausen G, Bibikova T, Gilroy S. Ca2+ Regulation of Transmembrane Proton Fluxes and ROS Production: A Common Mechanism in Tip Growth and Mechanosensing?; Annual Meeting of American Society of Plant Biologists; Boston, USA. 2006. Abstract #M0401. [Google Scholar]
- Nakagami H, Pitzschke A, Hirt H. Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 2005;10:339–346. doi: 10.1016/j.tplants.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Nurnberger T, Scheel D. Signal transmission in the plant immune response. Trends Plant Sci. 2001;6:372–379. doi: 10.1016/s1360-1385(01)02019-2. [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Hirt H. Mitogen-activated protein kinases and reactive oxygen species signaling in plants. Plant Physiol. 2006;141:351–356. doi: 10.1104/pp.106.079160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 2003;15:326–331. doi: 10.1016/s0955-0674(03)00048-6. [DOI] [PubMed] [Google Scholar]
- Rochaix JD. Postranscriptional control of chloroplast gene expression. From RNA to photosynthetic complex. Plant Physiol. 2001;125:142–144. doi: 10.1104/pp.125.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi M, Fluhr R. Superoxide production by plant homologues of the gp91(phox) NADPH oxidase. Modulation of activity by calcium and by tobacco mosaic virus infection. Plant Physiol. 2001;126:1281–1290. doi: 10.1104/pp.126.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi M, Fluhr R. Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 2006;141:336–340. doi: 10.1104/pp.106.078089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF. Calcium at the crossroads of signaling. Plant Cell. 2002;14:S401–S417. doi: 10.1105/tpc.002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala L, Cuin TA, Newman IA, Shabala S. Salinity-induced ion flux patterns from the excised roots of Arabidopsis sos mutants. Planta. 2005;222:1041–1050. doi: 10.1007/s00425-005-0074-2. [DOI] [PubMed] [Google Scholar]
- Shi H, Bressan RA. RNA extraction. In: Salinas J, Sanchez-Serrano JJ, editors. Arabidopsis Protocols. Totowa, New Jersey: Humana Press; 2006. pp. 345–348. [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl Acad. Sci. U.S.A. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell. 2002a;14:465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Xiong L, Stevenson B, Lu T, Zhu JK. The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell. 2002b;14:575–588. doi: 10.1105/tpc.010417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Lee BH, Wu SJ, Zhu JK. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2003a;21:81–85. doi: 10.1038/nbt766. [DOI] [PubMed] [Google Scholar]
- Shi H, Kim YS, Guo Y, Stevenson B, Zhu JK. The Arabidopsis SOS5 locus encodes a cell surface adhsion protein and is required for normal cell expansion. Plant Cell. 2003b;15:19–32. doi: 10.1105/tpc.007872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Bressan R, Hasegawa PM, Zhu JK. Sodium. In: Broadley M, White P, editors. Plant Nutritional Genomics. London: Blackwell Publishing; 2005. pp. 127–149. [Google Scholar]
- Shim J, Karin M. The control of mRNA stability in response to extracellular stimuli. Mol. Cells. 2002;14:323–331. [PubMed] [Google Scholar]
- Shin R, Schachtman DP. Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc. Natl Acad. Sci. U.S.A. 2004;101:8827–8832. doi: 10.1073/pnas.0401707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Plas F, Rusterucci C, Milat ML, Humbert C, Montillet JL, Blein JP. Active oxygen species production in tobacco cells elicited by cryptogein. Plant Cell Environ. 1997;20:1573–1579. [Google Scholar]
- Torres MA, Dangl JL. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 2005;8:397–403. doi: 10.1016/j.pbi.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Tourrière H, Chebli K, Tazi J. mRNA degradation machines in eukaryotic cells. Biochimie. 2002;84:821–837. doi: 10.1016/s0300-9084(02)01445-1. [DOI] [PubMed] [Google Scholar]
- Uchida A, Jagendorf AT, Hibino T, Takabe T, Takabe T. Effects of hydrogen peroxide and nitric oxide on both salt and heat stress tolerance in rice. Plant Sci. 2002;163:515–523. [Google Scholar]
- Wilusz CJ, Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nature Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- Wu XF, Block ML, Zhang W, Qin L, Wilson B, Zhang WQ, Veronesi B, Hong JS. The role of microglia in paraquat-induced dopaminergic neurotoxicity. Antioxid. Redox Signal. 2005;7:654–661. doi: 10.1089/ars.2005.7.654. [DOI] [PubMed] [Google Scholar]
- Xiong L, Gong Z, Rock CD, Subramanian S, Guo Y, Xu W, Galbraith D, Zhu JK. Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell. 2001;1:771–781. doi: 10.1016/s1534-5807(01)00087-9. [DOI] [PubMed] [Google Scholar]
- Xiong L, Lee H, Ishitani M, Tanaka Y, Stevenson B, Koiwa H, Bressan RA, Hasegawa PM, Zhu JK. Repression of stress-responsive genes by FIERY2, a novel transcriptional regulator in Arabidopsis. Proc. Natl Acad. Sci. U.S.A. 2002;99:10899–10904. doi: 10.1073/pnas.162111599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Zhao J, Davis LC, Verpoorte R. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 2005;23:283–333. doi: 10.1016/j.biotechadv.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.