Abstract
The spindle checkpoint prevents cell cycle progression in cells that have mitotic spindle defects. Although several spindle defects activate the spindle checkpoint, the exact nature of the primary signal is unknown. We have found that the budding yeast member of the Aurora protein kinase family, Ipl1p, is required to maintain a subset of spindle checkpoint arrests. Ipl1p is required to maintain the spindle checkpoint that is induced by overexpression of the protein kinase Mps1. Inactivating Ipl1p allows cells overexpressing Mps1p to escape from mitosis and segregate their chromosomes normally. Therefore, the requirement for Ipl1p in the spindle checkpoint is not a consequence of kinetochore and/or spindle defects. The requirement for Ipl1p distinguishes two different activators of the spindle checkpoint: Ipl1p function is required for the delay triggered by chromosomes whose kinetochores are not under tension, but is not required for arrest induced by spindle depolymerization. Ipl1p localizes at or near kinetochores during mitosis, and we propose that Ipl1p is required to monitor tension at the kinetochore.
Keywords: Ipl1/Aurora protein kinase, spindle checkpoint, budding yeast, Mps1 protein kinase, kinetochores, tension
The accurate propagation of genetic information depends on faithful chromosome segregation. Accurate chromosome segregation depends on the precise coordination of events in the chromosome cycle. When chromosomes replicate during S phase, linkage between the sister chromatids (cohesion) is established and must be maintained while chromosomes condense and align on the mitotic spindle. Chromosomes attach to the mitotic spindle by their kinetochores, specialized protein structures that are assembled on centromeric DNA sequences. Once all the chromosomes are correctly aligned on the mitotic spindle, sister chromatid cohesion must dissolve promptly at anaphase to allow the sister chromatids to segregate rapidly to opposite poles of the mitotic spindle. Defects in any of these steps can result in aneuploidy, a hallmark of tumor cells and some birth defects (Lengauer et al. 1997, 1998).
The spindle checkpoint prevents cells from separating their sister chromatids until chromosome alignment is complete. The conserved components of the checkpoint include the Mad (Mad1–Mad3) proteins, Bub1 and Bub3, Mps1 (a protein kinase), and Cdc55 (Hoyt et al. 1991; Li and Murray 1991; Minshull et al. 1996; Weiss and Winey 1996; Wang and Burke 1997). A separate control, the Bub2-dependent checkpoint, monitors a second aspect of chromosome segregation, the delivery of DNA or a spindle pole body into the daughter cell (Alexandru et al. 1999; Fesquet et al. 1999; Fraschini et al. 1999; Li 1999). Spindle checkpoint defects are associated with genetic instability, and some human cancers contain mutant spindle checkpoint genes (Cahill et al. 1998; Takahashi et al. 1999)
The spindle checkpoint monitors the interaction between kinetochores and microtubules. Spindle checkpoint proteins localize to kinetochores (Chen et al. 1996; Taylor and McKeon 1997; Bernard et al. 1998), and all known kinetochore and spindle defects that activate the checkpoint affect the interaction between kinetochores and microtubules (Wang and Burke 1995; Pangilinan and Spencer 1996; Wells and Murray 1996; Hardwick et al. 2000). How does the checkpoint monitor kinetochore alignment? Some experiments suggest that it senses the tension that microtubule-dependent forces exert on the kinetochore (Li and Nicklas 1995), whereas others suggest it senses microtubule attachment to kinetochores (Rieder et al. 1995; Waters et al. 1998). However, because tension affects microtubule attachment to the kinetochore (King and Nicklas 2000), the roles of tension versus attachment in the checkpoint signal are not easily separable. In budding yeast, the spindle checkpoint can detect defects in tension in meiosis (Shonn et al. 2000) and in mitosis (Stern and Murray 2001).
The spindle checkpoint arrests the cell cycle by inhibiting the separation of sister chromatids that leads to anaphase, the combination of chromosome segregation and spindle elongation. The regulated step in anaphase is the ubiquitin-mediated proteolysis of securin, an inhibitor of separase, the protease that cleaves Scc1/Mcd1p, a component of the cohesin complex holding sisters together (Cohen-Fix et al. 1996; Funabiki et al. 1996; Uhlmann et al. 1999, 2000), and the spindle midzone protein Slk19 (Sullivan et al. 2001). Securin (Pds1p) is ubiquitinated by the anaphase-promoting complex (APC), which is activated by Cdc20p (Visintin et al. 1997). The ability of the spindle checkpoint to inhibit Pds1 destruction depends on the binding of Mad2p to Cdc20p (Hwang et al. 1998; Kim et al. 1998).
The yeast Ipl1 and the Drosophila Aurora A proteins are the founding members of a conserved serine/threonine kinase family (Ipl1/Aurora) whose members are key regulators of chromosome segregation and cytokinesis (Chan and Botstein 1993; Glover et al. 1995). Budding and fission yeast contain a single Ipl1p/Aurora homolog, whereas multicellular eukaryotes have multiple homologs. The human aurora 1 and aurora 2 genes are oncogenes that are amplified in many colorectal and breast cancer cell lines, suggesting that the kinase is critical to maintaining genomic stability (Sen et al. 1997; Bischoff et al. 1998; Tanaka et al. 1999). The Aurora kinases contain conserved C-terminal catalytic domains and divergent N-terminal domains and are classified into three families, A, B, and C (for review, see Nigg 2001). The Aurora B kinases interact with the conserved inner centromere protein (INCENP) (Kim et al. 1999; Adams et al. 2000, 2001; Kaitna et al. 2000). Defects in INCENP localization disrupt Aurora B localization, suggesting that at least one function of the interaction may be to localize Aurora B to mitotic structures (Adams et al. 2000). Although the precise localization patterns of the Aurora kinases differ, they generally associate with mitotic structures such as the spindle, spindle midzone, centrosome, and kinetochore. Defects in Ipl1p function lead to severe chromosome segregation defects with many pairs of sister chromatids traveling to a single pole instead of segregating to opposite poles (Chan and Botstein 1993; Biggins et al. 1999; Kim et al. 1999). Experiments in vitro suggest that this phenotype is due to altered binding of microtubules to kinetochores in the ipl1 mutant cells, suggesting Ipl1p functions at kinetochores (Biggins et al. 1999).
Here we show that Ipl1p is needed for kinetochores that are not under tension to delay cells in mitosis, suggesting that Ipl1p may have a specific role in monitoring forces at kinetochores.
Results
ipl1 mutant cells do not activate the spindle checkpoint
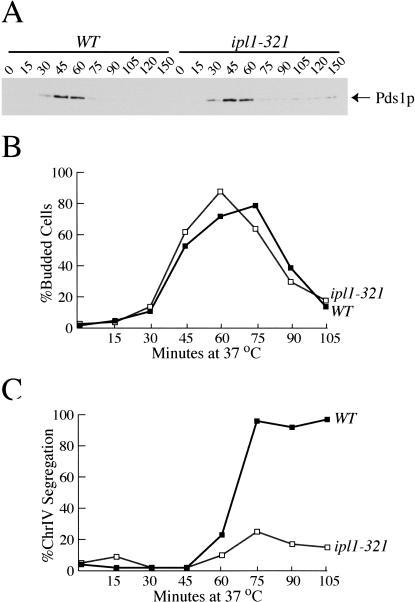
We previously isolated alleles of the IPL1 gene in a screen that identified mutants defective in sister chromatid separation or segregation and determined that the ipl1 mutant cells are defective in regulating microtubule binding to kinetochores (Biggins et al. 1999, 2001). Although Ipl1p is required for kinetochore function (Biggins et al. 1999), ipl1 mutant cells do not arrest in mitosis, suggesting that they do not activate the spindle checkpoint (Chan and Botstein 1993; Biggins et al. 2001). To confirm this suggestion, we analyzed the levels of Pds1p. Because the spindle checkpoint inhibits APC activation, Pds1p levels are stabilized when the spindle checkpoint is active. Wild-type and ipl1-321 temperature-sensitive mutant cells containing epitope-tagged Pds1-myc18 protein were arrested in G1 with α-factor, and then released to the nonpermissive temperature (37°C) in the absence of α-factor. Pds1p levels cycled similarly in ipl1-321 and wild-type cells (Fig. 1A), whereas if the spindle checkpoint were activated, Pds1p should have been stabilized. The budding and cell division of ipl1-321 cells is also similar to wild-type cells: both strains undergo budding and cytokinesis with similar kinetics (Fig. 1B). We analyzed the segregation of chromosome IV in wild-type and ipl1-321 strains in the same experiment to ensure that the ipl1 mutant allele was inactivated. Chromosome IV was visualized by binding of a GFP-lactose repressor (GFP-lacI) to an array of lactose operators integrated at the TRP1 locus, 12 kb from the centromere (Straight et al. 1996). Whereas chromosome IV sister chromatids always segregated to opposite poles in wild-type cells, they segregated to opposite poles in only 15% of the ipl1-321 cells, as we have previously shown (Fig. 1C; Biggins et al. 1999). Therefore, ipl1-321 cells do not activate the spindle checkpoint despite defects in kinetochore behavior that give rise to a severe chromosome segregation defect.
Figure 1.
ipl1 mutants do not activate the spindle checkpoint despite a chromosome segregation defect. Wild-type (SBY818) and ipl1-321 cells (SBY819) containing Pds1-myc18 were arrested in G1 with α-factor at the permissive temperature (23°C) and released to the non-permissive temperature (37°C). α-factor was added back when small buds formed to prevent cells from entering the next cell cycle. (A) Lysates were prepared at the indicated time points and immunoblotted with anti-myc antibodies to analyze Pds1-myc protein levels. Pds1p levels cycle in both wild-type cells and ipl1 mutant cells, indicating that the spindle checkpoint is not activated. Equal protein concentrations were loaded in all lanes as judged by Ponceau S staining (data not shown). (B) The percentage of budded cells in the same experiment was quantified by microscopy and shows that wild-type (filled squares) and ipl1-321 cells (open squares) undergo budding and then cytokinesis with similar kinetics. (C) In the same experiment, the percent chromosome IV segregation was monitored as the fraction of cells that had segregated two GFP-marked copies of chromosome IV to opposite poles of the spindle. Although chromosome IV segregates in wild-type cells (filled squares), there is a severe chromosome segregation defect in the ipl1-321 cells (open squares).
Ipl1p is required for Mps1 overexpression-induced checkpoint arrest
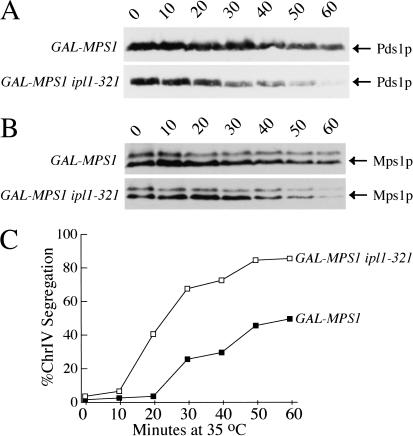
There are two possible explanations for the failure of ipl1 mutant cells to activate the spindle checkpoint: (1) Ipl1p function is required for the spindle checkpoint, or (2) the ipl1 kinetochore defect does not activate the checkpoint. To see if Ipl1p is part of the spindle checkpoint, we tested whether Ipl1p is required for the arrest induced by Mps1p overexpression, which constitutively activates the spindle checkpoint, arresting cells in metaphase with a bipolar spindle (Hardwick et al. 1996). We arrested ipl1-321 cells in mitosis by overexpressing Mps1p from the GAL1 promoter at the permissive temperature and then shifted them to 35°C to inactivate Ipl1p. We monitored metaphase arrest by analyzing Pds1p levels and cytokinesis. Pds1p levels started to decline in the GAL-MPS1 ipl1-321 cells after 20 min at the nonpermissive temperature, whereas there was little Pds1p degradation in GAL-MPS1 cells for at least 1 h (Fig. 2A). Several GAL-MPS1 cells exit the checkpoint arrest because galactose induction does not work as well at high temperatures. However, the GAL-MPS1 ipl1-321 cells exit the checkpoint arrest much faster, indicating that Ipl1p has a role in maintaining the checkpoint-dependent arrest caused by Mps1p overexpression. We monitored cytokinesis in the same experiment and found similar results: After 40 min at the nonpermissive temperature, 10% of the GAL-MPS1 cells had cytokinesed compared with 40% of the GAL-MPS1 ipl1-321 cells (data not shown).
Figure 2.
Ipl1p is required to maintain the GAL-MPS1 checkpoint arrest. GAL-MPS1 (SBY679) and GAL-MPS1 ipl1-321 (SBY680) cells were arrested in galactose for 3.5 h and then released to the nonpermissive temperature (35°C) to inactivate ipl1. (A) Pds1-myc protein levels were monitored by immunoblotting with anti-myc antibodies. Pds1 levels decline faster in the GAL-MPS1 ipl1-321 cells compared with the GAL-MPS1 cells, indicating that Ipl1p is required for full maintenance of the GAL-MPS1 checkpoint arrest. (B) Mps1 levels were monitored in the same experiment by immunoblotting with anti-myc antibodies. The levels of Mps1 protein decline in both strains but are unstable in the ipl1-321 cells. Equal protein concentrations were loaded in all lanes as judged by Ponceau S staining (data not shown). (C) Chromosome IV segregation was monitored by microscopy of GFP-marked chromosome IV. This chromosome segregated normally as GAL-MPS1 cells (filled squares) and GAL-MPS1 ipl1-321 cells (open squares) left the checkpoint arrest.
To determine whether the strains expressed different levels of Mps1 protein, we analyzed Mps1–myc protein levels by immunoblotting (Fig. 2B). The Mps1 protein levels were similar in both strains for at least 30 min, and then the levels started falling in both strains. Because the GAL-MPS1 ipl1-321 strain did not maintain Mps1p levels as high as the GAL-MPS1 strain, we considered the possibility that Ipl1p may affect Mps1p stability. To test this, we analyzed the stability of Mps1p in wild-type and ipl1-321 cells that were arrested in metaphase using nocodazole and found no difference in Mps1p stability between the strains (data not shown). Therefore, Ipl1p does not regulate Mps1p stability. Instead, it is likely that Mps1 protein becomes unstable as cells exit mitosis; cells arrested in G1 with α-factor had much less Mps1p relative to metaphase-arrested cells (data not shown). Taken together, these data suggest that Ipl1p is required for full maintenance of the GAL-MPS1 checkpoint arrest and that Mps1 protein levels decline as cells exit mitosis.
Mutations that completely abolish kinetochore function destroy the spindle checkpoint (Gardner et al. 2001). We ruled out the possibility that ipl1-321 was having such an effect by monitoring the segregation of GFP-marked chromosome IV in the experiment above. The two copies of chromosome IV segregated to opposite poles as the GAL-MPS1 cells escaped from the checkpoint-dependent arrest at 35°C. After 30 min at the nonpermissive temperature, 24% of the GAL-MPS1 and 66% of the GAL-MPS1 ipl1-321 cells had segregated chromosome IV to opposite poles (Fig. 2C). From the fact that chromosome IV segregated to opposite poles in the ipl1-321 cells, we conclude that Ipl1p function is not required to maintain the function of kinetochores once a bipolar spindle has been established but is needed to maintain a checkpoint-dependent arrest. Therefore, this experiment identifies a function for Ipl1p in spindle checkpoint maintenance that is temporally separable from its function in chromosome segregation.
Ipl1p is not required for the spindle checkpoint arrest induced by nocodazole
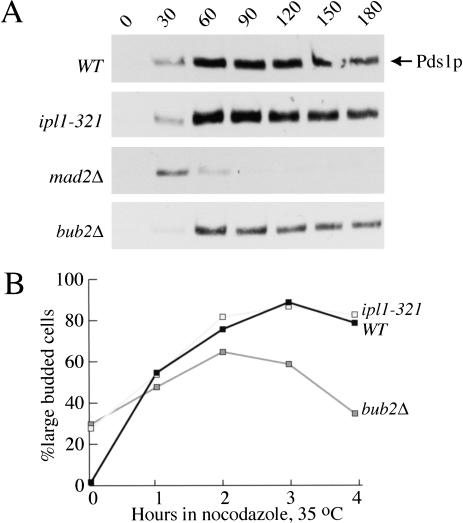
Next, we tested whether Ipl1p is required for the spindle checkpoint arrest induced by the drugs nocodazole and benomyl, which depolymerize the microtubules that make up the spindle. We arrested wild-type, ipl1-321, and mad2Δ mutant cells in G1 with α-factor and then released them into a mixture of nocodazole and benomyl at the nonpermissive temperature (35°C) to inactivate ipl1. We monitored Pds1p levels and found that wild-type and ipl1-321 cells activated the spindle checkpoint and arrested in nocodazole plus benomyl with high Pds1p levels (Fig. 3A). In contrast, mad2Δ cells did not maintain high Pds1p levels because the spindle checkpoint was not activated. Therefore, IPL1 behaves differently from known spindle checkpoint genes because it is required for full maintenance of the GAL-MPS1-induced arrest but is not required for the arrest induced by nocodazole. In addition, because functional kinetochores are required to activate the checkpoint in response to spindle depolymerization (Gardner et al. 2001), this experiment shows that the kinetochores in the ipl1 mutant cells are competent to activate the checkpoint in the absence of a spindle.
Figure 3.
(A) Ipl1p is not required for the checkpoint arrest induced by nocodazole. Wild-type (SBY818), ipl1-321 (SBY819), mad2Δ (SBY920), and bub2Δ (SBY934) cells containing Pds1-myc18 were arrested in G1 with α-factor at the permissive temperature (23°C). They were released into nocodazole and benomyl at the nonpermissive temperature (37°C), and α-factor was added back when small buds appeared, to prevent cells from entering the next cell cycle. Pds1p levels were analyzed by immunoblotting and show that wild-type, ipl1-321, and bub2Δ mutant cells activate the spindle checkpoint because they maintain high Pds1 levels. Pds1p levels cycle in mad2Δ mutant cells, which lack the spindle checkpoint. Equal protein concentrations were loaded in all lanes as judged by Ponceau S staining (data not shown). (B) IPL1 does not function in the BUB2-dependent checkpoint pathway. Wild-type (SBY214), ipl1-321 (SBY322), and bub2Δ (SBY432) mutant cells were released into nocodazole plus benomyl at the nonpermissive temperature. The percentage of large budded cells was monitored and shows that wild-type (filled squares) and ipl1-321 cells (shaded squares) arrest as large budded cells whereas bub2Δ cells (gray squares) rebud, indicating that Ipl1p is not in the same pathway as Bub2p.
The addition of nocodazole can inhibit mitotic exit by either activating the spindle checkpoint or the BUB2-dependent pathway that monitors delivery of a spindle pole body to the daughter cell. Although the spindle checkpoint stabilizes Pds1p, the BUB2-dependent pathway does not (Alexandru et al. 1999). We confirmed this by analyzing Pds1p levels in bub2Δ cells that were released from G1 into nocodazole at the nonpermissive temperature in the experiment described above (Fig. 3A). Because ipl1 mutants behaved like bub2 mutant cells in maintaining Pds1p levels in the presence of nocodazole, we tested whether Ipl1p was a component of the Bub2-dependent pathway instead of the spindle checkpoint. Wild-type, ipl1-321, and bub2Δ double mutant cells were released into nocodazole and benomyl at the nonpermissive temperature (35°C), and the percentage of large budded cells was monitored for 4 h (Fig. 3B). Although wild-type and ipl1-321 cells arrested as large budded cells, the bub2Δ cells did not maintain a large budded cell arrest. It was recently shown that cells would rebud in nocodazole only if both the spindle checkpoint and BUB2-dependent checkpoint are defective (Alexandru et al. 1999; Fesquet et al. 1999). However, we detected rebudding in bub2Δ cells, probably because nocodazole does not work as effectively at high temperatures. Although the bub2Δ cells rebud, we did not detect rebudding in the ipl1-321 mutant cells, suggesting that they behave differently than bub2Δ cells. Therefore, Ipl1p likely acts in a separate pathway from Bub2p because ipl1-321 arrests in nocodazole at high temperatures, a condition that allows bub2Δ cells to leave mitosis after a short delay.
Ipl1p is required for the spindle checkpoint delay induced by kinetochore tension defects
In multicellular eukaryotes, the checkpoint appears to monitor both microtubule attachment to the kinetochore and tension generated at the kinetochore (Li and Nicklas 1995; Rieder et al. 1995). We considered the possibility that Ipl1p monitors kinetochore tension but not attachment in budding yeast and tested this hypothesis in an experiment wherein microtubule attachment occurred but tension was not generated. Because sister chromatids are linked to each other, attempting to pull the sister kinetochores to opposite poles generates tension on the kinetochores and the linkage between them. In the absence of DNA replication, tension cannot be generated because kinetochores lack sisters. DNA replication can be prevented by repressing the CDC6 gene that is required for the initiation of replication (Piatti et al. 1996), without affecting the interaction between microtubules and kinetochores (Piatti et al. 1995). In these cells Pds1p is stabilized in a spindle checkpoint-dependent manner (Stern and Murray 2001).
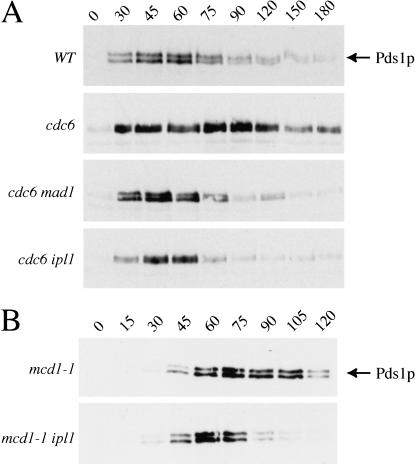
We used this manipulation to ask if Ipl1p is needed to sense the absence of kinetochore tension. We compared wild-type cells with three strains that failed to replicate their DNA when grown on glucose-containing medium: cells that have an intact spindle checkpoint (GAL-CDC6); cells lacking Mad1p, a known spindle checkpoint component (GAL-CDC6 mad1Δ); and cells with mutant Ipl1p (GAL-CDC6 ipl1-321). Cells depleted of Cdc6 protein were arrested in G1 with α-factor, released into conditions that inactivated Ipl1p (37°C) and repressed CDC6 (glucose-containing medium), and Pds1p levels were monitored by immunoblotting as they proceeded through the cell cycle (see Materials and Methods for details). Although Pds1p levels fall as wild-type cells enter anaphase, they are stabilized for at least 1 h in Cdc6-depleted cells containing unreplicated DNA (Fig. 4A). The stabilization of Pds1p requires the spindle checkpoint because it is eliminated in GAL-CDC6 mad1Δ cells. We found that Pds1p levels are also not stabilized in GAL-CDC6 ipl1-321 cells, indicating that Ipl1p is required for the spindle checkpoint to delay cells whose kinetochores are not under tension.
Figure 4.
Ipl1p is required for the checkpoint arrested induced by kinetochore tension defects. (A) Cells depleted of the Cdc6 protein were grown at the permissive temperature (23°C) and arrested in G1 with α-factor. Cells were then released from G1 to the nonpermissive temperature (37°C) in glucose to keep GAL-CDC6 repressed; Pds1p levels were analyzed by immunoblotting. Pds1p levels cycle in wild-type cells (SBY818) but are stabilized in GAL-CDC6 cells grown in repressing media (SBY772). Pds1p levels are not stabilized in GAL-CDC6 mad1Δ (SBY762) and GAL-CDC6 ipl1-321 (SBY771) mutant cells, indicating that the checkpoint is not activated. (B) mcd1-1 (SBY870) and mcd1-1 ipl1-321 cells (SBY871) were arrested in G1 with α-factor at the permissive temperature. They were released to the nonpermissive temperature (37°C) in the absence of α-factor, and Pds1-myc18 protein levels were monitored by immunoblotting. There is a delay in the degradation of Pds1p in the mcd1/scc1 mutant cells that is eliminated in the mcd1-1 ipl1-321 cells, indicating that Ipl1p is required for the spindle checkpoint induced by defects in sister chromatid cohesion. Equal protein concentrations were loaded in all lanes as judged by Ponceau S staining (data not shown).
We confirmed the role of Ipl1p by looking at a mutant that destroys tension at the kinetochore by a different mechanism. Mcd1p/Scc1p is a component of the cohesin complex that holds sister chromatids together (Guacci et al. 1997; Michaelis et al. 1997). In its absence, kinetochores can still attach to microtubules (Tanaka et al. 2000), but because sister chromatids are not linked to each other; there is no tension at these attachments. We arrested mcd1-1 and mcd1-1 ipl1-321 mutant cells in G1 with α-factor at the permissive temperature and then released them to the nonpermissive temperature (37°C) to inactivate the mutant alleles. There is a delay in the degradation of Pds1p in mcd1-1 cells, indicating that a checkpoint is activated (Fig. 4B). This delay is abolished in the mcd1-1 ipl1-321 double mutant cells, indicating that Ipl1p is required for the spindle checkpoint to delay cells whose kinetochores have been relaxed by a different mechanism.
Ipl1p localizes to kinetochores at metaphase
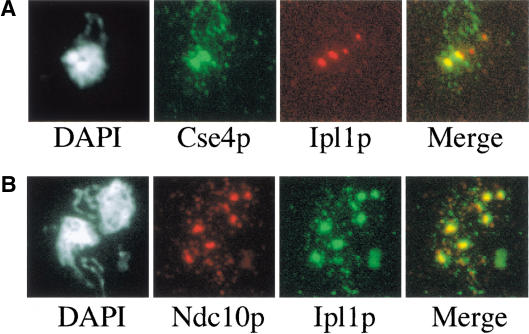
Because most known spindle checkpoint proteins localize to kinetochores, we analyzed Ipl1p localization in metaphase-arrested cells with or without spindle checkpoint activation. Yeast nuclei are small, making it impossible to see individual kinetochores by standard immunofluorescence techniques. Therefore, we examined chromosome spreads, the detergent-insoluble residue of yeast spheroplasts (Loidl et al. 1998). We used one epitope tag to see Cse4p or Ndc10p, two known kinetochore components, and another to see Ipl1p. To obtain metaphase-arrested cells, we used a deletion in the CDC26 gene that is required for APC activity at 37°C (Hwang and Murray 1997). cdc26Δ mutant cells containing epitope-tagged Cse4p and Ipl1p were shifted to the nonpermissive temperature for 3 h to arrest cells in metaphase. Immunofluorescence was performed on chromosome spreads and revealed that Ipl1p colocalized with the kinetochore protein Cse4p (Fig. 5A). Ipl1p did not colocalize with the spindle pole body (SPB) when we analyzed the SPB component Spc42p (data not shown). In addition, the Ipl1p localization is dependent on functional kinetochores because it disappears in the ndc10-1 mutant that abolishes all kinetochore function (data not shown). We also analyzed Ipl1p localization in cells arrested in metaphase with the spindle checkpoint activated. Cells were released into nocodazole for 3 h, and immunofluorescence on chromosome spreads revealed that Ipl1p colocalized with another kinetochore component, Ndc10p (Fig. 5B). Because the resolution of chromosome spreads is limited, we cannot distinguish whether Ipl1p localizes to the kinetochore itself or to an adjacent, kinetochore-dependent structure.
Figure 5.
Ipl1p localizes to kinetochores at metaphase. (A) Ipl1p localizes to kinetochores during a metaphase arrest. Cdc26Δ cells containing Cse4p–myc13 and Ipl1p–HA3 (SBY961) were arrested in metaphase by shifting cells to the nonpermissive temperature (37°C ) for 3 h. Chromosome spreads were performed and stained with DAPI to recognize the DNA (left panel), and with anti-myc and anti-HA antibodies to recognize Cse4p and Ipl1p, respectively (middle panels). The merged image (right panel) shows that there is colocalization of Ipl1p and Cse4p during a metaphase arrest. (B) Ipl1p localizes to kinetochores during a checkpoint arrest. Cells containing Ndc10p–HA3 and Ipl1p–myc12 (SBY596) were arrested in nocodazole for 3 h at 23°C. Chromosome spreads were performed and stained with DAPI to recognize the DNA (left panel), and with anti-HA and anti-myc antibodies to recognize Ndc10p and Ipl1p, respectively (middle panels). The merged image (right panel) of the Ndc10p and Ipl1p images shows that there is a colocalization of Ipl1p with Ndc10p to kinetochores during a checkpoint arrest.
Discussion
We found that the Ipl1/Aurora protein kinase has a role in the spindle checkpoint in budding yeast that is temporally separate from an earlier role in aligning chromosomes on the spindle. Ipl1p distinguishes between the lack of tension at kinetochores that are attached to microtubules, and kinetochores without bound microtubules. We suggest that Ipl1p is specifically required to monitor defects in kinetochore tension.
Functions of the Ipl1/Aurora kinase family
Members of the Aurora protein kinase family have functions in chromosome segregation, condensation, and cytokinesis (for review, see Bischoff and Plowman 1999). The chromosome segregation defect in ipl1 mutants results in pairs of sister chromatids traveling to a single spindle pole instead of opposite spindle poles, resulting in severe aneuploidy (Chan and Botstein 1993; Biggins et al. 1999; Kim et al. 1999). In Drosophila, depletion of the Aurora B by double-stranded RNA interference in cultured Drosophila cells results in polyploidy, a phenotype similar to the budding yeast ipl1 mutant phenotype (Adams et al. 2001; Giet and Glover 2001). In Caenorhabditis elegans, similar chromosome segregation defects are observed when AIR-2, the Aurora B homolog, is depleted by RNA interference (Kaitna et al. 2000). The exact role of Aurora B in chromosome segregation is not clear. In budding yeast, it appears to control kinetochore behavior, because extracts prepared from ipl1 mutant cells produce abnormally regulated microtubule interactions with kinetochores in vitro (Biggins et al. 1999). In Drosophila, Aurora B is required for normal metaphase chromosome alignment, kinetochore disjunction in anaphase (Adams et al. 2001), normal chromosome condensation, and the recruitment of the Barren condensin protein to chromosomes (Giet and Glover 2001). Aurora B phosphorylates histone H3 in budding yeast and C. elegans, an event that is correlated with chromosome condensation (Hsu et al. 2000). In yeast, however, there is no phenotype associated with a lack of H3 phosphorylation, suggesting that Ipl1p must have additional targets.
In budding yeast, Ipl1p is required to sense kinetochores that are not under tension, revealing yet another function for this protein kinase family. Despite defects in chromosome segregation, mutants in Aurora B do not result in cell cycle arrest in any organism, suggesting that this kinase may play a role in the spindle checkpoint. We confirmed this possibility by temporally separating the roles of Ipl1p in chromosome alignment and the spindle checkpoint. Overexpressing Mps1p activates the checkpoint, arresting cells in metaphase with apparently normal bipolar spindles (Hardwick et al. 1996), although the status of kinetochore tension when Mps1 is overexpressed is not known. When ipl1-321 cells overexpressing Mps1p are shifted to the nonpermissive temperature to inactivate Ipl1p, most cells exit the cell cycle and segregate their chromosomes normally. Therefore, Ipl1p is required to maintain the spindle checkpoint arrest, and this function is temporally independent of an earlier and essential role in chromosome segregation. In addition, this experiment shows that the essential role of Ipl1p in chromosome segregation must occur before or during spindle assembly. The lack of cell cycle arrest associated with defects in Aurora B in other organisms may be owing to a similarly defective spindle checkpoint.
Ipl1p localizes to kinetochores during metaphase
The Ipl1p kinase localizes at or near kinetochores during a metaphase arrest. We did not detect the kinetochore localization of Ipl1p previously by immunofluorescence techniques on fixed whole cells (Biggins et al. 1999). However, by performing immunofluorescence on chromosome spreads of insoluble nuclear material, we were able to detect the kinetochore localization of Ipl1p at metaphase, suggesting that Ipl1p is in the Aurora B family. Our localization results are similar to those recently published (He et al. 2001). Finding Ipl1p at kinetochores is consistent with its role in the spindle checkpoint. Some checkpoint components, such as Mad2p, are found at kinetochores that lack microtubules but are absent from metaphase chromosomes (Chen et al. 1996). This is consistent with Mad2p being recruited to arrest cells that have already detected defects at their kinetochores. Other checkpoint proteins, such as Bub1p, Bub3p (Hoffman et al. 2001), and Ipl1p, are present at kinetochores whether the checkpoint is active or not, suggesting that they may monitor the status of kinetochore–microtubule interactions.
Ipl1/Aurora monitors tension
Ipl1p appears to have a specific role in the spindle checkpoint. It is needed to respond to kinetochores that are not under tension, but dispensable for detecting those that are not attached to microtubules. We used two manipulations that reduce tension at the kinetochore and should not affect attachment to microtubules: preventing DNA replication or sister chromatid cohesion. Ipl1p is required for both of these checkpoint-induced arrests but not for the arrest induced by complete depolymerization of the spindle. A study in HeLa cells also suggests that separate branches of the checkpoint monitor tension and attachment (Skoufias et al. 2001).
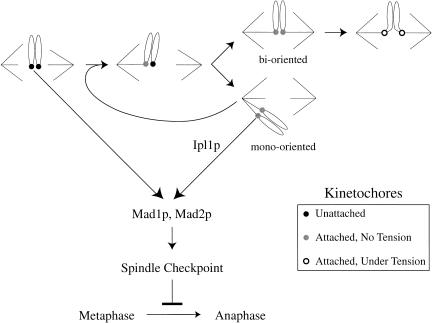
What is the advantage of monitoring both tension and attachment at the kinetochore? One possibility is that monitoring tension is the only way the cell can tell a pair of sister chromatids whose kinetochores are attached to the same pole (mono-orientation) from one whose kinetochores are attached to opposite poles (biorientation; Fig. 6).Both pairs of sisters have their kinetochores attached to microtubules, but the mono-oriented one will lead to aneuploid progeny unless the cell can detect this defect and delay anaphase until it has corrected it. Correction is difficult in budding yeast, where each kinetochore binds a single microtubule. To correct mono-orientation, one of the sister kinetochores must release its microtubule and then attach to a microtubule that originates from the opposite spindle pole (Fig. 6). More than 30 years ago, Nicklas and Koch showed that chromosome reorientation depended on kinetochore tension: kinetochore–microtubule linkages that are tense are stable; those that are not are unstable (Nicklas and Koch 1969). One possibility is that Ipl1p helps to destabilize microtubule attachments to kinetochores that are not under tension. In support of this model, we found previously that ipl1 mutant extracts are defective in the ATP-dependent release of microtubules in vitro and that this defect could be rescued by the addition of recombinant Ipl1 protein (Biggins et al. 1999). More recently, Tanaka and Nasmyth have found that ipl1 mutants appear to be unable to reorient chromosomes that are not under tension (T. Tanaka and K. Nasmyth, pers. comm.). These data suggest that the ATP-dependent loss of kinetochore–microtubule interactions in vitro may mimic an Ipl1p-dependent release of microtubules from kinetochores that are not under tension in vivo.
Figure 6.
Model for the role of Ipl1p in monitoring tension during spindle assembly. Telocentric chromosomes are shown for simplicity in illustrating kinetochore orientation. Early in mitosis, kinetochores are neither attached to microtubules nor under tension (black-filled kinetochores). Attaching both sister chromatids to the same pole (mono-orientation) produces kinetochores that are not under tension, but are bound to microtubules (gray-filled kinetochores). Once a bioriented spindle is established, tension can be generated on the kinetochores (open kinetochores). The spindle checkpoint must monitor defects in both attachment and tension to ensure bipolar spindle assembly. Our experiments suggest that Ipl1p has a specific role in monitoring tension but not attachment.
There are two ways in which Ipl1p could allow microtubule-bound kinetochores that are not tense to activate the spindle checkpoint. The first is by destabilizing microtubule attachment, thus producing naked kinetochores, which recruit proteins like Mad2 to activate the checkpoint. The second is by activating the checkpoint at kinetochores that are still attached to microtubules. We believe that both mechanisms exist. The evidence for the second is the ability of ipl1-321 to overcome the arrest caused by overexpression of Mps1p, coupled with the observation that Mps1p overexpression can activate the checkpoint in the absence of functional kinetochores (B. Stern and A.W. Murray, unpubl.). If the sole function of Ipl1p in the checkpoint was to create naked kinetochores, the absence of this protein should not affect an arrest that does not depend on the presence of kinetochores. If Ipl1p monitors tension, it may be the kinase that produces the phospho-specific 3F3/2 epitope found at kinetochores that are not under tension (Campbell and Gorbsky 1994; Nicklas et al. 1995).
Whether it activates the checkpoint directly or indirectly, Ipl1p seems to function upstream of the other known checkpoint components. The Mad1 and Mad2 proteins are required to respond to the absence of tension or the lack of bound microtubules at kinetochores. The role of other checkpoint proteins in responding to tension has not been tested, but all of them (Mps1p, Bub1p, Bub3p, and Cdc55p) are required to respond to kinetochores that are not attached to microtubules. Because Ipl1p is not required to respond to this defect, it must function upstream of other known checkpoint proteins, at least if the checkpoint is a simple linear pathway (see Fig. 6). The simplest interpretation of the ability of ipl1-321 to overcome the arrest caused by Mps1p overexpression is that Mps1 can be activated by two or more protein kinases: Ipl1p in response to the absence of tension, and another kinase, perhaps Bub1, in response to naked kinetochores. If the constitutive, basal activity of Ipl1p were sufficient to allow overexpressed Mps1 to arrest cells with normal spindles, inactivating Ipl1p would relieve the arrest. Although we do not detect any changes in the mobility of Mps1 protein by immunoblotting in the ipl1 mutant cells, the kinase activity of Mps1 in ipl1 mutant cells needs to be analyzed. Another possibility is that in response to defects in tension but not attachment, Ipl1p inhibits Cdc20p function in a manner similar to the Mad2 checkpoint protein. In Xenopus egg extracts, Aurora A interacts directly with Cdc20p, the activator of the APC that the spindle checkpoint inhibits (Farruggio et al. 1999).
One important alternative to the interpretation that Ipl1p is only required to detect certain kinetochore defects is that the effect of ipl1-321 is quantitative rather than qualitative: the mutant can still respond to a strong defect, but not to a weak one. This interpretation implies that the lack of tension generates a weak signal for the checkpoint, whereas the combined lack of tension and microtubule attachment generates a strong signal. Because IPL1 is an essential gene, it is impossible to know whether the null phenotype would have a stronger phenotype. The isolation of additional alleles of IPL1 may aid in testing these hypotheses.
Several studies have shown that the human Aurora genes are oncogenes. The aurora2 gene is amplified in many colorectal and breast cancer cell lines (Sen et al. 1997; Bischoff et al. 1998; Tanaka et al. 1999), and aurora2 maps to the 20q13 amplicon that is common to many human malignancies and is correlated with poor prognosis (Tanner et al. 1995; Sen et al. 1997). In addition, expression of activated Aurora2 can transform Rat1 fibroblasts and NIH3T3 cells in vitro and cause tumors in nude mice (Bischoff et al. 1998). These data suggest that defects in the regulation of the Ipl1/Aurora kinases can lead to genomic instability. Although our studies deal with loss of Ipl1 function, the overexpression of Aurora kinases may result in similar phenotypes. Because defects in checkpoint genes are associated with oncogenesis, it will be interesting to determine whether the human Aurora B kinase is needed for cells to delay when their kinetochores are not under tension. If so, it will be important to understand whether the genomic instability associated with defects in the kinase are caused by defects in chromosome alignment, the spindle checkpoint, or both.
Materials and methods
Microbial techniques and yeast strain constructions
Media and genetic and microbial techniques were essentially as described (Sherman et al. 1974; Rose et al. 1990). All experiments where cells were released from a G1 arrest were carried out by adding 1 μg/mL α-factor at the permissive temperature (23°C) for 3 h, washing cells twice in α-factor-free media, and resuspending them in prewarmed media. In all experiments studying synchronous cell cycles, α-factor was added back to 1 μg/mL after cells had budded to prevent cells from entering the next cell cycle. All experiments were repeated at least twice with similar results, and at least 100 cells were counted at each time point. Galactose was used at a final concentration of 4% in all experiments. Because galactose induction is somewhat temperature-sensitive, all experiments with galactose were performed at 35°C instead of 37°C. Stock solutions of inhibitors were made in DMSO and stored at −20°: 30 mg/mL benomyl (DuPont), 10 mg/mL nocodazole (Sigma), 10 mg/mL α-factor (Biosynthesis). For benomyl plus nocodazole experiments, cells were released into 30 μg/mL benomyl and 15 μg/mL nocodazole at 35°C because these drugs do not work as effectively at high temperatures. To visualize sister chromatids, copper sulfate was added to media at a final concentration of 0.25 mM to induce the GFP–lacI fusion protein that is under the control of the copper promoter.
The GAL-CDC6 experiment was carried out as follows to generate a synchronized G1 population of cells depleted of the Cdc6 protein. First, cells grown in galactose were arrested in G1 with α-factor at the permissive temperature (23°C). They were then released into galactose media for 20 min and then washed once into glucose to repress the CDC6 gene; α-factor was added when small buds formed to rearrest cells in the next cell cycle. To inactivate ipl1-321, the cells were released from the arrest at the nonpermissive temperature (37°C) in the presence of glucose to keep CDC6 repressed, and Pds1 levels were monitored during this cell cycle.
Yeast strains are listed in Table 1 and were constructed by standard genetic techniques. Diploids were isolated on selective media at 23°C and subsequently sporulated at 23°C. All strains containing PDS1-myc18:LEU2 were created by integration of a plasmid that was a gift of K. Nasmyth (Shirayama et al. 1998).
Table 1.
Yeast strains used in this study
| Strain
|
Genotype
|
|---|---|
| SBY214 | MATa ura3-1 leu2,3-112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 ade2-1 can1-100 bar1Δ lys2Δ |
| SBY322 | MATa ura3-1 leu2,3-112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 ade2-1 can1-100 bar1Δ lys2Δ ipl1-321 |
| SBY432 | MATa ura3-1 leu2,3-112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 ade2-1 can1-100 bar1Δ lys2Δ bub2ΔLEU2 |
| SBY596 | MATa ura3-1:IPL1-myc12:URA3 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ NDC10:HA3:KAN |
| SBY679 | MATa ura3-1:pGAL-MPS1-myc:URA3 leu2,3-112 ade2-1 can1-100 bar1Δ lys2Δ his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 PDS1-myc18:LEU2 |
| SBY680 | MATa ura3-1:pGAL-MPS1-myc:URA3 leu2,3-112 ade2-1 can1-100 bar1Δ lys2Δ his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 PDS1-myc18:LEU2 ipl1-321 |
| SBY762 | MATa ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ madlΔHIS3 cdc6:pGAL-UBI-R-CDC6:URA3 PDS1-myc18:LEU2 |
| SBY771 | MATa ura3-1 leu2,3-112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 ade2-1 lys2Δ can1-100 bar1Δ cdc6:pGAL-UBI-R-CDC6:URA3 PDS1-myc18:LEU2 ipl1-321 |
| SBY772 | MATa ura3-1 leu2,3-112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 ade2-1 can1-100 bar1Δ cdc6:pGAL-UBI-R-CDC6:URA3 PDS1-myc18:LEU2 |
| SBY818 | MATa ura3-1 leu2,3-112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 ade2-1 can1-100 bar1Δ lys2Δ PDS1-myc18:LEU2 |
| SBY819 | MATa ura3-1 leu2,3-112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 ade2-1 can1-100 bar1Δ lys2Δ PDS1-myc18:LEU2 ipl1-321 |
| SBY870 | MATa ura3-1 leu2,3-112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 ade2-1 can1-100 bar1Δ lys2Δ mcd1-1 PDS1-myc18:LEU2 |
| SBY871 | MATa ura3-1 leu2,3-112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 ade2-1 can1-100 bar1Δ lys2Δ mcd1-1 PDS1-myc18:LEU2 ipl1-321 |
| SBY920 | MATa ura3-1 leu2,3-112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 ade2-1 can1-100 bar1Δ lys2Δ PDS1-myc18:LEU2 mad2ΔURA3 |
| SBY934 | MATa ura3-1 leu2,3-112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 ade2-1 can1-100 bar1Δ lys2Δ PDS1-myc18:LEU2 bub2ΔKAN |
| SBY961 | MATa ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ IPL1-HA3:HIS3 cdc26ΔKAN CSE4:CSE4-myc13:URA3 |
All strains are isogenic with the W303 background.
Protein and immunological techniques
Protein extracts were made and immunoblotted as described (Minshull et al. 1996). 9E10 antibodies were obtained from Covance and used at a 1:10,000 dilution. For all time-course experiments, the optical density of each culture was measured at the beginning and at the end of the experiment, and samples were normalized in sample buffer accordingly. Equal protein concentrations were loaded in all lanes as judged by Ponceau S staining (data not shown).
Microscopy
Microscopy to analyze sister chromatids was performed as described (Biggins et al. 1999). Indirect immunofluorescence was carried out as described (Rose et al. 1990). DAPI was obtained from Molecular Probes and used at 1 μg/mL final concentration. Chromosome spreads were performed as described (Loidl et al. 1991; Michaelis et al. 1997). Lipsol was obtained from Lip Ltd. (Shipley, England). 12CA5 antibodies that recognize the HA tag were used at a 1:1000 dilution and obtained from Covance. A-14 c-myc rabbit antibodies (Santa Cruz Biotechnology) were used at a 1:1000 dilution to recognize the myc tag. Cy3 secondary antibodies were obtained from Jackson Immunoresearch and used at a 1:2000 dilution. FITC secondary antibodies were obtained from Jackson Immunoresearch and used at a 1:500 dilution.
Acknowledgments
We are especially grateful to Bodo Stern, whose work on the checkpoint and generosity made much of this work possible. We are grateful to Tomo Tanaka and Kim Nasmyth for sharing data prior to publication. We thank Stéphanie Buvelot, Rachel Howard-Till, Shelly Jones, Ben Pinsky, Marion Shonn, Bodo Stern, Sean Tatsutani, and Mark Winey for critical reading of the manuscript and discussions. We thank past and present members of the Murray and Morgan labs at UCSF for stimulating conversations and advice, especially Marion Shonn, Adam Rudner, Sue Jaspersen, Hiro Funabiki, and Needhi Bhalla. We thank the following people for strains and plasmids: Bodo Stern, Adam Rudner, and Kim Nasmyth. This work was supported by Jane Coffin Childs and American Cancer Society postdoctoral fellowships and a Sidney Kimmel Research Scholar grant to S.B. as well as grants from the National Institutes of Health and the Human Frontiers in Science Program to A.W.M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sbiggins@fhcrc.org; FAX (206) 667-6526.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.934801.
References
- Adams RR, Wheatleya SP, Gouldsworthy AM, Kandels-Lewis SE, Carmena M, Smythe C, Gerloff DL, Earnshaw WC. INCENP binds the aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr Biol. 2000;10:1075–1078. doi: 10.1016/s0960-9822(00)00673-4. [DOI] [PubMed] [Google Scholar]
- Adams RR, Maiato H, Earnshaw WC, Carmena M. Essential roles of Drosophila inner centromere protein (incenp) and aurora B in histone h3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J Cell Biol. 2001;153:865–880. doi: 10.1083/jcb.153.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandru G, Zachariae W, Schleiffer A, Nasmyth K. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J. 1999;18:2707–2721. doi: 10.1093/emboj/18.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Hardwick K, Javerzat JP. Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J Cell Biol. 1998;143:1775–1787. doi: 10.1083/jcb.143.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes & Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Bhalla N, Chang A, Smith DL, Murray AW. Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics. 2001;159:453–470. doi: 10.1093/genetics/159.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff JR, Plowman GD. The Aurora/Ipl1p kinase family: Regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 1999;9:454–459. doi: 10.1016/s0962-8924(99)01658-x. [DOI] [PubMed] [Google Scholar]
- Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- Campbell MS, Gorbsky GJ. Microinjection of mitotic cells with the 3F3/2 anti-phosphoepitope antibody delays the onset of anaphase. J Cell Biol. 1994;129:1195–1204. doi: 10.1083/jcb.129.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Botstein D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 1993;135:677–691. doi: 10.1093/genetics/135.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R-H, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes & Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Farruggio DC, Townsley FM, Ruderman JV. Cdc20 associates with the kinase aurora2/Aik. Proc Natl Acad Sci. 1999;96:7306–7311. doi: 10.1073/pnas.96.13.7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesquet D, Fitzpatrick PJ, Johnson AL, Kramer KM, Toyn JH, Johnston LH. A Bub2p-dependent spindle checkpoint pathway regulates the Dbf2p kinase in budding yeast. EMBO J. 1999;18:2424–2434. doi: 10.1093/emboj/18.9.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R, Formenti E, Lucchini G, Piatti S. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J Cell Biol. 1999;145:979–991. doi: 10.1083/jcb.145.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yangida M. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- Gardner RD, Poddar A, Yellman C, Tavormina PA, Monteagudo MC, Burke DJ. The spindle checkpoint of the yeast Saccharomyces cerevisiae requires kinetochore function and maps to the CBF3 domain. Genetics. 2001;157:1493–1502. doi: 10.1093/genetics/157.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R, Glover DM. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol. 2001;152:669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover DM, Leibowitz MH, McLean DA, Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Johnston RC, Smith DL, Murray AW. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J Cell Biol. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Rines DR, Espelin CW, Sorger PK. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at ptk1 kinetochores. Mol Biol Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Trotis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Hwang LH, Murray AW. A novel yeast screen for mitotic arrest mutants identifies DOC1, a new gene involved in cyclin proteolysis. Mol Biol Cell. 1997;8:1877–1887. doi: 10.1091/mbc.8.10.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast Cdc20: A target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Kaitna S, Mendoza M, Jantsch-Plunger V, Glotzer M. Incenp and an aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr Biol. 2000;10:1172–1181. doi: 10.1016/s0960-9822(00)00721-1. [DOI] [PubMed] [Google Scholar]
- Kim JH, Kang JS, Chan CS. Sli15 associates with the Ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1999;145:1381–1394. doi: 10.1083/jcb.145.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: An effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- King JM, Nicklas RB. Tension on chromosomes increases the number of kinetochore microtubules but only within limits. J Cell Sci. 2000;113 Pt 21:3815–3823. doi: 10.1242/jcs.113.21.3815. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- ————— Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Li R. Bifurcation of the mitotic checkpoint pathway in budding yeast. Proc Natl Acad Sci. 1999;96:4989–4994. doi: 10.1073/pnas.96.9.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li X, Nicklas RB. Mitotic forces control a cell cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- Loidl J, Nairz K, Klein F. Meiotic chromosome synapsis in a haploid yeast. Chromosoma. 1991;100:221–228. doi: 10.1007/BF00344155. [DOI] [PubMed] [Google Scholar]
- Loidl J, Klein F, Engebrecht J. Genetic and morphological approaches for the analysis of meiotic chromosomes in yeast. Methods Cell Biol. 1998;53:257–285. doi: 10.1016/s0091-679x(08)60882-1. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Minshull J, Straight A, Rudner A, Dernburg A, Belmont A, Murray AW. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr Biol. 1996;6:1609–1620. doi: 10.1016/s0960-9822(02)70784-7. [DOI] [PubMed] [Google Scholar]
- Nicklas RB, Koch CA. Chromosome manipulation III. Induced reorientation and the experimental control of segregation in meiosis. J Cell Biol. 1969;43:40–50. [Google Scholar]
- Nicklas RB, Ward SC, Gorbsky GJ. Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. J Cell Biol. 1995;130:929–939. doi: 10.1083/jcb.130.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Cell division mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- Pangilinan F, Spencer F. Abnormal kinetochore structure activates the spindle assembly checkpoint in budding yeast. Mol Biol Cell. 1996;7:1195–1208. doi: 10.1091/mbc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Bohm T, Cocker JH, Diffley JF, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes & Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Heiter P. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Sen S, Zhou H, White RA. A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene. 1997;14:2195–2200. doi: 10.1038/sj.onc.1201065. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink G, Lawrence C. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1974. [Google Scholar]
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonn MA, McCarroll R, Murray AW. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science. 2000;289:300–303. doi: 10.1126/science.289.5477.300. [DOI] [PubMed] [Google Scholar]
- Skoufias DA, Andreassen PR, Lacroix FB, Wilson L, Margolis RL. Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc Natl Acad Sci. 2001;98:4492–4497. doi: 10.1073/pnas.081076898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern BM, Murray AW. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr Biol. 2001;11:1462–1467. doi: 10.1016/s0960-9822(01)00451-1. [DOI] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Sullivan M, Lehane C, Uhlmann F. Orchestrating anaphase and mitotic exit: Separase cleavage and localization of Slk19. Nat Cell Biol. 2001;3:771–777. doi: 10.1038/ncb0901-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Haruki N, Nomoto S, Masuda A, Saji S, Osada H. Identification of frequent impairment of the mitotic checkpoint and molecular analysis of the mitotic checkpoint genes, hsMAD2 and p55CDC, in human lung cancers. Oncogene. 1999;18:4295–4300. doi: 10.1038/sj.onc.1202807. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Kimura M, Matsunaga K, Fukada D, Mori H, Okano Y. Centrosomal kinase AIK1 is overexpressed in invasive ductal carcinoma of the breast. Cancer Res. 1999;59:2041–2044. [PubMed] [Google Scholar]
- Tanaka T, Fuchs J, Loidl J, Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- Tanner MM, Tirkkonen M, Kallioniemi A, Holli K, Collins C, Kowbel D, Gray JW, Kallioniemi OP, Isola J. Amplification of chromosomal region 20q13 in invasive breast cancer: Prognostic implications. Clin Cancer Res. 1995;1:1455–1461. [PubMed] [Google Scholar]
- Taylor SS, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernek W, Poupart M-A, Koonin E, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: A family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Wang Y, Burke DJ. Checkpoint genes required to delay cell division in response to nocodazole respond to impaired kinetochore function in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6838–6844. doi: 10.1128/mcb.15.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Cdc55p, the B-type regulatory subunit of protein phosphatase 2A, has multiple functions in mitosis and is required for the kinetochore/spindle checkpoint in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:620–626. doi: 10.1128/mcb.17.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Salmon ED. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E, Winey M. The S. cerevisiae SPB duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells WAE, Murray AW. Aberrantly segregating centromeres activate the spindle assembly checkpoint in budding yeast. J Cell Biol. 1996;133:75–84. doi: 10.1083/jcb.133.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]