Abstract
Objectives
(a) To determine the extent and range of errors and issues in the Systematised Nomenclature of Medicine – Clinical Terms (SNOMED CT) hierarchies as they affect two practical projects. (b) To determine the origin of issues raised and propose methods to address them.
Methods
The hierarchies for concepts in the Core Problem List Subset published by the Unified Medical Language System were examined for their appropriateness in two applications. Anomalies were traced to their source to determine whether they were simple local errors, systematic inferences propagated by SNOMED's classification process, or the result of problems with SNOMED's schemas. Conclusions were confirmed by showing that altering the root cause and reclassifying had the intended effects, and not others.
Main results
Major problems were encountered, involving concepts central to medicine including myocardial infarction, diabetes, and hypertension. Most of the issues raised were systematic. Some exposed fundamental errors in SNOMED's schemas, particularly with regards to anatomy. In many cases, the root cause could only be identified and corrected with the aid of a classifier.
Limitations
This is a preliminary ‘experiment of opportunity.’ The results are not exhaustive; nor is consensus on all points definitive.
Conclusions
The SNOMED CT hierarchies cannot be relied upon in their present state in our applications. However, systematic quality assurance and correction are possible and practical but require sound techniques analogous to software engineering and combined lexical and semantic techniques. Until this is done, anyone using SNOMED codes should exercise caution. Errors in the hierarchies, or attempts to compensate for them, are likely to compromise interoperability and meaningful use.
Keywords: Knowledge bases, knowledge representations, methods for integration of information from disparate sources, knowledge acquisition and knowledge management, developing and refining EHR data standards (including image standards), data models, data exchange, controlled terminologies and vocabularies, communication, integration across care settings (inter- and intraenterprise), ontologies, terminology, EHRs
Introduction
The Systematized Nomenclature of Medicine–Clinical Terms (SNOMED CT) is now mandated in the USA, UK, and several other countries for coding of clinical problems. The SNOMED identifiers provide a stable reference point for coding diseases. There have been numerous studies of the coverage of SNOMED and its comparison with other coding systems, for example Cornet and De Keizer,1 and some studies of issues with reliability or unreliability of coding and mapping using SNOMED, for example.2 3
However, there have been few studies of the SNOMED hierarchies, despite the fact that, according to SNOMED's declared description logic semantics, they manifest the logical meaning of the codes. When doctors apply SNOMED codes to a patient, they are stating that those codes and all their ancestors in the hierarchy apply to that patient. When researchers use codes in queries, they are querying for those codes and all of their descendants. When software interprets postcoordinated expressions, it depends on the hierarchies to give those expressions their correct meaning.
This paper reports attempts to use the SNOMED hierarchies in two practical applications:
as a contributor to the ‘ontological component’ of the eleventh revision of the International Classification of Diseases (ICD-11);
as part of the documentation tools for a commercial clinical information system.
By contrast with most previous studies, we are concerned here only with inferences that are incorrect or misleading clinically. We are not concerned with whether or not SNOMED complies with some upper ontology4 5 or other set of desirable principles.6 7 We also omit issues around ‘Situations with explicit context,’i which we have discussed elsewhere.8 The paper has some analogies with Ceusters et al's analysis9 but focuses on issues that arose in practice rather than on discrepancies between linguistic analysis of SNOMED names and the formal representation of the corresponding concepts.
Background
Unlike classifications, such as ICD, SNOMED CT hierarchies are formulated using a subset of first-order logic known as ‘description logic’ that specifies their semantics.10 11 SNOMED hierarchies are comprehensive and universal. All and only concepts satisfying the definition of a higher-level ‘ancestor’ concept are classified under it as ‘descendants,’ and everything said within SNOMED about any concept applies to all of its descendants (see online appendix V).
Because it is formulated in a description logic, SNOMED hierarchies are created in two steps:
A ‘stated form’ that defines each concept is asserted manually by SNOMED's authors in the description logic.
A software ‘classifier’ is then used to organize the concepts logically into hierarchies based on their stated definitions. The classified hierarchies are then used to generate the distribution files. By comparison with a programming language, the ‘stated form’ is analogous to the source code; the distribution files are analogous to the compiled program.
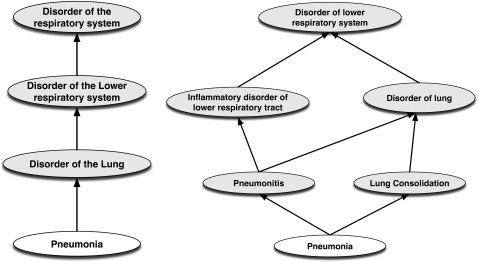
For example, figure 1 shows a graphical comparison of the stated and classified hierarchies for pneumonia. As can be seen, much of the detail is only apparent in the classified form.
Figure 1.
Comparison of stated (left) and classified (right) hierarchies for ‘pneumonia.’
Although using a logic-based formalism makes it possible to manage a larger terminology than could be managed manually, it also brings with it new sources of error:
Information propagates, whether it is correct or incorrect, so that a single statement can have wide-ranging, unintended effects.
Errors in the modeling schema can give rise to widespread errors in the resulting classification.
Attempting to correct errors without tracing them to their root source rarely works. If the incorrect statement at the root of the error is not corrected, the erroneous classification will be inferred again the next time the classifier is run. If attempts are made to correct widespread consequences rather than tracing them to their root, it is unlikely that all affected codes will be found. The result is ‘helter-skelter’ hierarchies where some codes are classified one way and some another.
Until recently, independent study of the SNOMED hierarchies has been difficult for two reasons:
SNOMED did not release the stated form, so that it was impossible to trace anomalies to their root source.
SNOMED is very large—nearly 500 000 concepts—which was beyond the range of most previously available generic tools that might be used for independent analysis.
These two barriers can now be largely overcome:
SNOMED now releases the stated form as part of its standard distribution.
The Unified Medical Language System is maintaining a ‘Core Problem List Subset’ii consisting of around 9000 concepts. New technologies make it possible to identify all other concepts that affect the classification of members of the subset consisting of fewer than 40 000 concepts.
Improvements in software and hardware make it possible to manage even the entire SNOMED corpus with standard tools.
Materials and methods
Content
This study used the SNOMED Core Problem List Subset as available in August 2010 and the 31 January 2010 IHTSDO distribution of SNOMED CT. The ‘stated form’ of the SNOMED was converted to the standard syntax most widely used by description logic tools, OWL,12 using the scripts provided in the SNOMED distribution. A module, the ‘Extended Core Subset,’ was then extracted using methods in the OWL APIiii. These methods guarantee that the classification will be the same in the module as in the complete SNOMED.13
Tools
For examining and visualizing the OWL, we used Protégé 4.0iv 14 with the SNOROCKET classifierv,15 which is SNOMED's preferred classifier. The results were further checked with the standard classifiers bundled with Protégé: FaCT++ and Pellet. For checking findings against the complete 31 January 2010 IHTSDO release of SNOMED, we used the SNOBvi browser supplemented by the CliniClue XPlore browser.vii SNOB was used because its ‘center’ function makes it easy to look upwards in the hierarchies as well as down (details in on-line appendix VI).
Methods
Identifying starting issues
This was a study of opportunity stimulated by the desire to use SNOMED in two applications: development of a commercial clinical system and as a contributor to the new 11th revision of ICD. The starting-points were taken from conditions important to those two applications, and focused on cardiovascular and respiratory diseases, and diabetes mellitus.
Our primary method to identify issues was to look first upwards and then downwards in the hierarchies from the selected starting codes. Most SNOMED codes have multiple parents and many ancestors; only those that raised issues are reported here. Most issues were identified looking upwards because this gives a smaller space to examine. Issues raised were discussed with our collaborators in our commercial development and our colleagues in the ICD revision process. Except where noted, there was overall consensus that the issues raised represented problems for the applications. Because of the degree of consensus, it was decided that an analysis of the anomalies was of higher priority than formal tests of inter-rater agreement, although methodologies for such studies are being piloted.
All findings were confirmed against the complete SNOMED release of 31 January 2010.
Analysis by repair
The source of each anomaly was sought using the method we term ‘Analysis by repair.’ Because SNOMED uses a relatively simple subset of description logic, it is usually possible to identify the source of an anomaly relatively easily by inspection and experimentation. To confirm the source, changes were made to the OWL version of the stated form. The modified model was reclassified to confirm that the changes had the intended consequences. The ‘usage’ view in Protégé was then used to find concepts that referenced each changed concept. Referenced concepts, or a significant sample of them, were checked to test if the changes had entailed any unintended consequences. Applicable tools to do a definitive semantic ‘diff’ between the original and altered models are not currently available, so tests for unintended consequences were systematic but not exhaustive. All changes were limited to the standard SNOMED formalism (‘EL++’ also known as ‘OWL-EL’).
Results
There were some areas of SNOMED, notably respiratory disorders, that raised few issues. However, we found seven major types of problems in the SNOMED hierarchies related to the description logic modeling and classification process, most of which had widespread consequences. These are summarized in table 1 and discussed below. More detailed explanations of issues, solutions, methods, and tools used can be found in the online appendices, along with the original screen shots from which the figures were derived.
Table 1.
Major types of problems related to description logic modeling in the Systematized Nomenclature of Medicine
| Problem type | Examples | |
| 1 | Simple error or omission, some with widespread consequences |
|
| 2 | Incomplete modeling | Myocardial infarction not kind of Ischemic heart disease |
| 3 | Issues with site and resulting inferences |
|
| 4 |
|
|
| 5 |
|
|
| 6 | Lack of distinction of structure, function |
|
| 7 | Inconsistent modeling of complications |
|
Errors and omissions with propagation and helter-skelter modeling
Diabetes and the pancreas
Diabetes mellitus (including types 1 and 2) is stated to have site endocrine pancreas. Diabetes type 2 is associated with a lack of response to insulin, rather than with malfunction of the pancreas. The relation between diabetes and the pancreas should be moved to refer only to diabetes type 1.
Autoimmune and allergic disorders
Of all autoimmune disorders, exactly three appear under ‘Allergic disorders by site’viii: ‘Graves’ disease’ix, ‘Myasthenia gravis,’x and ‘Idiopathic thrombocytopenic purpura’xi (ITP). On investigation, the first two inferences occur because the concepts are classified under ‘Antibody-mediated activation/inactivation’xii which is itself classified under ‘Immune hypersensitivity disorder’xiii which turns out to be the fully specified name for the concept with preferred term ‘Allergic Disorder.’ The third, ITP, follows from the fact that one of its ancestors ‘Antibody-mediated cytolysis’xiv is also a descendant of ‘Immune hypersensitivity disorder’/‘Allergic disorder.’
The simplest alternative is to move the two offending concepts up from ‘Allergic disorder’ to ‘Immune disorder.’ After this change, all autoimmune disorders are classified analogously, and the list of allergic disorders contains no obvious anomalies.
Skin and subcutaneous tissues
‘Skin and subcutaneous tissue,’xv and hence ‘Skin’xvi itself, are not classified as ‘Soft tissues.’xvii Because the classifier organizes disorders and injuries following the anatomical model, this means that injuries and disorder of skin are not inferred by the classifier to be included under ‘Disorder of soft tissue.’xviii
To add to the confusion, some disorders of the skin have been stated manually to be disorders of soft tissue, while others have not. For example, cuts of the anklexix are classified as ‘Injuries to soft tissue,’ but cuts of the ‘Lower limb’xx are not, even though the lower limb includes the ankle.
A single assertion that ‘Skin and subcutaneous tissue’ is a kind of ‘Soft tissue’ solves this problem systematically. All other assertions that injuries or disorders of the skin relate to soft tissue are then redundant.
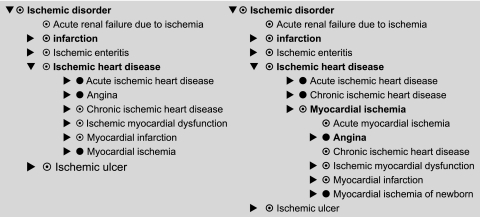
Incomplete modeling: myocardial infarction and ischemic heart disease
SNOMED does not classify ‘Myocardial infarction’xxi as a kind of ‘Ischemic heart disease’xxii although all references and experts consulted do. On investigation, the problem is that ‘Myocardial infarction,’ ‘Infarction’ and ‘Ischemic heart disease’ are all only partially defined, so that there is no logical connection between them. Hence, the classifier cannot infer the linkage. The root problem is that ‘Infarction’xxiii is not defined as being due to ‘Ischemia,’xxiv contrary to reference works and collaborators.
To solve this problem, all three concepts must be fully defined. To make the link between infarction and ischemia within SNOMED's formalism, there are two potential solutions. The first involves using a ‘right identity’ or ‘property path’ to link the attribute for ‘morphology’xxv to the attribute ‘due to.’xxvi The second requires stating directly that any disorder with morphology infarction is due to ischemia. The resulting hierarchy in either case is the same and shown on the left-hand side of figure 2. If, more radically, one makes ‘Myocardial ischemia’ fully defined and asserts that ‘Angina’ has the site myocardium rather than just heart, then one obtains the more compact structure in right-hand half of figure 2. Which is more appropriate is a matter for discussion among medical experts (for details, see online appendix I).
Figure 2.
Two alternative hierarchies for ‘Disorder due to Ischemia’ (the Systematized Nomenclature of Medicine has no concept for ‘Disorder due to ischemia’ per se).
Issues with sites of systemic disorders
SNOMED's policy is to give endocrine disorders the site of the organ responsible for secreting the responsible hormone, even when the effects are systemic. Specific sites are also given to other systemic disorders. This can lead to unexpected classifications.
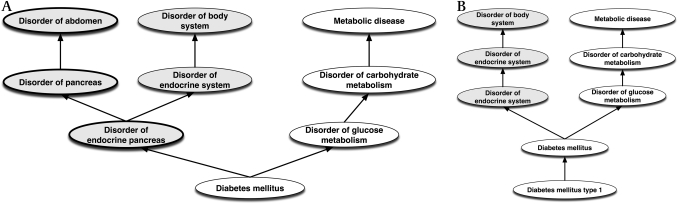
Is ‘Diabetes type 1’ a ‘Disorder of the abdomen’?
If we give diabetes, or even diabetes type 1, the site endocrine pancreas, then since the endocrine pancreas is part of the pancreas, which is part of the abdomen, the classifier infers that diabetes is a disorder of the abdomen.
A simple alternative is to make the site of diabetes simply a ‘Structure of endocrine system.’xxvii The relation between type 1 diabetes and the pancreas can then be made via another attribute. The inference that it is a disorder of the abdomen is therefore avoided. The hierarchies before and after the change are shown in figure 3.
Figure 3.
(A) Diabetes upwards hierarchy as is in the Systematized Nomenclature of Medicine showing classification as a disorder of the abdomen. (B) Alternative Diabetes type 1 upwards hierarchy with link to pancreas removed (and represented by other means not shown).
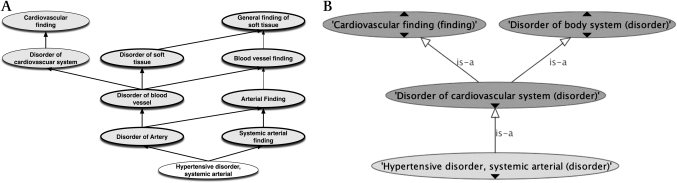
Is hypertension a disease of arteries? Of soft tissues?
Is ‘Systemic hypertension’xxviii a disease of arteries analogous to arteritis, atherosclerosis, arterial thromboses, etc? Or should it be treated simply as a disorder of the cardiovascular system? To say that all forms of hypertension are sited in arteries leads to odd results for some—for example, endocrine hypertension.xxix Furthermore, arteries are classified as soft tissues. This leads to the inference that ‘hypertension’ is a ‘disease of soft tissues,’ a conclusion to which all our experts objected.
An alternative is to make the site of hypertension ‘cardiovascular system’ or, almost equivalently, to assert directly that it is a kind of cardiovascular disorder. The original and modified hierarchies are shown in figure 4.
Figure 4.
(A) Existing upwards classification of Hypertension (fully specified name ‘Hypertensive disorder, systemic arterial’). (B) Hypertensive disorder site reduced to ‘Entire cardiovascular system’.
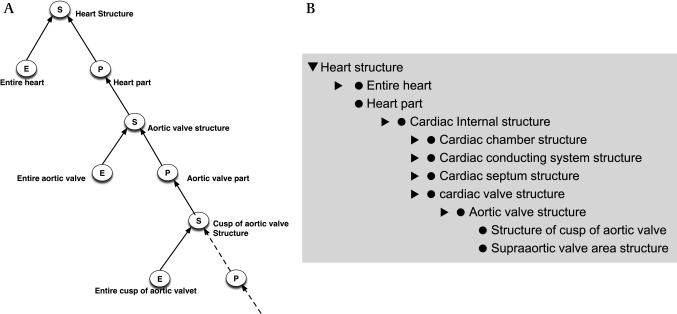
Errors in modeling anatomy: Structure-Entire-Part (SEP) triples and the ankle in the abdomen
In the past few years, SNOMED has changed its representation of anatomy, adopting a variant of the ‘Structure-Entire-Part (SEP)’ triple mechanism developed by Hahn and Schultz.16 17 SEP triples are a means to avoid the use of transitive properties and to make clear which disorders and procedures apply to an entire structure and which to the structure and/or its parts. For example, ‘total nephrectomy’ refers to the entire kidney, whereas ‘kidney operation’ refers to any operation on the kidney and/or any of its parts.
In the SEP triple representation, each anatomic structure is represented by a triple of concepts:
the Structure (S) concept—for the structure and all of its parts—for example, the heart and all its parts (‘Heart structure’xxx);
the Entire (E) concept—for the entire structure—for example, the entire heart (‘Entire heart’xxxi);
the Parts (P) concept—that represents the proper parts excluding the structure itself (‘Heart part’xxxii).
The graphical schema and an example from the hierarchy for ‘Heart structure’ are shown in figure 5A. ‘Cusp of aortic valve structures’ are kinds of ‘Aortic valve structures’ are, after several steps, kinds of ‘Heart parts,’ which are kinds of ‘Heart structure.’ SNOMED typically abbreviates the format slightly by omitting unused nodes, as shown in figure 5B.
Figure 5.
(A) Theoretical structure of Structure-Entire-Part (SEP) triple for heart and aortic valve. (B) Systematized Nomenclature of Medicine's implementation of SEP triple for heart aortic valve omitting unused nodes. Note also the inconsistent naming of structure (S) nodes for the right and left popliteal artery.
Used correctly, SEP triples lead to correct inferences. Unfortunately, there are two problems with SNOMED's implementation.
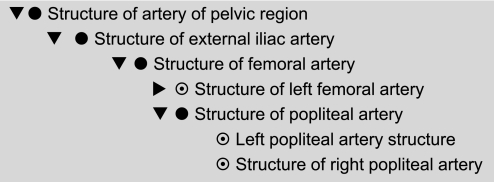
Inconsistent and underspecified naming
Even in the small example in figure 5, it is obvious that SNOMED sometimes uses the naming convention ‘X structure’ (eg, ‘Heart structure’) and sometimes ‘Structure of X’ (eg, ‘Structure of cusp of aortic valve’). In the extreme, left and right cases use different conventions, as shown in the last two lines of figure 6.
Figure 6.
Existing (Incorrect) Systematized Nomenclature of Medicine hierarchy for ‘Structure of artery of the pelvic region’ including ‘Popliteal artery structure’.
The problem is more serious where no indication is given in names referring to anatomy as to whether the ‘Entirety’ (E) node or ‘Structure’ (S) node is intended. This leads to oddities such as ‘Optic disc swelling’xxxiii is a kind of ‘Eye swelling.’xxxiv It is clear from the logical definitions that this means: ‘Optic disc swelling is a kind of swelling of a structure of the eye.’ However, this meaning is not conveyed by the fully specified names.
Incorrect modeling of branches
A much more serious problem is that branches are modeled analogously to parts as kinds of the structure (S) node rather than kinds of the entirety (E) node. The result is that everything said about the structure is inferred to be true of the branch. The portion of the hierarchy ‘Structure of artery of the pelvic region’xxxv to ‘Structure of the popliteal artery’xxxvi is shown in figure 6. In the full release, the pattern continues all the way to the dorsalis pedis artery.xxxvii This leads to inferences such as: ‘Injuries of the dorsalis pedis artery’xxxviii is a kind of ‘Injury of the abdomen’xxxix and ‘Injury to the pelvis.’xl It also results in the classification of ‘Deep thrombosis of the profunda femoris vein’xli as a ‘Thrombosis of the vena cava’xlii and hence a ‘Disorder of the trunk.’ Such errors are not isolated. For example, in the full release they lead to the conclusion that ‘Thrombophlebitis of breast’xliii is a disorder of the abdomen and lower extremity.
Correcting this requires correcting the modeling of all branches of nerves, veins, arteries, and lymphatic vessels. There are at least two alternative modeling schemas that retain the transitivity of the branching relation, both of which have been confirmed to work with the SNOROCKET reasoner both practically and theoretically18 (see online appendix II).
Some care must be taken because clinical language does not always map literally to logic. For example, ‘blockage of a branch of the aorta’ does not constitute a ‘blockage of the aorta,’ but ‘blockage of the anterior descending branch of the left coronary artery’ might well be considered a ‘blockage of the left coronary artery.’ Despite its name, therefore, the anterior descending branch of the left coronary artery might well be considered, logically, to be a part rather than a branch. Whether a structure should be modeled logically as a branch or a part depends on how it is used, not how it is named.
Overgeneralized concepts with underspecified ‘fully specified names’
Should there be a simple concept for intracranial subdural hemorrhage? Hematoma?
The concepts in the Core Problem List Subset mentioning ‘subdural’ are shown in table 2. Of the nine concepts, six are not classified under intracranial hemorrhage,xliv including the most used, which is simply ‘Subdural hemorrhage.’ Extracranial (spinal) subdural hemorrhages undoubtedly occur, but are rare. It seems likely that most ‘Subdural hemorrhages’ referred to intracranial hemorrhages. Certainly, our clinical systems collaborators felt it unsafe for subdural (or subarachnoid) hemorrhages not to be classed as intracranial by default.
Table 2.
Concepts mentioning ‘subdural’ in the Core Problem List Subset classed or not classed as intracranial
| Classified as Intracranial | Not classified as intracranial |
| Chronic intracranial subdural hematoma* | Subdural hemorrhage† |
| Subdural hemorrhage following injury with moderate loss of consciousness‡ | Traumatic subdural hemorrhage§ |
| Subdural hemorrhage following injury with brief loss of consciousness¶ | Subdural hemorrhage nontraumatic** |
| Nontraumatic subdural hematoma†† | |
| Traumatic subdural hematoma‡‡ | |
| Closed fracture of vault of skull with subarachnoid, subdural AND/OR extradural hemorrhage§§ |
Chronic intracranial subdural hematoma (disorder) | 304831001.
Subdural hemorrhage (disorder) | 35486000.
Subdural hemorrhage following injury without open intracranial wound AND with moderate loss of consciousness (1–24 h) (disorder) | 63323000.
Traumatic subdural hemorrhage (disorder) | 209987007.
Subdural hemorrhage following injury without open intracranial wound AND with brief loss of consciousness (less than 1 h) (disorder) | 26205001.
Subdural hemorrhage—nontraumatic (disorder) | 195176009.
Non-traumatic subdural hematoma (disorder) | 410064000.
Closed fracture of vault of skull with subarachnoid, subdural AND/OR extradural hemorrhage (disorder) | 57998008.
Closed fracture of vault of skull with subarachnoid, subdural AND/OR extradural hemorrhage (disorder) | 57998008.
However, to our surprise, there are no codes in SNOMED for intracranial subdural or subarachnoid hemorrhage per se. We would propose adding these concepts and making the fully specified names explicit:
‘Subdural hemorrhage, intracranial AND/OR extracranial’;
‘Subdural hemorrhage, intracranial.’
Most applications would then refer to ‘Subdural hemorrhage, intracranial,’ probably via a synonym of simply ‘Subdural hemorrhage.’ The same applies to subarachnoid hemorrhage.
Broad and narrow usages of ‘soft tissue’
A related problem concerns that ambiguity of the phrase ‘Soft tissues.’xlv Some authorities define soft tissues as anything that is not bone; other usages, such as ‘soft tissue injury,’ seem to imply a more restrictive definition of skin, muscle, connective tissue, etc, not including blood vessels, nerves, or internal organs. SNOMED uses a relatively broad definition, including vessels and nerves, but not internal organs. This leads to a number of classifications that most users considered to be wrong: for example that neuropathy and aortic aneurysm are soft-tissue disorders.xlvi Providing both a broad and narrow concept for soft tissue with suitable fully defined names makes it possible to capture both clinical intuitions unambiguously (see online appendix III).
Lack of distinction between structure and function
Is papilledema a kind of neuropathy?
Neuropathy is defined by SNOMED as a disorder of nerve.xlvii This appears to correspond to some reference definitions but gives rise to surprising inferences—for example that ‘Optic nerve edema’ (Papilledema),xlviii tumors, and injuries to nerves are classified as kinds of ‘Neuropathy.’
The simplest solution is to create two distinct concepts: ‘Disorder of nerve’ and a child concept ‘Neuropathy’ covering just the functional disorders usually expected by most of our collaborators.
Structure, function, and hormonal action in endocrine disorders
In the case of endocrine disorders, there is the further complication that dysregulation of the level of the hormone is not always the result of dysfunction of the corresponding organ—it may, for example, be iatrogenic or due to a tumor. There are, however, occasions in which a general heading is needed for all three—structural, functional, and regulatory.
The distinction between function and regulation is made in SNOMED's existing release for Diabetes mellitus, as shown in figure 7. However, the pattern is not carried through for other endocrine disorders; nor is there a common parent in the case of diabetes (see online appendix IV).
Figure 7.
Existing Systematized Nomenclature of Medicine classification of glucose metabolism and regulation.
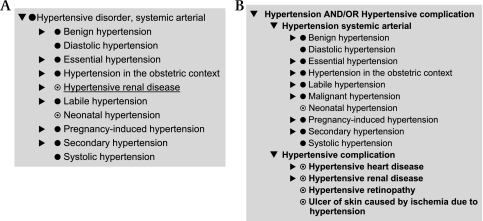
Inconsistent modeling of complications: hypertensive disorders
For ‘Diabetes mellitus,’ there is a separate class ‘Diabetic complication,’xlix formulated so that the logic guarantees that anything that is stated to be associated withl diabetes will be classified as a kind of ‘Diabetic complication.’
Unfortunately, this pattern is not carried through for other disorders. Consider hypertension.li ‘Hypertension’ is a synonym for ‘Hypertensive disorder, systemic arterial.’ The disorders classified under it include various forms of hypertension, ‘Hypertensive renal disease’lii and ‘Hypertensive encephalopathy’liii (in the full release) but not ‘Hypertensive retinopathy,’liv ‘Hypertensive heart disease’lv or ‘Ulcer of skin caused by ischemia due to hypertensive disease.’lvi There is no distinction between the disorder and its complications.
There are two problems here. The first is the incomplete modeling of some complications, which can easily be found by lexical search. This needs to be corrected.
The second, and more serious, issue is the lack of a generic notion of ‘complication of hypertension’ separate from ‘Hypertension.’ This problem also occurs for other conditions. To address it, we recommend that all disorders with complications follow a consistent pattern:
A parent concept: the disorder AND/OR its complications
-
Two child concepts:
the disorder;
the complications of the disorder.
The comparison of the original classification for hypertension/hypertensive disorder and the suggested alternative are shown in figure 8A,B.
Figure 8.
(A) Existing hierarchy of hypertensive disorder in Extended Core Subset showing only Hypertensive renal disease of potential complications. (B) Suggested revised hierarchy for Hypertension and its Complications, including complications found by lexical search.
Conclusions
This study has five classes of outcome:
On the SNOMED hierarchies. There are sufficient anomalies in the hierarchies that they cannot be used without significant modification in our applications. More generally, we question whether clinicians entering codes or researchers retrieving information understand their implications. As postcoordination relies on accurate classification, it is doubtful that applications using postcoordination will behave predictably.
On the use of description logic in SNOMED. Using a description logic is both part of the problem and part of the solution. The response to the issues raised here is not to abandon SNOMED's description logic but to use it more effectively. Using a description logic means that the correcting root errors found in modules will usually repair analogous problems throughout SNOMED.
On the possibility of quality assurance of SNOMED. Given modern tooling and computer power, the barriers quality assurance of SNOMED can now be overcome, although no well-integrated toolset is yet available.
On practicality of quality assurance of SNOMED. This was a preliminary study and not exhaustive, but it required less than three person-months using poorly integrated tools. Given an integrated toolset, we estimate that a thorough quality assurance of the Core Problem List Subset would require a small team under 2 years, probably less. This would cover a high fraction of all uses of SNOMED. Most changes would be propagated automatically by the description logic into the full SNOMED corpus. Applying these methods to the remainder of the SNOMED findings would require further resources, but they would be minor by comparison with the effort already devoted to SNOMED's development, let alone to those that will be required for its implementations.lvii
-
On methods required. Using a description logic requires staff who understand both medical content and description logics. It requires adapting the techniques of software engineering to tracing and managing errors. Space does not permit setting out a detailed methodology.lviii However, key maxims should include:
Start from clinically important concepts—use clinical intuition.
Focus on the classified hierarchies—reclassify after every change.
Work in small modules—so that reclassification is quick.
Look upwards first and then downwards—there are fewer ancestors than descendants.
Trace all errors to their root cause—avoid local ‘kluging.’
Look for analogous errors and repair using consistent patterns—for example, complications and sites.
Reformulate problematic sections systematically rather than attempting to repair them—for example, head injury and branches in anatomy.
Use a combination of lexical and semantic methods—as first suggested by Campbell et al19 and now made straightforward using Ontology Patterns Preprocessing Language (OPPL).20
Test systematically—maintain a suite of ‘unit tests’ covering all issues identified; include tests for unintended consequences of changes; run test suite after every major set of changes and before each release.
Some might argue that many of the erroneous classifications reported here are several steps removed from the original concept in the hierarchies and would be ignored by clinicians. However, the semantics of the description logic underpinning SNOMED is unambiguous. Software and queries must follow them literally. Likewise, the reliability of postcoordination is a function of the reliability of the classifier, which is best determined by its manifestation in the hierarchies.
Until comprehensive quality assurance has been undertaken, anyone using, or mandating, SNOMED should be aware that the hierarchies contain serious anomalies. Should a ‘Reference terminology’ classify diabetes as a disease of the abdomen; fail to classify myocardial infarction as ischemic heart disease; place the arteries of the foot in the abdomen?
Without further quality assurance, clinicians may not realize the implications of what they are saying; researchers may not realize what their queries should retrieve, and postcoordination cannot be expected to be reliable. Interoperability, and therefore meaningful use, will be limited.
Supplementary Material
Acknowledgments
The team working on the Manchester-Siemens collaboration is gratefully acknowledged. Particular thanks to M Lawley at his team at Australian E-Health Research Centre (http://aehrc.com/hie/snorocket.html) for providing SNOROCKET in a form that works with Protégé and their prompt and helpful technical support. The input of the WHO ICD-11 revision team is also gratefully acknowledged including the Topic Advisory Groups (TAGs) and the Revision Steering Group (RSG). The authors also express their gratitude to the International Health Standards Development Organization (IHTSDO) for making available the stated form of SNOMED CT along with the Perl script to convert it to OWL syntax.
Footnotes
Funding: This work was supported in part by Siemens Health Solutions.
Competing interests: None.
Provenance and peer review:Not commissioned; externally peer reviewed.
Situation with explicit context (situation) | 243796009.
http://www.owlapi.sourceforge.net/. There appears to be some issue with version, because approximately 80 concepts from the Unified Medical Language System subset were not found in the version of the SNOMED CT Stated Form used. However, this merely reduces the size of the module and does not in any way affect the validity of any statement in this paper. The scripts used for the setup can be obtained from http://owl.cs.manchester.ac.uk/modproj/snomedmod/.
Allergic disorder by body site affected (disorder) | 421095001.
Grave's disease (disorder) | 353295004.
Myasthenia gravis (disorder) | 91637004.
Idiopathic thrombocytopenic purpura (disorder) | 32273002.
Antibody-mediated activation/inactivation (disorder) | 362985009.
Immune hypersensitivity disorder (disorder) | 421668005.
Antibody-mediated cytolysis (disorder) | 362986005.
Disorder of skin AND/OR subcutaneous tissue (disorder) | 80659006.
Skin structure (body structure) | 39937001.
Soft tissues (body structure) | 87784001.
xviii: Disorder of soft tissue (disorder) | 19660004.
Cut of ankle (disorder) | 283438007.
Cut of lower limb (disorder) | 283430000.
Myocardial infarction (disorder) | 22298006.
Ischemic stroke (disorder) | 422504002.
xxiii: Infarct (morphologic abnormality) | 55641003.
Ischemia (disorder) | 52674009.
Associated morphology (attribute) | 116676008.
Due to (attribute) | 42752001.
xxvii: Structure of endocrine system (body structure) | 113331007.
xxviii: Hypertensive disorder, systemic arterial (disorder) | 38341003.
Endocrine hypertension | 59997006 (Not in Core Problem List Subset).
Heart structure (body structure) | 80891009.
Entire heart (body structure) | 302509004.
xxxii: Heart part (body structure) | 119202000.
xxxiii: Optic disc swelling (finding) | 248487006.
xxxiv: Eye swelling (finding) | 45177002.
Structure of artery of pelvic region (body structure) | 116373007.
xxxvi: Structure of popliteal artery (body structure) | 43899006.
xxxvii: Structure of dorsalis pedis artery (body structure) | 86547008.
xxxviii: Injury of dorsalis pedis artery (disorder) | 285738005.
xxxix: Injury of abdomen (disorder) | 128069005.
Pelvic injury | 282771003.
Deep venous thrombosis of profunda femoris vein (disorder) | 427775006.
Thrombosis of vena cava (disorder) | 83938003.
xliii: Thrombophlebitis of breast’ (disorder) | 69954004.
Intracranial hemorrhage | 1386000.
Soft tissues (body structure) | 87784001.
Disorder of soft tissue (disorder) | 19660004.
xlvii: ‘Disorder of nervous system (disorder)’ that RoleGroup some (‘Finding site (attribute)’ some ‘Nerve structure (body structure)’).
xlviii: Optic disc edema (disorder) | 423341008.
Diabetic complication (disorder) | 74627003.
Associated with (attribute) | 47429007.
Hypertensive disorder, systemic arterial (disorder)—the fully specified name. ‘Hypertension’ is a synonym.
Hypertensive renal disease (disorder) | 38481006.
Hypertensive encephalopathy (disorder) | 50490005.
Hypertensive retinopathy (disorder) | 6962006.
Hypertensive heart disease | 64715009.
Ulcer of skin caused by ischemia due to hypertensive disease (disorder) | 95343001.
There are, of course, other issues of quality of assurance of SNOMED not covered by quality assurance of the description logic model alone.
lviii: Detailed QA methodologies for SNOMED and for OWL/DL ontologies more generally will be the subject of separate papers.
References
- 1.Cornet R, De Keizer N. Forty years of SNOMED: a literature review. BMC Med Inform Decis Mak 2008;8(Suppl 1):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews JE, Patrick TB, Richesson RL, et al. Comparing heterogeneous SNOMED CT coding of clinical research concepts by examining normalized expressions. J Biomed Inform 2008;41:1062–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vikström A, Skånér Y, Strender LE, et al. Mapping the categories of the Swedish primary health care version of ICD-10 to SNOMED CT concepts: Rule development and intercoder reliability in a mapping trial. BMC Med Inform Decis Mak 2007;7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulz S, Suntisrivaraporn B, Baader F. SNOMED CT's Problem List: Ontologists' and Logicians' Therapy Suggestions. Proc Medinfo 2007. Brisbane, Australia: IOS Press, 2007:802–6 [PubMed] [Google Scholar]
- 5.Spackman KA, Reynoso G. Examining SNOMED from the perspective of formal ontological principles: Some preliminary analysis and observations. Whistler, Canada: Proc KR-MED-04, 2004:81–7 [Google Scholar]
- 6.Schulz S, Suntisrivaraporn B, Baader F, et al. SNOMED reaching its adolescence: Ontologists' and logicians' health check. Int J Med Inform 2009;78:S86–94 [DOI] [PubMed] [Google Scholar]
- 7.Bodenreider O, Smith B, Kumar A, et al. Investigating subsumption in SNOMED CT: An exploration into large description logic-based biomedical terminologies. Artif Intell Med 2007;39:183–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rector A, Brandt S. Why do it the hard way? The case for an expressive ontological schemas for SNOMED. J Am Med Inform Assoc 2008;15:744–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ceusters W, Smith B, Kumar A, et al. Mistakes in medical ontologies: where do they come from and how can they be detected? Stud Health Technol Inform 2004;102:145–64 [PubMed] [Google Scholar]
- 10.Spackman KA. Normal forms for description logic expressions of clinical concepts in SNOMED RT. Washington, DC: Proceedings of the AMIA Symposium, 2001:627–31 [PMC free article] [PubMed] [Google Scholar]
- 11.Baader F, Calvanese D, McGinness DL, et al., eds. The Description Logic Handbook. Cambridge: Cambridge University Press, 2003:555 [Google Scholar]
- 12.W3C OWL Working Group OWL2 web ontology language document overview. http://www.w3.org/TR/owl2-overview/
- 13.Grau BC, Horrocks I, Kazakov Y, et al. Modular reuse of ontologies: Theory and practice. Journal of Artificial Intelligence Research 2008;31:273–318 [Google Scholar]
- 14.Horridge M, Drummond N, Goodwin J, et al. The Manchester OWL syntax. 2006; OWL Experiences and Directions (OWLED–06). Athens, GA: CEUR; http://sunsite.informatik.rwth-aachen.de/Publications/CEUR-WS//Vol-216/submission_9.pdf [Google Scholar]
- 15.Lawley MJ. Exploiting Fast Classification of SNOMED CT for Query And Integration Of Health Data. KR-MED, 2008. Phoenix, AZ: CEUR-WS 2008:8–14 [Google Scholar]
- 16.Hahn U, Schulz S, Romacker M. Partonomic reasoning as taxonomic reasoning in medicine. Proceedings of the 16th National Conference on Artificial Intelligence & 11th Innovative Applications of Artificial Intelligence (AAAI-99/IAAI-99). Orlando, FL: AAAI Press/MIT Press, 1999:271–6 [Google Scholar]
- 17.Schulz S, Hahn U, Romacker M. Modeling anatomical spatial relations with description logics. AMIA Fall Symposium (AMIA-2000). Los Angeles, CA: Hanly & Belfus, 2000:799–803 [PMC free article] [PubMed] [Google Scholar]
- 18.Horrocks I, Sattler U. The decidability of SHIQ with complex role inclusion axioms. Artif Intell 2004;160:79–104 [Google Scholar]
- 19.Campbell KE, Tuttle MS, Spackman KA. A ‘lexically-suggested logical closure’ metric for medical terminology maturity. Proc AMIA Fall Symposium (AMIA 1988). Orlando, FL: IEEE Computer Society Press, 1988:785–9 [PMC free article] [PubMed] [Google Scholar]
- 20.Iannone L, Aranguren ME, Rector A, et al. Augmenting the Expressivity of the Ontology Pre-Processor Language. OWL Experiences and Directions (OWLEd 2008). Karlsruhe, Germany: OWL Experiences and Directions, 2008 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.