Abstract
Objective
As biomedical technology becomes increasingly sophisticated, researchers can probe ever more subtle effects with the added requirement that the investigation of small effects often requires the acquisition of large amounts of data. In biomedicine, these data are often acquired at, and later shared between, multiple sites. There are both technological and sociological hurdles to be overcome for data to be passed between researchers and later made accessible to the larger scientific community. The goal of the Biomedical Informatics Research Network (BIRN) is to address the challenges inherent in biomedical data sharing.
Materials and methods
BIRN tools are grouped into ‘capabilities’ and are available in the areas of data management, data security, information integration, and knowledge engineering. BIRN has a user-driven focus and employs a layered architectural approach that promotes reuse of infrastructure. BIRN tools are designed to be modular and therefore can work with pre-existing tools. BIRN users can choose the capabilities most useful for their application, while not having to ensure that their project conforms to a monolithic architecture.
Results
BIRN has implemented a new software-based data-sharing infrastructure that has been put to use in many different domains within biomedicine. BIRN is actively involved in outreach to the broader biomedical community to form working partnerships.
Conclusion
BIRN's mission is to provide capabilities and services related to data sharing to the biomedical research community. It does this by forming partnerships and solving specific, user-driven problems whose solutions are then available for use by other groups.
Keywords: Genomics, statistical genetics, bioinformatics, complex traits, data, machine learning, data sharing, information integration, data mediation, data security, data management, knowledge engineering
Introduction
As biomedical technology becomes increasingly sophisticated, researchers can begin to probe ever more subtle effects. Often these projects require large amounts of data to gain the statistical power needed to tease out the phenomenon of interest. In biomedicine, studies to discover small effects often rely upon data acquired at multiple sites. The requirement for multiple sites often arises from the inability of a single site either to generate enough data over a reasonable period of time or to recruit the needed subjects from a single geographical area.
In principle, the data flow for large-scale biomedical projects is simple: data are collected at multiple sites, combined, and processed. In practice, however, there are myriad difficulties, both technological and sociological, that must be overcome for data to be accessible to the outside world and for the end result to be useful. For instance, data in clinical settings are generated behind firewalls, and moving those data to the outside world can be difficult, given that the institution must protect all data that contain private health information (PHI). Also, researchers who have generated the funding and acquired the data are often unwilling to give the data to a central storage location not under their control.
Despite these obstacles, data sharing in the biomedical realm is becoming more common due in part to new rules put in place by funding agencies and by a growing realization that data acquired for one purpose often have value outside the original intent. Another major reason for the increase in data sharing is the recognition that subtle effects (eg, genetic associations) require such large amounts of information that it is infeasible for a single site to acquire the necessary data. Therefore, to probe these small effects, consortia of like-minded researchers will often naturally arise. Such consortia have a common set of tasks: acquisition protocols must be standardized across sites; users must be authenticated and managed; incoming data must be checked for quality; data must be uploaded for processing and stored in a queryable repository; and the repository must be sent to processing streams.
The goal of the Biomedical Informatics Research Network is to address the challenges inherent in many of the stages of data sharing outlined above. BIRN has a toolset which is grouped into ‘capabilities’ that allow researchers to conduct a multisite study. BIRN tools are designed to be modular—a conscious decision that acknowledges that researchers often have pre-existing tools at their disposal. Within this structure, BIRN users can choose the capabilities most useful for their application, while not having to ensure their project conforms to a monolithic architecture.
The capabilities that BIRN is charged with creating are new and require exploratory work and experiments to validate their utility in the scientific enterprise. The strategy BIRN has elected to follow consists of forming partnerships with actively-engaged end users who have immediate needs for data management and sharing. The goal is to form a joint understanding of needs and requirements, and to construct solutions that users can begin using as quickly as possible. In addition, those solutions can then be used by other projects having similar needs. We believe that this approach of building up user-driven capabilities over a period of years and following best practices for quality improvement will result in tools and services that are of high quality and genuinely useful to a large number of projects. In the following text, we outline the structure of BIRN and its capabilities, and give exemplars to demonstrate the capability model.
Background
Overview of BIRN
The management structure of BIRN is organized as a set of interacting, domain-specific working groups overseen by both steering and executive committees. Currently, there are five working groups: Data Management, Security, Information Integration, Knowledge Engineering, and Operations. The chair of each working group, members of external user groups, and members of BIRN's outreach effort form a Steering Committee, which is charged with resource allocation and ongoing management of current projects. The Executive Committee sets policy and the overall direction of the program. Each of the working groups is discussed in the ‘System description’ section.
Overview of BIRN technology
BIRN has recently moved from a hardware-based architecture, in which identical hardware and software stacks were distributed to each site involved in sharing data, to a layered software-based approach. The hardware-based architecture had the advantage that each node on the grid was identical and therefore it was easy to test and deploy software changes. The disadvantage of this model, however, is that hardware becomes obsolete quickly and replacement is often expensive. The layered software-based approach is shown in figure 1. The layered architectural model is well established, and has distinct advantages. First, modifications are more easily implemented since the entire code base does not need to be modified; second, separate security implementations can be applied to different levels; and third, moving to a layered service-oriented architectural concept allows BIRN to incorporate emerging standards, and thus to integrate new devices and methodologies rapidly into the overall data-sharing environment without changing the underlying infrastructure. In summary, this layered approach allows BIRN to adapt to new usage scenarios in a more time- and cost-effective manner.
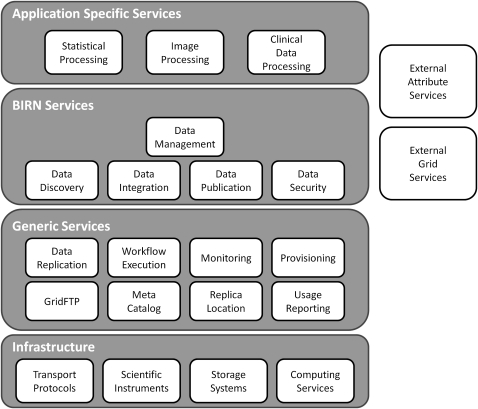
Figure 1.
A schematic of the layered architecture approach used in BIRN. The lowest infrastructure level of the architecture consists of the core infrastructure, that is, networks and associated protocols for generating, accessing, storing, and moving data. Above this layer are the Globus Toolkit1 generic data management services that are format and content agnostic. The BIRN services layer contains common standard services that are more specialized for biomedicine, but not for any specific biomedical domain. Finally, the top layer consists of application-specific services and tools that address the specific requirements of a particular research activity or specialized research domain. These are often supplied by the end users. One advantage of the layered approach is that it allows BIRN capabilities to be incorporated into the existing technology of the end user.
The lowest infrastructure level of the architecture consists of the core infrastructure, that is, networks and associated protocols for accessing and moving data, storage systems and databases for storing the data, computing services for processing the data, and finally, the equipment that generates the data, such as MRI scanners, CT machines, or microarrays. Above this layer are the Globus Toolkit1 generic data management services (which include data movement), replication management, access control and authorization. These services are format and content agnostic. More information on these services is given below in the section describing the Data Management Working Group. The BIRN services layer contains common standard services that are more specialized for the biomedical domain, but not for any specific use case. For example, publishing and discovering MRI data in DICOM or NIfTI format could be useful in either neuroscience or cardiology research. This layer also provides higher-level biomedical services that layer on top of the standard biomedical services or directly use the generic services. Finally, the top layer consists of application-specific services and tools that address the specific requirements of a particular research activity or specialized research domain. The end users often supply these application-specific resources. In the following section we describe the capabilities residing in the generic and BIRN services layers, organizing the discussion by BIRN working group.
System description
BIRN capabilities—working groups
Data management
The members of the BIRN Data Management Working Group are computer scientists, software developers, system administrators, and biomedical scientists representing both BIRN itself and participating user groups. Focus areas for the working group include remote file sharing, secure data access, data mirroring and replication, data synchronization, and metadata management.
The Data Management Working Group is responsible for formulating strategies for the use cases of data publication, sharing, and access within and among user communities. A typical data-sharing scenario is illustrated in figure 2, in which multiple data storage sites need to transfer data among sites through firewalls. Capabilities are available to address the challenges inherent in this scenario in several areas.
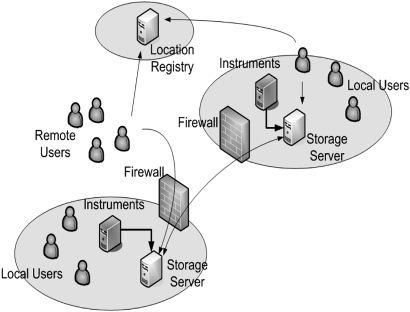
Figure 2.
An example of federated data sharing using BIRN. The figure depicts two sites federating local storage systems and sharing with local and remote users. Each site (large circled) is composed of a storage server, data producing instruments, local users of data, and institutional firewalls. Typically, instruments rapidly produce large volumes of data and store the data on the local storage server. As data are stored, the system registers file locations at a location registry, often at a third-party location (small circled). When accessing data, client software queries the location registry to find the storage locations of the data. Users retrieve data from local or remote storage systems transparently. All communication between services and clients is secured with cryptographically strong methods that ensure authenticity, privacy, and integrity.
Data access and movement
BIRN users can access remote data storage services, primarily for access to file-based data, and perform storage, retrieval, and deletion operations on individual files, multiple files, and individual directories. Data access service operations are secured by user authentication and file access permissions required to authorize the operation, by integrating BIRN security capabilities. One tool that BIRN provides for data access is the Globus Toolkit's1 GridFTP Server,2 a widely deployed data transfer service that provides high-performance, reliable, and secure data transfers for many scientific Grid environments. GridFTP3 provides interfaces to a wide range of storage platforms including local file systems, parallel file systems such as GPFS, archival storage systems such as HPSS, and emerging ‘cloud’ storage systems such as Amazon S3. BIRN supports additional file transfer protocols including HTTP/S, Secure Copy (scp), Secure FTP, and others.
Data replication
In a federated system, datasets are acquired and hosted at multiple locations. To ensure that datasets are always available, copies of data are often distributed among user sites. BIRN projects can use the Replica Location Service (RLS),4 5 a highly scalable distributed file registry, to keep track of the locations of replicated datasets and for data discovery. RLS catalogs may be deployed centrally or at multiple sites to monitor local replicas. Summary information about local catalogs is distributed to multiple index nodes, which can be queried for information about items stored in all the reporting catalogs. In addition to being highly scalable, the RLS has been shown to be extremely reliable.6 The RLS is used in the Function BIRN (FBIRN) collaboration to locate and manage data at all the sites in the federation.
Metadata access and management
This capability allows BIRN users to associate metadata with data objects, such as images. BIRN users use the Human Imaging Database (HID)7 and XNAT8 for storing and accessing image metadata, such as data acquisition parameters and data provenance. The HID was developed for the needs of the FBIRN multi-site federated research project. XNAT functions as a research PACS (picture archiving and communication system) to complement clinical PACS systems9 and supports a subset of DICOM (Digital Imaging and Communications in Medicine) protocols.10
In addition, the BIRN team has implemented a metadata catalog with a web service front end based on the Apache web server and the Django web framework for the BIRN Pathology project. This system associates detailed pathology metadata stored in a relational database back end with pathology images stored in a file system. By specifying desired metadata attributes through the web service front end, users can identify pathology images with those attributes and view the images either as thumbnail summaries or use a web browser-based viewer for full resolution images with progressive rendering and zoom-and-pan functionality.
DICOM Image Gateway service
Another service available to BIRN users is a DICOM Image Gateway service. This gateway can store images in the DICOM format that are pulled or pushed from a PACS system using standard protocols. The system automatically extracts image metadata from the DICOM file headers when the image files are stored on the gateway service. A simple graphical user interface (GUI), which can be customized based on specific project requirements, provides attribute-based retrieval and display of DICOM images. In addition, using the Mediator developed by the Information Integration Working Group (described below), users can issue queries across federated DICOM Image Gateway services.
Data security
The Security Working Group is responsible for providing security solutions to the BIRN community and serves as a forum for discussion of cross-cutting security issues. These solutions ensure that the data-sharing capabilities are secure, data policies are enforced, data integrity and privacy are preserved, and access to data is limited to authorized users. The working group provides hosting of common security services, tools for users to deploy, and a consulting service for security integration with community applications. The Security Working group serves as a peer group for service providers to share experiences, expertise, and best practices.
The Security Working Group also defines and develops security capabilities that are common across these user communities, defines policies for these capabilities, and works with the Operations Working Group to host such services for the user community, if desired. Currently, two such services are available: the User Identity Management Capability and the User Credential Management Capability. The User Identity Management Capability provides interfaces to register and vet new users and to issue user credentials (which allow users access to certain datasets). The Credential Management Capability allows users to securely manage their long-term credential and provides access to these credentials from multiple machines. Driven by requirements from user communities, the Security Working Group has constructed a Group Management Capability that allows user communities to manage their users and access to resources by creating groups of users. The Security Working Group also identifies and provides software and tools to interact with these common services that users can download and deploy.
The BIRN Security Working Group leverages several existing open source solutions to build and deploy these capabilities for the BIRN community. Deployment uses the MyProxy Service and Simple CA, tools that are developed and shipped as part of Globus Toolkit and which generate and manage security certificates for BIRN users. We also use Grid Grouper, a service that was developed by the caGrid project11 and that uses the Globus Toolkit and the Grouper system to provide a group management capability. BIRN has invested effort in integrating these services with various applications and building user accessible portals for BIRN communities.
Finally, the Security Working Group provides a process and system to report and handle potential security vulnerabilities in the software provided by BIRN. This process provides a mechanism for users to report issues to a secure list, staffed by BIRN personnel and interested users, to evaluate and resolve issues prior to publicly issuing an advisory.
Information integration
One of the core capabilities provided by BIRN is an information integration infrastructure that provides uniform, semantically-consistent query access to heterogeneous data sources.12–16 There are two main approaches to integrating data. The first, physical integration, creates a centralized warehouse where data from multiple sources are loaded and cleaned. The second, virtual integration, is a federated system in which a mediator provides live access to data stored in remote sources. These approaches have complementary properties. The warehouse approach provides more efficient query response for queries that require large amounts of data to be processed simultaneously. However, the drawback of the data warehouse is that the data may become stale; they are only as recent as the last update cycle. In contrast, the virtual integration approach always accesses the latest data from the sources. Therefore, this approach is used when data are updated often, when having the latest data is critical, or when organizational constraints prevent the incorporation of data sources into a warehouse. The third approach, of course, is to have some data stored locally and some accessed at run-time from remote sources. Fortunately, the computational machinery needed to integrate the data either in the virtual or the warehouse approach can be designed to overlap substantially. In what follows, we describe the BIRN virtual integration approach, but much of this machinery could be reused for data warehousing and mixed systems.
Over the last decade, research on data mediation has progressed rapidly.13 14 17 18 One of the challenges of data mediation is to provide accurate queries over data sources whose schema may contain different terms for the same objects. The mediation approach consists of defining a common model/schema/ontology at the user level that captures the semantics of the application domain and then defining formal logical mappings between the schemas of the data sources and the domain model.
The typical behavior of a data integration system is as follows: the user poses a query using the domain model; the mediator rewrites the user query into a query over the set of terms used by each source; and the mediator query engine optimizes the execution of the rewritten query, issuing sub-queries to the sources as appropriate and composing the results. The mediation approach has three key properties. First, data reside and are maintained at the original data sources; no changes are required to any of the data sources. Second, all that is required for integration is knowledge of the information content and access to the data source via an application programming interface or other method. Third, integrated data access is provided via a mediator, which is software that can reside on any server and that contacts the various data sources at query-time to obtain the information requested.
The BIRN mediator follows the classical virtual data integration architecture,16 as shown in figure 3. However, a novel feature is that our architecture is built upon grid computing technologies19 20 in order to leverage their scalability and security infrastructure.
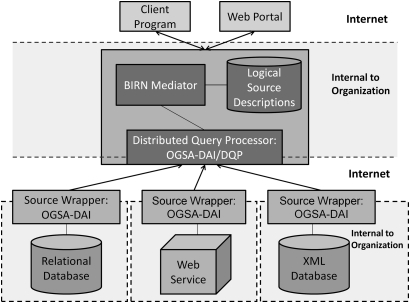
Figure 3.
A schematic of the BIRN Mediator. Users are presented with a single database view that is achieved through a uniform semantically-consistent domain model of all of the data. No data are stored at the mediator; the data remain at the sources. The mediator reconciles the semantic discrepancies among the sources by using a set of declarative logical descriptions of the contents of the sources. The user poses queries to the mediator using terms from the domain model. The mediator then uses the source descriptions to identify the sources relevant to the user query and to rewrite the domain-level user query, expressed in terms of the domain model, into a source-level query, expressed in terms of the source schemas. The Mediator architecture is designed in a modular fashion, so that any mediation approach that produces source queries in a language supported by the query evaluation engine can be used by the system.
The data integration system has three major components, as illustrated in figure 3. The Mediator component presents a uniform, semantically-consistent schema (the domain model) to users of the system. To users, the system looks like a single database. However, this is a virtual database, as no data are stored at the mediator; the data remain at the sources. Our current mediator is based on the relational Global-as-View model.21–23 However, we plan to support other mediation approaches such as Local-as-View17 18 and more expressive languages such as OWL2-QL.24 The Distributed Query Evaluation Engine component evaluates source-level queries after they are generated by the Mediator. This component is based on the grid-enabled OGSA-DAI/DQP project.20 25 26 Finally, the Source Wrapper components wrap the actual data sources as OGSA-DAI resources. OGSA-DAI includes a library of connectors to common data sources, such as relational databases, and provides a common extensible framework to add new types of data sources.
The data integration infrastructure is the same for all application domains. To integrate data sources in a new application domain, the developer needs only to define a declarative domain model and source descriptions. Occasionally, if the domain includes a novel type of source not previously encountered, then the developer needs to define a wrapper for the new source type, which is then added to the library of wrappers and can be reused in future applications. One important point in understanding the BIRN mediator is that it relies on a domain model rather than a full ontology. Using a domain model is a pragmatic stance that enables immediate data sharing within a collaboration even when there are no fully agreed-upon ontologies in a given scientific domain or community.
Security is a critical design requirement for biomedical applications in which PHI is involved. Our data integration system leverages the Grid Security Infrastructure27 which provides encryption of transmitted data using the industry standard TLS/SSL protocol and public key infrastructure to authenticate users, sources, and servers.
Knowledge engineering
BIRN's activities are mainly focused on sharing information already available in a digital form, for example images and files containing metadata. If we consider ‘knowledge engineering’ to refer to the development of information processing systems in which any form of available, actionable knowledge plays a role,28 then we must also consider other sources of information. This directly pertains to researchers attempting to build their own bioinformatics resources from scratch in a well-ordered and rigorous way. This may require the curation of information from the natural language text of published articles or the construction of data repositories from local laboratory archives that have not been designed for sharing with a broader audience. For example, a group may wish to annotate a locally-curated in-house database with standardized terms from a community-curated ontology. Or, a group may need assistance constructing a domain schema that represents its experiments and the resulting data, while linking to published ontologies so that the data can be shared with the wider community.
The Knowledge Engineering Working Group develops technology to support our users' interactions with biomedical knowledge within the context of the overall scientific process. The schematic diagram in figure 4 illustrates our modeling approach, centering on the development of generic, ontological models of experimental design29 that leverage community-driven ontologies.30 The Knowledge Engineering Working Group consists of bioinformatics developers, biologists, and computer scientists working in the areas of bio-ontologies,31 natural language processing,32 information-integration systems,33 bioinformatics systems-development,34 and knowledge representation and reasoning.29 This working group supports the process of generating knowledge conforming to the model shown in figure 4 and providing ontological support for knowledge-based activities within BIRN.
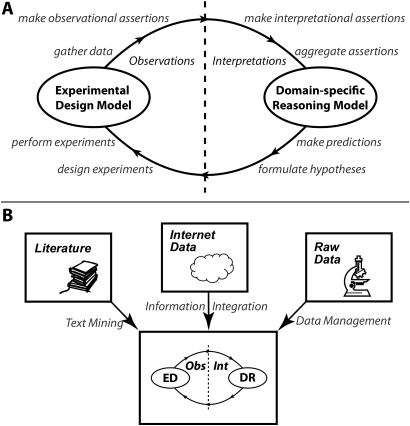
Figure 4.
Knowledge processing in biomedicine is based on cycles of formulation—experimentation—interpretation. (A) BIRN's knowledge modeling methodology is based on a generic knowledge model for experimental design (EDlabel>) and specialized models for domain reasoning (DR). (B) We are developing technology to populate this modeling approach from the published literature (using a text mining infrastructure), from online databases (using BIRN's information integration technology) and raw data (using our modeling tools directly within database development systems).
Originally, BIRN mediation systems were dependent on the development of global vocabularies in the form of large-scale, centrally-curated ontologies (such as BIRNLex,35 which is now incorporated into NIFSTD). This strong coupling between information integration systems and external ontologies can be problematic, since the criteria for acceptance of terms to globally-accessible ontologies are much more stringent than those required for developing functional domain models. In the current project, we decouple these activities. The Knowledge Engineering Working Group is developing capabilities that follow the high-level tasks described in figure 4. We are building a text mining infrastructure to speed up curation of information from published sources into databases.32 We are actively supporting the Information Integration Working Group by providing support to link both source and mediated models to the terminology being developed within the OBO-Foundry31 ontology community development effort. Finally, we are developing generic data repositories that permit scientists to store their data based on a relatively simplified model of experimental design without any explicit database administration or design.29 34
Operations
The Operations Working Group oversees the processes and practices for operating the BIRN collaboration, including services provided by BIRN itself, by BIRN user teams, and by third-party service providers. The Operations Working Group focuses on three areas: assisting BIRN service providers with operational issues, establishing community-wide operational standards, and prioritizing known system issues. The working group also helps the BIRN Steering Committee identify potential user needs based on reported operational experiences.
The Operations Working Group provides four primary services to the BIRN community: a central software repository, the BIRN system definition, a system information service, and a verification and validation service.
The central software repository36 is a web-accessible library of software code, tools, and documentation that have been developed by the BIRN community. It provides the community with an authoritative location for finding and obtaining software that has been developed as part of the BIRN program. In addition, BIRN makes its neuroimaging-related software available through the Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC).37
The most critical element in operating any system is clarity regarding the system definition. A public directory of the system's intended capabilities, its components, and the intended state of each element is vitally important to keeping the system in its intended state. The Operations Working Group works with all of BIRN's working groups to define the capabilities they provide to BIRN users in sufficient detail to set measurable operational targets. Using these definitions, the working group establishes goals for system quality, including the state of individual components, delivery and deployment mechanisms, user and administrator documentation, and testing criteria. The Operations Working Group maintains these definitions on behalf of the BIRN community and uses them as the basis for other quality control services. The clarity that is provided by our system definition is critical to understanding how the BIRN system is supposed to behave, to directing appropriate tests of the system, and to interpreting the results of those tests, particularly in the event of a test failure.
Capability and component definitions are one of several critical elements stored in BIRN's system information service. The system information service38 is an online description of the BIRN system and its capabilities. It can be browsed using web browsers or accessed by software to auto-configure tools and applications. In addition to capability definitions, the system information service also stores the locations and configurations of deployed capabilities for BIRN's user teams and any other detail that is needed to understand and access the system.
Based on data in the system information service, BIRN's verification and validation (V&V) system continuously monitors the state of registered components, deployments, and system capabilities. The V&V service is a framework for executing pre-defined system tests. These tests are designed specifically to verify that the capabilities listed in the information service work as expected wherever scientists have deployed them for use. The Operations Working Group uses the V&V service to monitor the system, identify issues (ideally before users encounter them), notify responsible parties, and prioritize corrective action.
Working with BIRN
The tools and services that BIRN is charged with bringing to bear on biomedical research are new, requiring exploratory work and experiments to validate their utility in the scientific enterprise. There is no well known, reliable blueprint for creating a biomedical informatics research network. The strategy that BIRN has elected to follow consists of forming partnerships with scientific teams with immediate needs for data management, processing, and sharing. The goal is to form a joint understanding of needs and requirements, construct solutions that users can begin using as quickly as possible, and build on these initial successes. In addition, these solutions are then offered to new projects with similar needs. BIRN's belief is that this approach, taken for a period of years and following best practices for quality improvement, will produce tools and services that are of high quality and genuinely useful to a large number of projects. The tangible products of this program will be a series of capabilities that can be utilized by teams who have never worked with BIRN.
Prospective users of BIRN can begin the process by requesting more information through BIRN's website. Contact will be made by representatives of the Collaborative Tools Research Network (CTSN), the outreach arm of BIRN (which is also funded by the National Center for Research Resources). The goal of the CTSN is to determine the needs of prospective users, educate them about BIRN's areas of expertise, and connect them to the appropriate working groups. Once the initial needs are determined, the BIRN Steering Committee members are informed of the request, and the appropriateness of the project, the amount of resources required, and the required timelines are discussed.
In working with BIRN, users will be first asked to identify a small-scale project to address an immediate need. This has three advantages. First, BIRN and the user group can establish a working relationship; second, the success of the initial project can be used to ‘sell’ the collaboration to the larger user group; and third, the very act of having to define an initial project often helps the users understand their project's organization and priorities in a new way. The initial use case will initiate an assessment of the project's goals, priorities, needs, opportunities, and risks. The purpose of this assessment is to identify the joint activities that would be most likely to provide benefits to the project without overextending the project's resources: personnel, skills, time, and ability to use the results. The BIRN Steering Committee is in charge of managing the project and ensuring its resource needs are met. After the initial project is completed, an assessment of the group's future needs is conducted, and plans are formulated to meet those needs in the context of a now-working relationship with the user group.
Status report
BIRN is currently working with a number of groups who require the ability to share data. Some of these groups have an established audience within their domain, while others are beginning the process of creating a community that will freely share both data and research methodologies (eg, the establishment of standard research protocols). Often, newer groups will begin to build a community through simple collaborative services such as wikis to create a community consensus before moving on to larger-scale projects. We describe below one such large-scale project, whose goal is to provide a federation of functional MR images, metadata, genetic data, and clinical assessment results. A listing of other collaborators can be found on the BIRN website.39
Function BIRN
The Function BIRN (FBIRN) is a consortium of 13 sites, spanning the nation, and focused on developing fMRI tools and techniques in the context of schizophrenia research.40 Both clinical and neuroimaging data are made available to the consortium via a federation of databases containing neuroimaging, clinical, and genetic data accessed securely and queried transparently using the core infrastructure provided by BIRN.
The technological challenge of accessing fMRI datasets over geographically distributed sites is a formidable one. The datasets consist of thousands of small files per subject, an uncommon data scenario for most distributed and high-performance file systems. Additionally, each site maintains a subset of the overall imaging data, and access to the full dataset should be seamless to the user, appearing as a single storage system. To address these requirements, FBIRN uses a BIRN-developed integrated system consisting of a grid-based transport protocol (GridFTP), a location lookup service (Replica Location Service), a data mediation system based on a common schema, and a user/group/project management console. All components are tied to single sign-on grid credentials. This infrastructure has enabled the computer scientists in FBIRN to focus on developing domain specific tools (eg, HID, XCEDE, analysis tools), saving the consortium considerable resources. For example, FBIRN has developed image quality control standards and processes, implemented in software, that both monitor the reliability of the scanners and provide quick access to the processed data for review.41–44
In addition, FBIRN is using the BIRN mediator to provide an integrated (virtual) view of data sources stored in both the Human Imaging Database used by FBIRN and public data sources stored in XNAT Central.45
Summary
BIRN's mission is to provide capabilities and services related to data sharing to the biomedical research community. It does this by forming partnerships and solving specific, user-driven problems whose solutions are then available for use by other groups. The BIRN capability model avoids the need for the establishment of rigid standards that can be a barrier to adoption, focusing instead on providing solutions that can be generalized for the broader community. In addition, BIRN has recently implemented a software-based data-sharing model, in contrast to its previous incarnation in which identical hardware-based systems were deployed at each site. BIRN provides capabilities in data management, security, system operations, information integration, and knowledge engineering and is actively involved in outreach to the broader biomedical community.
Footnotes
Funding: BIRN is supported by grants from the National Center for Research Resources (NCRR) through the following grants: U24-RR025736, U24-RR021992, and U24-RR021760. The outreach portion of BIRN is supported through U24-RR026057-01. Some of the knowledge engineering work is supported though a grant from the National Institute of General Medical Sciences (NIGMS; RO1 GM083871) and the National Science Foundation (grant 0849977), and through the Kinetics and Michael J. Fox Foundations.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.The Globus Alliance The Globus Toolkit. http://www.globus.org/toolkit/
- 2.The Globus Alliance GridFTP. http://www.globus.org/toolkit/data/gridftp/
- 3.Allcock W, Bresnahan J, Kettimuthu R, et al. The Globus Striped GridFTP Framework and Server. SC05 Conference; 2005; Seattle, WA, 2005:54 [Google Scholar]
- 4.Chervenak AL, Schuler R, Ripeanu M, et al. The globus replica location service: design and experience. IEEE Trans Parallel Distrib Syst 2009;20:1260–72 [Google Scholar]
- 5.The Globus Alliance Replica Location Service (RLS). http://www.globus.org/toolkit/
- 6.Chervenak A, Schuler R, Kesselman C, et al. Wide area data replication for scientific collaborations. 6th IEEE/ACM Int'l Workshop on Grid Computing (Grid2005). Seattle, WA, 2005:1–8 [Google Scholar]
- 7.Human Imaging Database (HID). http://www.nitrc.org/projects/hid/
- 8.XNAT. http://www.xnat.org/
- 9.Greenes RA, Brinkley JF. Imaging systems in radiology. In.Shortliffe EH, Cimino JJ, eds. Biomedical Informatics. New York: Springer, 2006:626–59 [Google Scholar]
- 10.Radiological Society of North America (RSNA), DICOM http://www.rsna.org/Technology/DICOM/index.cfm [DOI] [PubMed]
- 11.caGrid. https://cabig.nci.nih.gov/workspaces/Architecture/caGrid
- 12.Dayal U, Hwang H-Y. View definition and generalization for database integration in a multidatabase system. IEEE Trans Softw Eng 1984;10:628–44 [Google Scholar]
- 13.Halevy AY, Rajaraman A, Ordille J. Data integration: the teenage years. International Conference on Very Large Databases (VLDB), 2006; Seoul, Korea;2006 [Google Scholar]
- 14.Levy A. Logic-based techniques in data integration. In: Minker J, ed. Logic Based Artificial Intelligence. Dordrecht, Kluwer Publishers, 2000 [Google Scholar]
- 15.Ullman JD. Information Integration Using Logical Views. Proceedings of the Sixth International Conference on Database Theory. Delphi, Greece, 1997:19–40 [Google Scholar]
- 16.Wiederhold G. Mediators in the architecture of future information systems. IEEE Computer 1992;25:38–49 [Google Scholar]
- 17.Halevy AY. Answering queries using views: a survey. VLDB J. 2001;10:270–94 [Google Scholar]
- 18.Lenzerini M. Data integration: a theoretical perspective. Proceedings of ACM Symposium on Principles of Database Systems. Madison, Winsconsin, 2002 [Google Scholar]
- 19.Foster I, Kesselman C, Tuecke S. The anatomy of the grid: enabling scalable virtual organizations. Int J High Performance Comput Appl 2001;15:200–22 [Google Scholar]
- 20.University of Edinburgh Open Grid Services Architecture Data Access and Integration (OGSA-DAI). 2008. http://www.ogsadai.org.uk/ [Google Scholar]
- 21.Adali S, Candan KS, Papkonstantinou Y, et al. Query caching and Optimization in Distributed Mediator Systems. SIGMOD Record (ACM Special Interest Group on Management of Data) 1996;25:137–48 [Google Scholar]
- 22.Florescu D, Raschid L, Valduriez P. Answering Queries Using OQL View Expressions. Workshop on Materialized Views: Techniques and Applications. Montreal, Canada: SIGMOD, 1996:84–90 [Google Scholar]
- 23.Garcia-Molina H, Papakonstantinou Y, Quass D, et al. The TSIMMIS approach to mediation: data models and languages. J Intell Inform Syst 1997;8:117–32 [Google Scholar]
- 24.Calvanese D, Giacomo GD, Lembo D, et al. Tractable reasoning and efficient query answering in description logics: the DL-Lite family. J Automat Reas 2007;9:385–429 [Google Scholar]
- 25.Alpdemir MN, Mukherjee A, Gounaris A, et al. Using OGSA-DQP to Support Scientific Applications for the Grid. Scientific Applications of Grid Computing: First International Workshop. Beijing, China: SAG, 2004 [Google Scholar]
- 26.Grant A, Antonioletti M, Hume AC, et al. OGSA-DAI: Middleware for Data Integration: Selected Applications. Fourth IEEE International Conference on eScience. Indianapolis, Indiana, USA, 2008 [Google Scholar]
- 27.Lang B, Foster I, Siebenlist F, et al. A Multipolicy Authorization Framework for Grid Security. Fifth IEEE International Symposium on Network Computing and Applications (NCA'06). 2006:269–72 [Google Scholar]
- 28.Schreiber G, Akkermans H, Anjewierden A, et al. Knowledge Engineering and Management, The CommonKADS Methodology. Cambridge, MA: MIT Press, 2000 [Google Scholar]
- 29.Burns G, Russ T. Biomedical Knowledge Engineering tools based on Experimental Design: a case study based on neuroanatomical tract-tracing experiments. K-Cap 2009: The Fifth International Conference on Knowledge Capture. Redondo Beach, California, USA, 2009 [Google Scholar]
- 30.Courtot M, Bug W, Gibson W, et al. The OWL of Biomedical Investigations. Karlsruhe, Germany: OWL: Experiences and Directions (OWLED), 2008 [Google Scholar]
- 31.Smith B, Ashburner M, Rosse C, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol 2007;25:1251–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramakrishnan C. Building the Scientific Knowledge Mine (SciKnowMine1): a Community-driven Framework for Text Mining Tools in Direct Service to Biocuration. Malta; Language Resources and Evaluation, 2010 [Google Scholar]
- 33.Thakkar S, Ambite JL, Knoblock CA. Composing, optimizing, and executing plans for bioinformatics web services. VLDB 2005;14:330–53 [Google Scholar]
- 34.Pittendrigh S, Jacobs G. NeuroSys: a semistructured laboratory database. Neuroinformatics 2003;1:167–76 [DOI] [PubMed] [Google Scholar]
- 35.Bug WJ, Ascoli GA, Grethe JS, et al. The NIFSTD and BIRNLex vocabularies: building comprehensive ontologies for neuroscience. Neuroinformatics 2008;6:175–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.BIRN Central Software Repository. http://software.nbirn.org/
- 37.Neuroimaging Informatics Tools and Resources Clearinghouse. http://www.nitrc.org/
- 38.BIRN Integrated Information Service. http://info.nbirn.org/
- 39.Biomedical Informatics Research Network. http://www.birncommunity.org
- 40.Keator DB, Grethe JS, Marcus D, et al. A national human neuroimaging collaboratory enabled by the Biomedical Informatics Research Network (BIRN). IEEE Trans Inf Technol Biomed 2008;12:162–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedman L, Glover GH. Reducing interscanner variability of activation in a multicenter fMRI study: controlling for signal-to-fluctuation-noise-ratio (SFNR) differences. Neuroimage 2006;33:471–81 [DOI] [PubMed] [Google Scholar]
- 42.Friedman L, Glover GH. Report on a multicenter fMRI quality assurance protocol. J Magn Reson Imaging 2006;23:827–39 [DOI] [PubMed] [Google Scholar]
- 43.Friedman L, Glover GH, Krenz D, et al. Reducing inter-scanner variability of activation in a multicenter fMRI study: role of smoothness equalization. Neuroimage 2006;32:1656–68 [DOI] [PubMed] [Google Scholar]
- 44.Friedman L, Stern H, Brown GG, et al. Test-retest and between-site reliability in a multicenter fMRI study. Hum Brain Mapp 2008;29:958–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The XNAT Group XNAT Central. http://central.xnat.org/