Abstract
Objective
The long-term goal of this work is the automated discovery of anaphoric relations from the clinical narrative. The creation of a gold standard set from a cross-institutional corpus of clinical notes and high-level characteristics of that gold standard are described.
Methods
A standard methodology for annotation guideline development, gold standard annotations, and inter-annotator agreement (IAA) was used.
Results
The gold standard annotations resulted in 7214 markables, 5992 pairs, and 1304 chains. Each report averaged 40 anaphoric markables, 33 pairs, and seven chains. The overall IAA is high on the Mayo dataset (0.6607), and moderate on the University of Pittsburgh Medical Center (UPMC) dataset (0.4072). The IAA between each annotator and the gold standard is high (Mayo: 0.7669, 0.7697, and 0.9021; UPMC: 0.6753 and 0.7138). These results imply a quality corpus feasible for system development. They also suggest the complementary nature of the annotations performed by the experts and the importance of an annotator team with diverse knowledge backgrounds.
Limitations
Only one of the annotators had the linguistic background necessary for annotation of the linguistic attributes. The overall generalizability of the guidelines will be further strengthened by annotations of data from additional sites. This will increase the overall corpus size and the representation of each relation type.
Conclusion
The first step toward the development of an anaphoric relation resolver as part of a comprehensive natural language processing system geared specifically for the clinical narrative in the electronic medical record is described. The deidentified annotated corpus will be available to researchers.
Keywords: Clinical natural language processing, coreference, relation extraction, information extraction, lexical resources, other methods of information extraction, natural-language processing, monitoring the health of populations, knowledge representation
Introduction
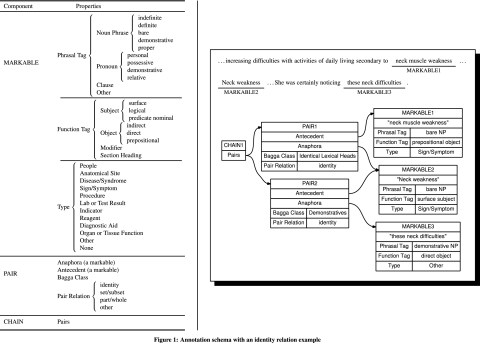
A substantial part of the information within the electronic medical record (EMR) is free-text. Natural language processing (NLP) techniques are therefore being used to expose that knowledge. A number of systems aim specifically at information extraction from it: Medical Language Extraction and Encoding System,1–3 Clinical Text Analysis and Knowledge Extraction System (cTAKES),4 5 Health Information Text Extraction,6 MedKAT/P,7 SymText and MPLUS,8–12 the systems from the National Center for Text Mining13 and JULIE lab,14 Cancer Tissue Information Extraction System.15 16 Almost all of them implement a named entity recognition (NER) module. The identification of named entities (NEs) referring to the same world object (‘coreference resolution’) is critical for comprehensive information extraction. ‘Anaphoric relations’ are relations between linguistic expressions where the interpretation of one linguistic expression (the anaphor) relies on the interpretation of another linguistic expression (the antecedent). Anaphoric relations define identity, set/subset, or part/whole relations between the participating linguistic expressions. ‘Coreferential relations’, or coreferences, are anaphoric relations of identity.17 For example, in the sentences ‘… increasing difficulties with activities of daily living secondary to neck muscle weakness… Neck weakness… She was certainly noticing these neck difficulties.’ to interpret the anaphor ‘these neck difficulties’, one needs to resolve its antecedent ‘Neck weakness’. Similarly, ‘neck muscle weakness’ and ‘Neck weakness’ constitute another coreference pair. The two pairs form one chain (figure 1).
Figure 1.
Annotation schema with an identity relation example. NP, noun phrase.
Background
In the general domain, there are two coreference datasets developed for the Message Understanding Conferences 6 and 7 (MUC-6 and MUC-7) used by the NLP community to develop and evaluate algorithms for coreference resolution.18 The MUC-7 annotation schema includes the annotation of identity relations between entities. The GNOME19 project extends the annotations to set/subset and part/whole relations. The ACE20 annotation schema adds to the identity relations links for appositive and predicative phrases.
Recognition of the importance of a coreference resolver in the biomedical domain is discussed by Castano et al.21 As pointed out by Gasperin et al22 ‘although anaphora resolution was identified as one of the new frontiers in biomedical text mining in the call for papers of a recent conference, there were no papers on the topic published in the proceedings.’ The organizers attribute this to lack of publicly available data.23 As discussed by Roberts et al,24 there are three purposes of such annotated data: clarifying the task's information requirements, serving as a resource for system development, and providing a much needed test bed.
In the biomedical scientific literature domain, several ongoing annotation efforts are focusing on coreference.21 22 25–27 Coreference annotations of clinical narrative are almost non-existent. Coden and colleagues7 describe work to develop a tool for extracting cancer characteristics from pathology notes. They manually annotated 302 Mayo Clinic pathology notes to serve as a gold standard. The annotation schema included coreference annotations for anatomical sites and histologies mapped to the International Classification of Diseases for Oncology (ICD-O).28 Two mentions that are exact strings and map to the same concept were annotated as coreferential. In addition, each mention is coreferenced with any instance of its parent anatomical site as defined by ICD-O. Roberts and colleagues24 describe their work on creating a multi-layered, semantically annotated corpus which includes coreference. Our work is closest to refs7 24 29 but differs from them in the scope of the annotated anaphoric relations. We go beyond identity relations of coreference to include relations of type set/subset, part/whole and other (a catch-all category for anaphoric relations that cannot be binned into the pre-defined categories). We do not limit the anaphoric relations to pronouns and noun phrases, but include a wide variety of linguistic constructs.
In this paper, we present the creation and quality of a gold standard set for clinical anaphoric relations building on our preliminary work,30 and describe high-level characteristics of that gold standard set. We intend to use this gold standard to develop and evaluate methodologies for coreference resolution in clinical text. Our long-term goal is to extend the open-source cTAKES with a clinical coreference resolver and incorporate it within the Ontology Development and Information Extraction (ODIE) platform.31 This project contributes to our long-term vision of a comprehensive, open-source, modular and extensible clinical NLP system. To our knowledge, the work presented here is the first to describe a clinical narrative corpus annotated for deep anaphoric relations.
Methods
Annotation guidelines
The annotation guidelines are based on the MUC-7 Coreference Task Definition17 (see online supplement 2 for the full guidelines). The modifications are in: (1) the types of NEs participating in anaphoric relations, specifically clinical concepts; (2) allowing relations other than identity to capture set/subset and part/whole; (3) including markables of grammatical categories beyond noun phrases and pronouns. Our goal is a broad investigation of anaphoric relations in the clinical narrative, and thus anaphoric relations within a given clinical note document, across its paragraphs and sections are annotated. A fully annotated example is shown in figure 1.
The ‘markable’ is the linguistic expression signifying a clinical concept belonging to one of the allowed semantic types (see semantic class attribute below). The markable participates in anaphoric relations within the given document across its paragraphs and sections and has several attributes:
Grammatical category—noun phrase (indefinite, definite, bare, demonstrative, proper), pronoun (personal, possessive, demonstrative, relative), clause. The ‘other’ captures phrasal tags not listed above. Example: MARKABLE1, MARKABLE2, and MARKABLE3 from figure 1 are bareNP, bareNP, and demonstrative NP, respectively.
Sentence function—subject (surface, logical, predicate nominal), object (indirect, direct, prepositional), modifier (to subjects and objects), section heading, other (for function tags not listed above). Example: MARKABLE1, MARKABLE2, and MARKABLE3 from figure 1 are prepositional object, surface subject, and direct object, respectively.
Semantic class—In addition to MUC's people type, we added biomedical types based on the Unified Medical Language System (UMLS)32: the semantic groups of anatomy, disorders (disease or syndrome in the tables and figures below) with a separate type for sign/symptom, and procedures and the individual semantic types of laboratory or test result; indicator, reagent, or diagnostic aid; and organ or tissue function (cf Bodenreider and McCray33 for semantic group inclusions). ‘Other’ is assigned if the markable type cannot be classified as any of the above. ‘None’ applies mostly to pronouns, which do not have a semantic type themselves but inherit one through coreference. Example: MARKABLE1, MARKABLE2, and MARKABLE3 from figure 1 are sign/symptom, sign/symptom, and other, respectively.
Each coreferring markable and its attributes are to be annotated. In this study, the markables were pre-annotated with existing gold standard annotations and, through an additional automated process, with pronouns and honorifics.
The ‘pair’ links two markables to represent the anaphor and the immediately preceding antecedent, which are the first two pair attributes. The third pair attribute, the Bagga class, is an indicator of the computational processing amount required to resolve the pair34:
Appositives: ‘Dr Smith, chair of Neurology’.
Syntactic equatives: ‘The patient is a homemaker.’
Proper names: ‘The patient underwent …. I reassured Mrs Smith that ….’
Pronouns: ‘Mr Smith is a 69-year-old-gentleman. He complains of…’
Quoted speech pronouns are pronouns used in quoted speech: ‘John said: ‘I am not feeling well.’’
Demonstratives: ‘She has noted change in her hand function and states that this is getting better.’
Exact matches: two coreferring markables with a text span of ‘colon cancer’
Substring matches: two coreferring markables of ‘staph bacteremia’ and ‘staph bacteremia infection’.
Identical lexical heads with different modifiers: ‘thickened aortic valve’ and ‘the aortic valve’.
Synonyms: ‘Patient complains of shortness of breath. The dyspnea…’
External world knowledge: in ‘John Doe. Social history: The patient…’, the reader must have knowledge that the patient is John Doe.
To the original Bagga set, we added another type, ontology knowledge, to utilize biomedical knowledge encoded in existing ontologies such as SNOMED CT and UMLS. An example of ontology knowledge is ‘The patient was found to have staph bacteremia. The patient was transferred for explantation of a pacemaker system felt to be involved by infection.’
The Bagga class assignment to a pair is determined by the relationship of the anaphor to the antecedent, anchored by the anaphor. For example, in ‘The patient complained of a sore throat. She has body aches as well.’, the anaphor is ‘she’ and the antecedent is ‘the patient’, thus the Bagga coreference class is pronouns.
The fourth pair attribute—the pair relation type attribute—describes the relation between the anaphor and its antecedent: identity, set/subset, part/whole, other. Two markables have an identity relation if they refer to one and the same discourse referent. For example:
Mr Smith complained of a headache. He had a sore throat.
In the set/subset relation, the pair comprises a set and a subset of entities of the same semantic type. The anaphor refers to a subset of a set of entities or is a superset of a previously mentioned linguistic expression in the discourse. For example:
The tumors have changed. Two are stage three.
The part/whole relation is a relation where one discourse referent is a part of another discourse referent. For example:
Her arm was scarred, but her hand was not.
This relation is different from the set/subset relation in that the entities that ‘part’ is referring to are not necessarily of the same type as those that ‘whole’ is referring to. However, in a set/subset relation, both ‘set’ and ‘subset’ refer to entities of the same type.
The ‘other’ relation category is a catch-all category for relations different than identity, part/whole, and set/subset.
There are two pairs in figure 1. PAIR1 is between MARKABLE1 (antecedent) and MARKABLE2 (anaphor) with a pair relation of type identity and a Bagga class of identical lexical heads; PAIR2 is between MARKABLE2 (antecedent) and MARKABLE3 (anaphor) with a pair relation of type identity and a Bagga class of demonstratives.
The chain takes all anaphoric pairs. In figure 1, PAIR1 (‘neck muscle weakness’ and ‘Neck weakness’) and PAIR2 (‘Neck weakness’ and ‘these neck difficulties’) form a chain. The grounding instance of an anaphoric chain is the first markable—that is, ‘neck muscle weakness.’
Corpus
Material
The corpus (105 082 tokens) consists of clinical notes from two institutions. This corpus has been part of previous clinical NLP studies and has layers of pre-existing gold standard annotations for linguistic and semantic concepts. We believe that (1) these pre-existing layered annotations would be of critical importance for coreference resolution as they provide gold standard features for training machine learning algorithms, and (2) the addition of the coreference layer to the pre-existing annotations would make the corpus a valuable lexical resource for further discourse-level annotations.
The Mayo set comprises 100 notes equally distributed between clinical (cc) and pathology (p) notes created following this procedure: 160 clinical notes and 302 colon cancer pathology notes were randomly selected from the Mayo Clinic Repository (for details, see refs4 7 35). We manually reviewed the notes and selected 100 documents that appeared to have anaphoric relations. The Mayo notes were pre-annotated with gold standard NEs of type disorders, signs/symptoms, procedures and anatomy created under a separate project for NE annotations.7 35
The University of Pittsburgh Medical Center (UPMC) set comprises 80 selected notes equally distributed among four types of narratives: emergency department notes (er), discharge summaries (ds), surgical pathology notes (sp), and radiology notes (rad). They were randomly selected from a larger set and manually pre-annotated with gold standard NEs for symptoms, signs, findings, and diagnoses36; they were manually examined and found to all have anaphoric relations.
The two datasets were programmatically pre-annotated for people type mentions of pronouns and for names preceded by ‘Mr’, ‘Mrs’ and ‘Ms’. The task of anaphoric relation annotations consists of marking a valid relation between given NEs. To isolate the anaphoric relation task from NER, the human annotators were asked to identify anaphoric relations between pre-annotated NEs. If a markable of an allowed semantic type was missed as a pre-annotation and it participated in an anaphoric relation, the human annotators were asked to add it in order to annotate the anaphoric relation. We show that there was no clear trend that pre-annotations affect inter-annotator agreement (IAA) for a NER task.35
Annotation flow
Knowtator37 was used as the annotation tool. The initial guidelines were created by three medical informaticists through an incremental and iterative annotation of five notes. After the initial round, three more annotators with medical backgrounds—a medical retrieval specialist/SNOMED CT terminology specialist (A1), a medical retrieval specialist (A2), and a knowledge engineer with a linguistics background (A3)—were trained on the annotation guidelines. Each annotator was given an initial 2 h training session and asked to annotate two notes from the initial guidelines development set of five and to then compare their annotations with the ‘correct’ ones generated by the developers. This training exercise generated many questions, resulting in a refined guidelines document. Over the next 4 weeks, the domain experts annotated weekly batches of a total of 21 clinical notes and 23 pathology notes. After the weekly assignments were completed, we computed IAA, resolved disagreements, and clarified the guidelines. The IAA stabilized in the mid 70s during the last 2 weeks. After the fourth week, we finalized the guidelines and proceeded with the closed annotations where no discussions were permitted. Each document was independently annotated by two annotators: the Mayo dataset by the annotator pairs of A1/A2, A1/A3 and A2/A3; the UPMC dataset by A1/A3, because A2 was not available during this phase of annotations.
The final gold standard annotations were produced by merging the individual experts’ annotations followed by adjudication of the mismatches. The jointly annotated training set notes are added to the gold standard but excluded from the final IAA computation.
Inter-annotator agreement
F-score (eqn 1) is a well-established metric in the information retrieval community. Hripcsak and Rothschild38 show that the F-score approximates the traditional κ39 40 (4) for many NLP tasks, as the number of the true negatives (TN) is very large.
| (1) |
where
| (2) |
| (3) |
and TP is true positives, FN is false negatives, FP is false positives. For the κ statistic, the number for TNs is computed as the possible markable permutations multiplied by the number of possible pair relations and adjusted for the discovered pairs. For example, if there were 10 markables in a document, then the possible pair permutations were Permutations(10,2)=90. Accounting for four possible types of pair relations, the number of TNs was 360. If the annotators identified six pairs distributed over TPs, FPs and FNs, then the TN number was 354. Computing κ follows (eqn 4).
| (4) |
where P(a) is the relative observed agreement among the annotators (eqn 5), and P(e) is the hypothetical probability of chance agreement in the assignment of positive and negative instances (eqns 6–8).
| (5) |
| (6) |
| (7) |
| (8) |
In the hypothetical example based on the previous description of 10 markables, the contingency table had two TPs, three FPs, one FN, and 354 TNs. Hence, P(a)=(2+354)/360=0.989; P(e)=((5/360)×(3/360))+((355/360)×(357/360))=0.978; κ was 0.495. In comparison, the F-score for the same example was 0.5 with precision of 0.40 and recall of 0.667.
We report the IAA on the pairs generated by (1) two annotators and (2) an annotator and the gold standard. The first shows the overlap between the annotators, and the second the contribution of each annotator to the gold standard.
Results
High-level corpus characteristics
The gold standard annotations resulted in 7214 markables, 5992 pairs, and 1304 identity chains. Each report averaged 40 anaphoric markables, 33 pairs and seven identity chains. Detailed corpus analysis will be presented in a separate paper. Here we present the high-level corpus characteristics. All semantic types in our schema occurred in the dataset. The most common type was people (51%), followed by anatomic site and disorders (both 14%). Identity was the dominant relation found in the gold standard set, accounting for 91% of pair relations. However, set/subset (5%) and part/whole (4%) relations also occurred and were more prevalent in pathology and radiology reports: 18% of the pairs in pathology reports showed a part/whole relationship, and 9% a set/subset; 15% in radiology reports were part/whole and 9% set/subset. Anaphoric expressions took a variety of phrasal types, including bare noun phrases (26%), personal pronouns (14%), definite noun phrases (11%), and possessive pronouns (6%). The Bagga class of pronouns was the most common (39%, or 2363 instances) followed by exact match (17%, or 1012 instances) and identical lexical head (12%, or 736 instances). The next most common Bagga relations were world knowledge (11%, or 660 instances) and ontological knowledge (5%, or 292 instances).
Inter-annotator agreement
The Mayo corpus was divided into 12 even batches. A1 and A3 annotated the first four sets; A1 and A2 annotated the second four sets; and A2 and A3 annotated the last four sets (table 1–5). The overall κ showed a substantial agreement (k=0.66). The κ for the clinical notes was high (0.67); the one for the pathology notes was moderate (0.58) (table 1). Table 2 presents the results for the agreement between the two annotators per set. The agreement between A1 and A2 was always substantial, ranging from 0.67 to 0.72, while the IAA between A3 and either A1 or A2 was lower, dropping to 0.34 for Mayo pathology set 6 (p6). The IAA between each annotator and the gold standard (table 3) is high reaching 0.92. Per type of anaphoric relation (table 4), κ is the strongest for identity relations (0.72) and fair for part/whole (0.32) and set/subset (0.23) relations. IAA results per markable type (table 5) show substantial agreement for anaphoric relations between people mentions and moderate agreement for anaphoric relations between anatomic sites, diseases or syndrome, procedures, and sign/symptoms.
Table 1.
Summary of pair-wise inter-annotator agreement results (Mayo dataset)
| All | Clinical | Pathology | |
| True positive | 846 | 730 | 116 |
| False positive | 393 | 314 | 79 |
| False negative | 476 | 389 | 87 |
| Precision | 0.6828 | 0.6992 | 0.5949 |
| Recall | 0.6399 | 0.6524 | 0.5714 |
| F-Score | 0.6607 | 0.6750 | 0.5829 |
| κ | 0.6607 | 0.6750 | 0.5828 |
Table 2.
Pair-wise inter-annotator agreement results per annotation subset (Mayo dataset)
| cc1 | cc2 | cc3 | cc4 | cc5 | cc6 | p1 | p2 | p3 | p4 | p5 | p6 | |
| True positive | 63 | 170 | 111 | 148 | 92 | 146 | 24 | 24 | 20 | 16 | 20 | 12 |
| False positive | 29 | 71 | 45 | 78 | 47 | 44 | 16 | 11 | 10 | 11 | 12 | 19 |
| False negative | 30 | 70 | 69 | 75 | 29 | 116 | 11 | 8 | 17 | 15 | 8 | 28 |
| Precision | 0.6848 | 0.7054 | 0.7115 | 0.6549 | 0.6619 | 0.7684 | 0.6000 | 0.6857 | 0.6667 | 0.5926 | 0.6250 | 0.3871 |
| Recall | 0.6774 | 0.7083 | 0.6167 | 0.6637 | 0.7603 | 0.5573 | 0.6857 | 0.7500 | 0.5405 | 0.5161 | 0.7143 | 0.3000 |
| F-Score | 0.6811 | 0.7069 | 0.6607 | 0.6592 | 0.7077 | 0.6460 | 0.6400 | 0.7164 | 0.5970 | 0.5517 | 0.6667 | 0.3380 |
| κ | 0.6808 | 0.7067 | 0.6605 | 0.6591 | 0.7076 | 0.6459 | 0.6396 | 0.7161 | 0.5965 | 0.5511 | 0.6662 | 0.3372 |
cc, clinical notes; p, pathology notes.
Table 3.
Inter-annotator agreement results for each annotator (A1, A2, and A3) and the gold standard (Mayo dataset)
| A1 and gold standard | A2 and gold standard | A3 and gold standard | |||||||
| All | Clinical | Pathology | All | Clinical | Pathology | All | Clinical | Pathology | |
| True positive | 653 | 557 | 96 | 722 | 613 | 109 | 839 | 720 | 119 |
| False positive | 247 | 204 | 43 | 300 | 251 | 49 | 117 | 77 | 40 |
| False negative | 150 | 120 | 30 | 132 | 113 | 19 | 65 | 40 | 25 |
| Precision | 0.7256 | 0.7319 | 0.6906 | 0.7065 | 0.7095 | 0.6899 | 0.8776 | 0.9034 | 0.7484 |
| Recall | 0.8132 | 0.8227 | 0.7619 | 0.8454 | 0.8444 | 0.8516 | 0.9281 | 0.9474 | 0.8264 |
| F-Score | 0.7669 | 0.7747 | 0.7245 | 0.7697 | 0.7711 | 0.7622 | 0.9022 | 0.9249 | 0.7855 |
| κ | 0.7669 | 0.7747 | 0.7244 | 0.7697 | 0.7710 | 0.7622 | 0.9021 | 0.9248 | 0.7854 |
Table 4.
Pair-wise inter-annotator agreement results per type of pair relation (Mayo dataset)
| Identity | Part/whole | Set/subset | |
| True positive | 806 | 19 | 21 |
| False positive | 281 | 48 | 59 |
| False negative | 345 | 33 | 85 |
| Precision | 0.7415 | 0.2836 | 0.2625 |
| Recall | 0.7003 | 0.3654 | 0.1981 |
| F-Score | 0.7203 | 0.3193 | 0.2258 |
| κ | 0.7202 | 0.3193 | 0.2258 |
Table 5.
Pair-wise inter-annotator agreement results per markable type (Mayo dataset)
| Anatomical site | Disease or syndrome | People | Procedure | Sign or symptom | Other | |
| True positive | 82 | 91 | 560 | 19 | 39 | 0 |
| False positive | 92 | 58 | 163 | 18 | 41 | 6 |
| False negative | 63 | 65 | 185 | 37 | 23 | 4 |
| Precision | 0.4713 | 0.6107 | 0.7746 | 0.5135 | 0.4875 | 0 |
| Recall | 0.5655 | 0.5833 | 0.7517 | 0.3393 | 0.6290 | 0 |
| F-Score | 0.5141 | 0.5967 | 0.7629 | 0.4086 | 0.5493 | 0 |
| κ | 0.5140 | 0.5967 | 0.7629 | 0.4085 | 0.5492 | −0.0006 |
The UPMC corpus was annotated by A1 and A3. As shown in table 6, the overall κ was moderate (0.41) as was the κ for the discharge summaries (0.56). The IAAs for the emergency room, radiology, and surgical pathology reports were fair (0.33, 0.23, and 0.35, respectively). The IAA between the annotators and the gold standard (table 7) points to substantial agreement (0.67 and 0.71). The identity relation type exhibits the highest IAA per relation type (table 8), whereas part/whole and set/subset varied from slight to fair. Pairs with markables of type people have the highest IAA by markable type, followed by disease or syndrome, sign or symptom, procedure, anatomic site, laboratory or test results, organ or tissue function, and none (table 9).
Table 6.
Summary of pair-wise inter-annotator agreement results (UPMC dataset)
| All | Discharge summaries | ER reports | Radiology reports | Surgical pathology reports | |
| True positive | 1032 | 496 | 396 | 39 | 101 |
| False positive | 1377 | 566 | 301 | 220 | 290 |
| False negative | 1628 | 205 | 1287 | 47 | 89 |
| Precision | 0.4284 | 0.4670 | 0.5681 | 0.1506 | 0.2583 |
| Recall | 0.3880 | 0.7076 | 0.2353 | 0.4535 | 0.5316 |
| F-Score | 0.4072 | 0.5627 | 0.3328 | 0.2261 | 0.3477 |
| κ | 0.4072 | 0.5626 | 0.3328 | 0.2260 | 0.3476 |
ER, emergency room.
Table 7.
Pair-wise inter-annotator agreement results for each annotator (A1 and A3) and the gold standard (UPMC dataset; all results shown)
| A1 and gold standard | A3 and gold standard | |
| True positive | 2134 | 2166 |
| False positive | 1526 | 1494 |
| False negative | 526 | 243 |
| Precision | 0.5831 | 0.5918 |
| Recall | 0.8023 | 0.8991 |
| F-Score | 0.6753 | 0.7138 |
| κ | 0.6753 | 0.7138 |
Table 8.
Pair-wise inter-annotator agreement results per type of pair relation (UPMC dataset)
| Identity | Part/whole | Set/subset | Other | |
| True positive | 1028 | 0 | 4 | 409 |
| False positive | 1155 | 94 | 108 | 288 |
| False negative | 1540 | 7 | 41 | 1274 |
| Precision | 0.4709 | 0.0000 | 0.0357 | 0.5868 |
| Recall | 0.4003 | 0.0000 | 0.0889 | 0.2430 |
| F-Score | 0.4328 | 0.0000 | 0.0510 | 0.3437 |
| κ | 0.4327 | 0.0000 | 0.0510 | 0.3437 |
Table 9.
Pair-wise inter-annotator agreement results per markable type (UPMC dataset)
| Anatomical site | Disease or syndrome | People | Procedure | Sign or symptom | None | Organ or tissue function | Laboratory or test results | |
| True positive | 20 | 60 | 790 | 36 | 25 | 0 | 0 | 1 |
| False positive | 352 | 179 | 406 | 160 | 68 | 9 | 10 | 9 |
| False negative | 27 | 64 | 1124 | 55 | 51 | 1 | 1 | 26 |
| Precision | 0.0538 | 0.2510 | 0.6605 | 0.1837 | 0.2688 | 0 | 0 | 0.1 |
| Recall | 0.4255 | 0.4839 | 0.4127 | 0.3956 | 0.3289 | 0 | 0 | 0.0370 |
| F-Score | 0.0955 | 0.3306 | 0.5080 | 0.2509 | 0.2959 | 0 | 0 | 0.0541 |
| κ | 0.0954 | 0.3305 | 0.5080 | 0.2508 | 0.2957 | −1.20E–05 | −0.0009 | 0.0534 |
Discussion
High-level corpus characteristics
Anaphoric reference in the corpus is quite common in clinical reports, with an average of 33 anaphoric pairs per report. The main type of anaphoric relation was the identity relation in which two mentions refer to the same entity, and a mean of seven chains per report ranged in chain length (ie, number of markables in the chain) from two to greater than 20. Slightly over half of the anaphoric markables were people—mainly the patient. It is not clear whether resolving patient references would benefit information extraction systems, which tend to focus on clinical conditions, medications, and procedures. Radiology (rad) and pathology reports (p and sp) showed similar characteristics in relation to the annotations when compared with discharge summaries (ds), clinical notes (cc), and ER reports (er), resulting in more anaphoric pairs for part/whole and set/subset relationships and showing higher frequency of markables for anatomic locations than for people. Based on an assessment of the annotations on the corpus, our schema was quite complete: very few of the annotations required the use of classes such as ‘other’, which we included in our schema to capture characteristics we had not foreseen.
We adopted the Bagga classes to facilitate comparison with general English texts annotated with these classes. The Bagga class annotations showed that the majority of the anaphoric pairs may in theory be resolved with common linguistic and textual cues, whereas a substantial portion of the pairs will require domain and world knowledge to resolve. However, the Bagga classes are limited in several ways. Classification to one class could sometimes be ambiguous—for example, ‘acute myocardial infarction’ and ‘myocardial infarction’ can be viewed as both ontology knowledge (of note, ontology knowledge is our addition to the original Bagga classes) and identical lexical heads. In addition, some of the Bagga classes had fewer than 200 instances—for example, appositives, syntactic equatives, demonstratives, and proper nouns—a number that is likely insufficient for experimenting with machine learning algorithms. Because only one annotator assigned Bagga class to the pairs, we have not shown that assignment of these categories can be agreed upon by annotators. However, the distribution of Bagga classes can be helpful in clarifying the information requirements of the task and in understanding different requirements for different report types.
Inter-annotator agreement
IAA was higher for the Mayo reports than for the UPMC reports. All the Mayo sets except pathology notes set 6 (p6 in last column of table 2) exhibit greater IAA than the UMPC notes. One explanation for this is that A1 and A2 are Mayo retrieval specialists very familiar with the Mayo dataset and its domain. Many of the disagreements were probably errors expected from any detailed annotation task with a substantial cognitive demand, such as missed anaphoricity relations between markables and annotation inconsistencies (of note, in most cases markables were pre-annotated). For instance, A1 missed some apparently easy pairs, such as exact coreferring matches of ‘central canal’ (see online supplement 1, A.1).
Other disagreements represented potential patterns that help understand the resulting gold standard corpus. First, differentiating the set/subset, part/whole, and identity relations was often difficult. For example, the ‘colon, sigmoid’ and ‘colonic’ mentions were related as identity by A1 and part/whole by A2. In the Mayo dataset, there was only one instance of the catch-all pair relation category ‘other’, which, after analysis, was relabeled identity. However, the annotators individually created many ‘other’ type pairs as implied by table 8. Almost all these pairs were later relabeled during the consensus phase. Tightening the definition of the relations—or perhaps even merging set/subset and part/whole—is likely to improve the IAA results.
The difference in domain background among the annotators caused disagreements, but ultimately contributed to a more complete annotated corpus, emphasizing the importance of a team with a mixture of linguistic, medical and computer science background to achieve maximum coverage. The most indicative results to support this are the difference between the pair-wise IAAs between the annotators and the IAAs between an annotator and the gold standard, which is very strong (tables 3 and 7). This result points to the complementary nature of the annotations produced by each domain expert because of their different knowledge backgrounds. Therefore, we argue that the important agreement is that between an annotator and the final gold standard, because that represents the completeness of the annotations. The complementary nature of the annotations is further supported by our error analysis (see online supplement 1, A.1).
Limitations and future steps
The process of building an effective gold standard relies on having a set of annotators who have a broad spectrum of knowledge. The annotation task presented a significant cognitive load and substantial resource commitments (1.5 h per document per annotator on average).
Although we built a diverse two-site corpus, the use of a pre-existing corpus might have introduced a bias in the agreement analysis. The overall generalizability of the guidelines and the schema will be further strengthened by annotations on data from additional sites, which we are actively working on within the Strategic Health Advanced Research Project Area 4 (http://www.sharpn.org) and the Informatics for Integrating Biology and the Bedside (http://www.i2b2.org) initiatives. This will further increase the representation of each relation type. A broader investigation of the definitions of Set/Subset, Part/Whole and Other relations is needed including possible category merging. A study comparing annotations produced by physician and non-physician annotators is likely to bring additional insights into the most optimal set of domain expertise. Future directions also include automatic pre-annotations of the values for the Bagga class attribute to alleviate some of the annotation burden. The future challenge of cross-document anaphoric relations will further advance information extraction across the entire patient's record.
The anaphoric relation corpus we have built is intended to become a community-shared lexical resource to bootstrap investigations of methods for anaphora resolution in clinical free-text. We developed our first version of a prototype coreference resolver trained on this corpus as part of cTAKES and ODIE.
Conclusion
We describe our efforts to build a manually annotated lexical resource for anaphoric relations in clinical free-text, which will become a community-shared lexical resource to further clinical NLP. This is a step toward developing a comprehensive NLP information extraction system specifically designed for the clinical narrative.
Supplementary Material
Acknowledgments
We are grateful to our annotators—Donna Ihrke, Pauline Funk, and Melissa Castine—and to Lynette Hirschman, Cheryl Clark, and Kevin Cohen for excellent feedback. The work was conducted under IRB 08-007020 and REN09050055/PRO07070252. Annotations from the UPMC reports are available for research purposes at http://www.dbmi.pitt.edu/blulab/nlprepository.html; those from the Mayo notes are available on an individual basis through a Data Use Agreement.
Footnotes
Funding: The work was funded by grant R01 CA127979.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Friedman C. Towards a comprehensive medical language processing system: methods and issues. Proc AMIA Annu Fall Symp 1997:595–9 [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman C. A broad-coverage natural language processing system. Proc AMIA Symp 2000:270–4 [PMC free article] [PubMed] [Google Scholar]
- 3.Hripcsak G, Kuperman GJ, Friedman C. Extracting findings from narrative reports: software transferability and sources of physician disagreement. Methods Inf Med 1998;37:1–7 [PubMed] [Google Scholar]
- 4.Savova GK, Masanz JJ, Ogren PV, et al. Mayo Clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. JAMIA 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.cTAKES Clinical Text Analysis And Knowledge Extraction System. http://www.ohnlp.org [DOI] [PMC free article] [PubMed]
- 6.Health Information Text Extraction (HITEx). https://www.i2b2.org/software/projects/hitex/hitex_manual.html [Google Scholar]
- 7.Coden A, Savova G, Sominsky I, et al. Automatically extracting cancer disease characteristics from pathology reports into a cancer disease knowledge representation model. J Bio Med Inform 2009;42:937–49 [DOI] [PubMed] [Google Scholar]
- 8.Haug P, Koehler S, Lau LM, et al. Experience with a mixed semantic/syntactic parser. Ann Symp Comp Appl Med Care 1995:284–8 [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen L, Haug P, Fiszman M. MPLUS: a probabilistic medical language understanding system. BioNLP 2002:29–36 [Google Scholar]
- 10.Fiszman M, Haug P, Frederick P. Automatic extraction of PIOPED interpretations from ventilation/perfusion lung scan reports. Proc AMIA Symp 1998:860–4 [PMC free article] [PubMed] [Google Scholar]
- 11.Fiszman M, Chapman W, Aronsky D, et al. Automatic detection of acute bacterial pneumonia from chest X-ray reports. J Amm Med Inform Assoc 2000;7:593–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trick W, Chapman W, Wisniewski M, et al. Electronic interpretation of chest radiograph reports to detect central venous catheters. Infect Control Hosp Epidemiol 2003:950–4 [DOI] [PubMed] [Google Scholar]
- 13.National Center for Text Mining (NaCTeM) http://www.nactem.ac.uk/index.php
- 14.JULIE Lab. http://www.julielab.de
- 15.caTIES. https://cabig.nci.nih.gov/tools/caties
- 16.Crowley RS, Castine M, Mitchell K, et al. caTIES - a grid based system for coding and retrieval of surgical pathology reports and tissue specimens in support of translational research. J Am Med Inform Assoc 2010;17:253–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MUC-7 MUC-7 Coreference Task Definition. 1997. http://www-nlpir.nist.gov/related_projects/muc/proceedings/co_task.html
- 18.Ng V, Cardie C. Improving Machine Learning Approaches To Coreference Resolution. Philadelphia, PA: 40th annual meeting Association for Computational Linguistics, 2002:104–11 [Google Scholar]
- 19.Poesio M. The Mate/Gnome Proposals For Anaphoric Annotations, Revisited. Cambridge, MA: SIGdial Workshop on Discourse and Dialogue, 2004:154–62 [Google Scholar]
- 20.ACE. http://projects.ldc.upenn.edu/ace/annotation/2005Tasks.html
- 21.Castano J, Zhang J, Pustejovsky J. Anaphora Resolution In Biomedical Literature. Spain, Alicante: International Symp on Reference Resolution for NLP, 2002 [Google Scholar]
- 22.Gasperin C, Karaminis N, Seal R. Annotation Of Anaphoric Relations In Biomedical Full-Text Articles Using A Domain-Relevant Schema. Logos, Portugal: DAARC-2007, 2007:19–24 [Google Scholar]
- 23.Zweigenbaum P, Demner-Fushman D, Yu H, et al. New Frontiers In Biomedical Text Mining: Session Introduction. Hawaii: Pacific Symposium of Biocomputing - PSB, 2007:205–8 [Google Scholar]
- 24.Roberts A, Gaizauskas R, Hepple M, et al. Building a semantically annotated corpus of clinical text. J Biomed Inform 2009:950–66 doi:10.1016/j.jbi.2008.12.013 [DOI] [PubMed] [Google Scholar]
- 25.GENIA2009. http://www-tsujii.is.s.u-tokyo.ac.jp/GENIA/home/wiki.cgi?page=GENIA+Project
- 26.Yang X, Su J, Zhou G, et al. An NP-Cluster Based Approach To Coreference Resolution. Geneva, Switzerland: COLING, 2004:226–32 [Google Scholar]
- 27.Sanchez-Graillet O, Poesio M, Kabadjov M, et al. What kind of problems do protein interactions raise for anaphora resolution? - a preliminary analysis. SMBM 2006:109–12 Jena. [Google Scholar]
- 28.Fritz A, ed. International Classification Of Diseases For Oncology. 3rd edn World Health Organization, Switzerland, 2000 [Google Scholar]
- 29.CLEF Coreference Annotation Guidelines. http://nlp.shef.ac.uk/clef/TheGuidelines/TheGuidelinesCoreference.html
- 30.Savova G, Chapman W, Zheng J, et al. Annotation schema for anaphoric relations in the clinical domain. San Francisco, CA: AMIA, 2009 [Google Scholar]
- 31.ODIE, Ontology Development and Information Extraction (ODIE) toolset. http://www.bioontology.org/ODIE [Google Scholar]
- 32.Unified Medical Language System (UMLS). http://www.nlm.nih.gov/research/umls/ [Google Scholar]
- 33.Bodenreider O, McCray A. Exploring semantic groups through visual approaches. J Biomed Inform 2003;36:414–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bagga A. Evaluation Of Coreferences And Coreference Resolution System: First Language Resource and Evaluation Conference, Granada, Spain. 1998:563–6 [Google Scholar]
- 35.Ogren P, Savova G, Chute C. Constructing Evaluation Corpora For Automated Clinical Named Entity Recognition. Marakesh, Morrocco: LREC,1 2008:3143–50 http://www.lrec-conf.org/proceedings/lrec2008/ [Google Scholar]
- 36.Harkema H, Thornblade T, Dowling J, et al. ConText: An algorithm for determining negation experiencer, and temporal status from clinical reports. J Biomed Inform 2009;42:839–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogren P. Knowtator: a Protégé Plug-In For Annotated Corpus Construction. New York, NY: Conference of the North American Chapter of the Association for Computational Linguistics, 2006:273–5 http://dx.doi.org/210.3115/1225785.1225791 [Google Scholar]
- 38.Hripcsak G, Rothschild A. Agreement, the F-measure, and reliability in information retrieval. J Am Med Inform Assoc 2005;12:296–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen J. A coefficient of agreement for nominal scales. Educational and Psychol Meas 1960;20:37–46 [Google Scholar]
- 40.Carletta J. Assessing agreement on classification tasks: the Kappa statistic. Comput Linguist 1996;22:249–54 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.