Abstract
Objective
DNA biobanks linked to comprehensive electronic health records systems are potentially powerful resources for pharmacogenetic studies. This study sought to develop natural-language-processing algorithms to extract drug-dose information from clinical text, and to assess the capabilities of such tools to automate the data-extraction process for pharmacogenetic studies.
Materials and methods
A manually validated warfarin pharmacogenetic study identified a cohort of 1125 patients with a stable warfarin dose, in which 776 patients were managed by Coumadin Clinic physicians, and the remaining 349 patients were managed by their providers. The authors developed two algorithms to extract weekly warfarin doses from both data sets: a regular expression-based program for semistructured Coumadin Clinic notes; and an advanced weekly dose calculator based on an existing medication information extraction system (MedEx) for narrative providers' notes. The authors then conducted an association analysis between an automatically extracted stable weekly dose of warfarin and four genetic variants of VKORC1 and CYP2C9 genes. The performance of the weekly dose-extraction program was evaluated by comparing it with a gold standard containing manually curated weekly doses. Precision, recall, F-measure, and overall accuracy were reported. Associations between known variants in VKORC1 and CYP2C9 and warfarin stable weekly dose were performed with linear regression adjusted for age, gender, and body mass index.
Results
The authors' evaluation showed that the MedEx-based system could determine patients' warfarin weekly doses with 99.7% recall, 90.8% precision, and 93.8% accuracy. Using the automatically extracted weekly doses of warfarin, the authors successfully replicated the previous known associations between warfarin stable dose and genetic variants in VKORC1 and CYP2C9.
Keywords: Automated learning, knowledge representations, discovery, text and data-mining methods, other methods of information extraction, natural-language processing, NLP, warfarin, old epass, Genetics, translational research—application of biological knowledge to clinical care, improving the education and skills training of health professionals, linking the genotype and phenotype
Introduction
Rapid growth in the use of large electronic health records (EHRs) has led to an unprecedented expansion in the availability of dense longitudinal datasets for observation research.1 2 Many efforts have linked large EHR databases with archived biological material, such as DNA, to accelerate clinical and genomic research. BioVU,3 the DNA data bank linked to deidentified EHRs at Vanderbilt University Hospital, currently contains over 100 000 DNA samples. In a previous study, we used BioVU and EHR-derived disease phenotypes to identify genetic variants contributing to common diseases and traits.4 An appealing vision, which has not been extensively explored, is to use the EHR-linked DNA biobanks for pharmacogenetic studies, which aim to identify associations between genetic variations and drug efficacy and toxicity.5 6 Recently, we replicated associations between steady-state warfarin weekly dose and variants in VKORC1 and CYP2C9 in BioVU.7 8 We found that manual extraction of weekly doses of warfarin was one of the most time-consuming steps. In this study, we developed an automated weekly dose calculation system based on an existing medication-information extraction system called MedEx, and applied it to datasets from the aforementioned warfarin pharmacogenetic study. Using automatically extracted warfarin weekly doses, we achieved similar p values for genetic associations to those from manual data extraction, indicating that such EHR-based pharmacogenetic studies could be done in an in silico fashion, with the help of informatics approaches.
Background
Personalized medicine aims at providing tailored medical care to individuals based on information such as genotypes and gene-expression profiles. Pharmacogenomics and pharmacogenetics contribute to personalized medicine by identifying associations between genetic variations and drug efficacy and toxicity. A number of studies have successfully identified genetic variants that contribute to the variability in response to drugs; examples include azathioprine dosing and TPMT variants,9 irinotecan and UGT1A1 variants,10 and clopidogrel and CYP2C19 variants.11
Unlike single-dose medications such as clopidogrel, therapeutic warfarin dose varies up to 10-fold between individuals based on an individual's composition, diet, gender, age, and other interacting medications. Warfarin has a narrow therapeutic window; improper dosing can lead to significant toxicity with either overanticoagulation including major gastrointestinal and intracranial bleeds or underanticoagulation including stroke and thrombosis. Thus, patients taking warfarin regularly have their blood checked to maintain the proper degree of anticoagulation, as measured by the International Normalized Ratio (INR). Despite such efforts, rates of major bleed in the initiation phase of therapy are between 16 and 25%.12 Studies have shown that genetic polymorphisms in CYP2C9 and VKORC1 contribute substantially to the variation observed for warfarin steady-state dose.13–15 Algorithms that integrate pharmacogenetics, demographic, and clinical factors have been shown to be more effective at predicting the therapeutic dose of warfarin than clinical algorithms alone.16–18 While these data suggest the potential to reduce adverse drug events, pharmacogenetic research has been hampered by small sample sizes for most prospective studies and the large amount of effort (in terms of cost and time) required for sample collection. The ability to perform pharmacogenetic studies in an efficient manner is critical for accelerating translation between genetic research and clinical practice.
An important potential enabling resource for pharmacogenetics is the combination of a DNA repository with EHR systems sufficiently robust to serve as resources for analysis of therapeutic outcomes across patient populations.6 A prominent example is the Electronic Medical Records & Genomics network,19 a consortium of five institutions which each have DNA data banks coupled to large EHRs. BioVU,3 the Vanderbilt DNA biobank, currently contains DNA samples from over 100 000 subjects. They are linked to longitudinal clinical data in the Synthetic Derivative (SD) database, a deidentified copy of the Vanderbilt EHR database with over 1.8 million subjects. Recently, we manually constructed a cohort of warfarin patients from BioVU and successfully replicated previously reported associations between steady-state warfarin weekly dose and variants in VKORC1 and CYP2C9.7 8
Drug data in EHRs usually exist as heterogeneous data types including both structured (eg, from e-prescribing systems) and unstructured (eg, within clinical notes) formats. Much of the detailed drug information is embedded in narrative text and is not immediately available for data analysis. We have developed a general information-extraction tool called MedEx, which could identify medication relevant information from clinical notes in Vanderbilt University Hospital with high performance.20 Later, it was extended to process clinical text from Partners Healthcare Systems by participating the 2009 i2b2 NLP challenge, in which it was ranked the second best system.21 A number of natural-language-processing (NLP) systems, including those that participated in the 2009 i2b2 NLP challenge,22 have focused on medication information extraction,21 23–26 including drug names and signature information (such as dose, frequency, and route). Studies have shown the uses of such informatics tools in clinical research, such as construction of statin dose–response relations.27 However, the challenges, issues, and effectiveness of using such medication information extraction systems for pharmacogenetic studies have not been investigated previously. In this study, we extended an existing tool called MedEx20 to automatically extract drug weekly doses from clinical text and applied it to a pharmacogenetic study of warfarin.
Methods
Defining a cohort of patients with stable warfarin doses
For each patient with at least one mention of warfarin or Coumadin keyword in the SD, we searched for a ‘stable dose’ window, which is defined as the first window in which a patient had a consistently therapeutic INR between 2 and 3 for at least 3 weeks (with no out-of-range INRs). We identified 1125 patients with a stable dose window of warfarin. Among them, 776 patients had ‘Coumadin Clinic’ notes, which indicated that their warfarin dose was managed by specialized pharmacists. The rest of the 349 patients did not have Coumadin Clinic notes, so they were managed by individual providers. Coumadin Clinic notes contain a semistructured warfarin dose using a table-like format, which lists doses for each day of a week. For clinical notes entered by providers, they usually contain a free text description of multiple dosing pieces, which are more difficult to extract (see examples in Column 1 in table 1).
Table 1.
Examples of warfarin dosing text and their calculations
| Text | MedEx output | Normalization | Calculation rational | Weekly dose |
| Coumadin 2.5 mg po dly except 5 mg qTu,Th |
|
|
DOSE_1×(DAYS_1–DAYS_2)+DOSE_2×DAYS _2 | 22.5 mg |
| Coumadin 5 mg alternate with 2.5 mg qod |
|
|
(DOSE_1+DOSE_2)×7/2 | 26.25 mg |
| Coumadin 6 mg po 4× week, 4 mg po 3× week |
|
|
DOSE_1×DAYS _1+DOSE_2×DAYS _2 | 36 mg |
DAYS, no of days in a week; DOSE, daily dosage.
Extracting the warfarin weekly dose from the clinical text
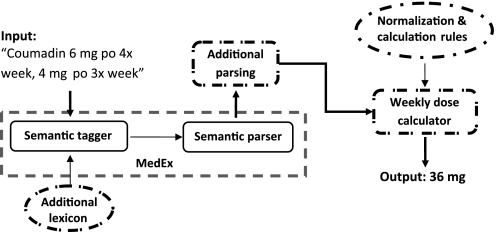
For Coumadin Clinic notes, we developed a regular expression (RegEx)-based script to extract warfarin doses of each day of a week, and then summed them up to obtain the weekly dose. For regular clinical notes, we developed a weekly dose extraction system, which consisted of two steps: (1) to extend a general medication information extraction system (MedEx) to accurately capture dosing-related findings (eg, dose, frequency) from clinical text; and (2) to build a weekly dose calculator, which will normalize textual dosing findings into numeric values and perform the calculation based on predefined rules. Figure 1 shows an overview of the automated weekly dose extraction system for warfarin.
Figure 1.
Overview of the warfarin weekly dose calculation system based on MedEx. Extensions to MedEx include the ‘Additional Lexicon’ knowledge base and the ‘Additional Parsing’ component. The ‘weekly dose calculator’ reads the outputs from the extended MedEx, and normalizes and calculates weekly doses using another knowledge base ‘Normalization & Calculation Rules.’
MedEx implements a semantic parsing approach, which labels words/phrases with semantic categories first and then uses a semantic grammar to parse medication findings into structured forms. The extension to MedEx included new lexicon entries (eg, adding new frequency terms representing irregular weekly combinations such as ‘qTu, Th’), and an additional parsing step, which relies on semantic patterns specifically for warfarin dosing text. Based on our observation, a total of 32 semantic rules (see online appendix) were used in the additional parsing step. This implementation architecture allowed rapid integration of drug-specific knowledge, without changing the core of MedEx.
As MedEx (as well as other medication extraction systems) outputs dose-related information such as strength and frequency as textual strings, they cannot be directly used to calculate the weekly dose. Therefore, outputs from MedEx (eg, dose and frequency) need to be normalized into numeric values, and specific rules for weekly dose calculation need to be defined for each pattern of warfarin dosing text. This process was carried out using a knowledge base containing rules for dose/frequency normalization and weekly dose calculation. Table 1 shows three examples of warfarin-dosing sentences, as well as the normalized fields and calculation rules.
Evaluation of warfarin weekly dose extraction programs
Within the cohort, sentences containing warfarin were extracted and randomly divided into a training set and a test set. The weekly dose extraction system was developed using the training set and evaluated using the test data set. We randomly selected 200 warfarin sentences from Coumadin Clinic notes and 500 sentences from providers' notes. Each sentence was manually reviewed to determine the correct weekly dose, if it contained dosing information. Precision, recall, F-measure, and accuracy were used to evaluate the performance of systems. Precision was defined as the ratio between the numbers of correctly extracted weekly doses (exactly the same) and all extracted weekly doses by the system. Recall was defined as the ratio between the number of extracted weekly doses by the system and the number of weekly doses identified in the gold standard. The F-measure was calculated as the harmonic mean of precision and recall, or 2×Precision×Recall/(Precision+Recall). Accuracy was defined as the ratio between the number of correct outputs by the system (it either generated the correct weekly dose or generated nothing when there was no dosing information in the sentence) and the total number of warfarin sentences.
Select patient-level weekly doses for analysis
Within a stable dose window, multiple weekly doses of warfarin were often extracted from different types of clinical notes at different time points. We named such weekly doses extracted from clinical text ‘document-level’ weekly doses. From these, we need to select one weekly dose for each patient and use it for genetic analysis, which was called the ‘patient-level’ weekly dose. We selected the median value of document-level weekly doses in a stable dose window as the patient-level weekly dose to avoid any undue influence of outliers in our automated data-extraction approach. In the manual data-extraction method, a physician determined the ‘patient-level’ weekly dose as follows: (1) reviewed all the clinical notes in a window to extract all the sentences containing warfarin dosing information; (2) manually calculated the weekly dose for each warfarin mention; and (3) selected the median weekly dose if more than one weekly dose was reported in the window unless text in clinical notes indicated unequivocally otherwise. We compared the automated approach with the manual method and conducted analyses on patient-level weekly doses generated by both approaches.
Genotyping and association analysis
Genotyping for the single-nucleotide polymorphisms (SNPs) was conducted by the Vanderbilt DNA Resources Core using Sequenom's iPLEX Gold assay coupled with Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS, Sequenom, Sequenom, San Diego, California). For this study, we used genotype data for SNPs that are known to be related to warfarin sensitivity (see table 2 for SNP rs numbers). Genotype calls were determined by investigators blinded to dose taken by subjects. Quality-control procedures included examination of marker and sample genotyping efficiency, allele frequency calculations, and tests of Hardy–Weinberg equilibrium. Warfarin weekly stable dose was log-transformed and regressed against VKORC1 and CYP2C9 genotypes assuming an additive genetic model. Linear models unadjusted and adjusted for sex, age, and body mass index were tested using the genetic analysis software PLINK version 1.07.28
Table 2.
p Values of associations between four single-nucleotide polymorphisms (SNPs) and warfarin stable weekly dose extracted by automated methods
| Gene | SNP | β-adjusted | p Value | SE |
| VKORC1 | rs9934438 | −0.3436 | 1.20×10−60 | 0.0217 |
| VKORC1 | rs2359612 | −0.3418 | 1.30×10−60 | 0.0216 |
| VKORC1 | rs9923231 | −0.3426 | 3.78×10−60 | 0.0217 |
| CYP2C9 | rs4917639 | −0.2916 | 9.42×10−29 | 0.0279 |
β is the effect size or increase in log-transformed warfarin dose per minor allele.
Results
The RegEx program achieved 100% recall, 97.4% precision, and 97.5% accuracy, on the annotated set of 200 warfarin sentences from Coumadin Clinic notes. The MedEx-based weekly dose extraction system achieved 99.7% recall, 90.8% precision, and 93.8% accuracy, on the test set of 500 physician-annotated warfarin sentences from clinical notes. Table 3 shows the details of the evaluation results on weekly dose calculation.
Table 3.
Evaluation results of warfarin weekly dose extraction programs developed in this study
| Type of notes | Weekly dose extraction method | No of warfarin sentences | No of warfarin sentences with dose information | Precision (%) | Recall (%) | F-measure (%) | Accuracy (%) |
| Coumadin Clinic notes | RegEx | 200 | 200 | 97.4 | 100 | 98.7 | 97.5 |
| Narrative providers' notes | MedEx-based calculator | 500 | 326 | 90.8 | 99.7 | 95.0 | 93.8 |
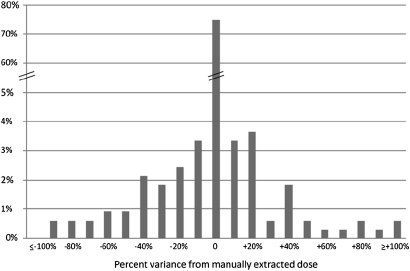
Coumadin Clinic notes contained semistructured weekly dose information on patients, and automatically generated median values were almost 100% correct. Therefore, our analysis was focused on the results from narrative providers' notes. Our automated method exactly matched the manually extracted weekly dose for 75% of the patients who were managed by their providers. Figure 2 shows the distribution of differences between the manually extracted weekly dose and the automatically derived median dose. Eighty-eight percent of all median doses were within 20% of the manually extracted doses. A total of 4.9% were more than 50% higher or lower than the manually extracted dose. We randomly selected 20 patients from the 25% mismatched population, and manually reviewed the differences between two approaches. Our analysis showed that the automated weekly dose system had extracted the incorrect dose for 11 of 20 patients. However, the manual approach extracted the incorrect weekly dose for the rest of the nine patients.
Figure 2.
Distribution of differences between automatically and manually extracted warfarin stable weekly doses, for patients who were managed by their providers. The x-axis shows the difference in weekly doses in percentage, and the y-axis shows the percentage of patients in the population.
Steady-state weekly warfarin doses extracted from both Coumadin Clinic and providers' notes were used for genetic association analysis. Our results showed that log-transformed patient-level steady-state warfarin dosage was strongly associated with VKORC1 variant rs9923231 (p≤2.5×10−54, βunadjusted=−0.36) and CYP2C9 poor metabolizers (rs4917639, tags both CYP2C9*2 and CYP2C9*3) (p≤3.0×10−27, βunadjusted=−0.31). Table 2 shows the details of p values and betas for each SNP, when the linear model was adjusted for sex, age, and body mass index. The p values from automatically derived weekly doses were very similar to those from manual abstraction.7 8 The VKORC1 variants explained ∼21% of dose variability, and CYP2C9 rs4917639 explained ∼11%.
Discussion
Recently, comprehensive EHRs linked with DNA biorepositories have demonstrated their value for genomic research, including identifying genetic variants contributing to diseases.4 29 30 Since EHRs contain a longitudinal record of medication exposure and response, EHR-linked genomic data may be a great resource for pharmacogenomic data as well. A recent study demonstrated this finding, replicating the associations between steady-state warfarin weekly dose and genetic variants in VKORC1 and CYP2C9 in BioVU.7 8 However, the most challenging step in this study was extracting drug-exposure and outcome information from the EHR. A manual chart review is costly and time-consuming, hampering the efficiency of conducting pharmacogenetic studies. In this study, we extended an existing medication extraction tool to automatically calculate weekly doses of warfarin from clinical text, and evaluated the ability to find known genetic associations with warfarin dose using only automatically extracted values. Evaluation showed that the system performed well in capturing weekly doses of warfarin. Moreover, the genetic association analysis successfully replicated previously known associations between steady-state warfarin weekly dose and variants in VKORC1 and CYP2C9 genes, using completely automated calculations of weekly dose. This demonstrates that informatics tools have the potential to simplify the data-extraction processes for EHR-based pharmacogenetic studies.
Determining medication doses is critical to many drug-related studies using EHR data, including pharmacogenetics. In inpatient settings, structured medication data can be obtained from computerized systems such as physician order entries and electronic medication administration records. However, for outpatients, detailed drug-dose information is often embedded in clinical text, often requiring costly manual abstraction. Therefore, NLP systems that can automatically extract and calculate daily or weekly doses of medications used in the outpatient setting are very useful. However, outputs from current medication extraction systems, such as those developed for the 2009 i2b2 NLP challenges, are textual strings of extracted information such as dose and frequency, and such information is not directly usable for daily or weekly dose calculation. Interpreting these data for real-world use is not a trivial task. In this study, we demonstrated that we could extend an existing NLP tool (MedEx) by adding new knowledge components to accurately capture weekly doses of warfarin, providing a good example of applying existing NLP tools for practical research uses.
Although MedEx performs well unchanged for most medications and dosing regimens, specific application of MedEx to warfarin extraction was challenging. We found that the dosing text of warfarin was much more complicated than average drugs (see examples in table 1). Our current implementation with new lexicons and an additional parsing step provides a generalizable solution to solve this problem, allowing MedEx to be customized to reach a desirable high performance for any specific drug. In addition, we conducted an experiment to assess if such modifications to MedEx affect its performance on other drugs. We randomly selected 1000 discharge summaries from the SD, and processed them using both the original and the modified MedEx for this study. Among all 42 563 medication entities (including associated signature information) recognized by the modified MedEx, 41 942 (98.54%) were identical to the outputs from the original MedEx. A manual analysis of 50 mismatched medication entities extracted by the modified MedEx showed that 53% of them were correctly identified by the original MedEx, and 47% were correctly identified by the modified MedEx. Such results indicated that the modification to MedEx for improving warfarin dose extraction did not significantly affect its performance on other drugs.
We also looked into errors in the weekly dose extraction, which could be categorized into two classes: (1) failures in capturing dose-related findings (eg, ‘1/4 tablet’ was not identified in the sentence ‘warfarin 2.5 mg PO 0.5 tabs daily ex ¼ tablet q Th’); and (2) failures in dose normalization and weekly dose calculation (eg, the sentence ‘Continue Coumadin 5 mg’ indicated a weekly dose of 35 mg, as it omitted the default frequency ‘daily’; but our program outputted 5 mg as its weekly dose). Manual analysis of 20 sentences with incorrect weekly doses revealed that 10% of errors were from normalization and calculation, and 90% of errors were from dose entity extraction, which also indicated the complexity of natural language expression (eg, we noticed different expressions for ‘every Monday and Wednesday’: ‘qM, W; q Mon, Wed; qMon, Wed; q Mondays and Wednesdays; qM&W … etc’). Our future work will include investigations on methods to capture variants of dosing entities.
Our genetic association algorithm selected the steady-state warfarin dose by finding a time period between 3 and 12 weeks in which the patient had stable INR values between 2 and 3 (in whom that was the goal range). Given that many people had stable doses for long periods of time with only slight adjustments in dose, we analyzed the ‘median’ dose during that time period. When we compared the patient-level stable weekly doses from the automated approach and those from manual review, 75% were exactly the same, and 88% were within 20% of the manually extracted doses. Manual review of 20 patients who were randomly selected from the 25% mismatched population showed that 11 of them (55%) had the incorrect stable weekly dose from the automated approach due to errors by the weekly dose calculation system. Seven patients (35%) had dose differences because the automated weekly dose extraction system identified more warfarin dosing mentions than found in the manual review. The remaining two patients had incorrect stable weekly doses by the manual review approach (ie, the reviewer calculated the weekly dose incorrectly). Such findings indicate that the weekly dose extraction system needs further improvement, but also that some of the differences were due to errors in the manual review process. We also noticed a fair number of patients had discordant drug-dosing information on the same day stored in EHRs. Six out of 20 patients (30%) reviewed had at least one discrepant pair of warfarin weekly doses from different notes that were recorded at the same date. A detailed analysis in the discrepancy is beyond the scope of this study but would be helpful to identify the correct weekly dose by deciphering such discrepant information.
Despite its success in replicating known warfarin pharmacogenetic associations, this study has limitations. The drug-outcome data in this study are about drug doses instead of drug responses such as adverse events or treatment efficacy, which involve accurate assessment of an event and timing in correlation with drug exposure, which could be more challenging for automated extraction. Additionally, assessing drug exposure based on medication mention in clinical text does not adjust for possible non-compliance issues, which can be common with many medications. Finally, such methods require the presence of a robust EHR that can be easily queried, and that has been linked to genomic information. The medication dose extraction tools developed in this study are valuable for clinical research, but they are not robust enough to support practical applications such as decision-support systems in the clinical settings.
Conclusion
In this study, we developed a weekly dose calculation system for warfarin by extending an existing medication information extraction system, which provides a general model for building high-performance drug-specific dose extraction systems for clinical research. Our study demonstrates that DNA repositories linked to NLP-derived drug outcome data in an EHR replicate previously known pharmacogenetic associations. Broader application of such methods in EHR-linked biobanks may enable rapid generation of very large datasets for pharmacogenetic discovery and validation.
Supplementary Material
Footnotes
Funding: This study was supported by NIH grants number RC2GM092618 and R01CA141307.
Competing interests: None.
Ethics approval: This study was approved by Vanderbilt University IRB.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Jha AK, DesRoches CM, Campbell EG, et al. Use of electronic health records in US hospitals. N Engl J Med 2009;360:1628–38 [DOI] [PubMed] [Google Scholar]
- 2.Shea S, Hripcsak G. Accelerating the use of electronic health records in physician practices. N Engl J Med 2010;362:192–5 [DOI] [PubMed] [Google Scholar]
- 3.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008;84:362–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ritchie MD, Denny JC, Crawford DC, et al. Robust replication of genotype–phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet 2010;86:560–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCarty CA, Wilke RA. Biobanking and pharmacogenomics. Pharmacogenomics 2010;11:637–41 [DOI] [PubMed] [Google Scholar]
- 6.Wilke RA, Xu H, Denny JC, et al. The emerging role of electronic medical records in pharmacogenomics. Clin Pharmacol Ther 2011;89:379–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramirez A, Xu H, Oetjens M, et al. Identifying Genotype–Phenotype Relations in Electronic Medical Record Systems: Application to Warfarin Pharmacogenomics. American Heart Association, 2010. http://circ.ahajournals.org/cgi/content/meeting_abstract/122/21_MeetingAbstracts/A19509 [Google Scholar]
- 8.Oetjens M, Havens-Ramirez A, Denny J, et al. Using Electronic Medical Records Linked to a Biorepository for Pharmacogenomics—Replication of the Genetic Predictors of Warfarin Maintenance Dose in the Community. Washington, DC: American Society of Human Genetics, 2010 [Google Scholar]
- 9.Gearry RB, Barclay ML. Azathioprine and 6-mercaptopurine pharmacogenetics and metabolite monitoring in inflammatory bowel disease. J Gastroenterol Hepatol 2005;20:1149–57 [DOI] [PubMed] [Google Scholar]
- 10.Innocenti F, Undevia SD, Iyer L, et al. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. J Clin Oncol 2004;22:1382–8 [DOI] [PubMed] [Google Scholar]
- 11.Mega JL, Close SL, Wiviott SD, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med 2009;360:354–62 [DOI] [PubMed] [Google Scholar]
- 12.Beyth RJ, Quinn L, Landefeld CS. A multicomponent intervention to prevent major bleeding complications in older patients receiving warfarin. A randomized, controlled trial. Ann Intern Med 2000;133:687–95 [DOI] [PubMed] [Google Scholar]
- 13.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin Pharmacol Ther 2008;83:460–70 [DOI] [PubMed] [Google Scholar]
- 14.Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med 2005;352:2285–93 [DOI] [PubMed] [Google Scholar]
- 15.Crawford DC, Ritchie MD, Rieder MJ. Identifying the genotype behind the phenotype: a role model found in VKORC1 and its association with warfarin dosing. Pharmacogenomics 2007;8:487–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein TE, Altman RB, Eriksson N, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med 2009;360:753–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gage BF, Eby C, Johnson JA, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther 2008;84:326–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sagreiya H, Berube C, Wen A, et al. Extending and evaluating a warfarin dosing algorithm that includes CYP4F2 and pooled rare variants of CYP2C9. Pharmacogenet Genomics 2010;20:407–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The eMERGE Network. 2006. https://www.mc.vanderbilt.edu/victr/dcc/projects/acc/index.php/Main_Page (accessed 18 Feb 2011). [Google Scholar]
- 20.Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc 2010;17:19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doan S, Bastarache L, Klimkowski S, et al. Integrating existing natural language processing tools for medication extraction from discharge summaries. J Am Med Inform Assoc 2010;17:528–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010;17:514–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, Liu F, Antieau L, et al. Lancet: a high precision medication event extraction system for clinical text. J Am Med Inform Assoc 2010;17:563–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meystre SM, Thibault J, Shen S, et al. Textractor: a hybrid system for medications and reason for their prescription extraction from clinical text documents. J Am Med Inform Assoc 2010;17:559–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamon T, Grabar N. Linguistic approach for identification of medication names and related information in clinical narratives. J Am Med Inform Assoc 2010;17:549–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spasic I, Sarafraz F, Keane JA, et al. Medication information extraction with linguistic pattern matching and semantic rules. J Am Med Inform Assoc 2010;17:532–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peissig P, Sirohi E, Berg RL, et al. Construction of atorvastatin dose–response relationships using data from a large population-based DNA biobank. Basic Clin Pharmacol Toxicol 2007;100:286–8 [DOI] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denny JC, Ritchie MD, Crawford DC, et al. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation 2010;122:2016–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kullo IJ, Ding K, Jouni H, et al. A genome-wide association study of red blood cell traits using the electronic medical record. PLoS One 2010;5:e13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.