Abstract
The meiotic mutant c(3)G (crossover suppressor on 3 of Gowen) abolishes both synaptonemal complex (SC) formation and meiotic recombination, whereas mutations in the mei-W68 and mei-P22 genes prevent recombination but allow normal SC to form. These data, as well as a century of cytogenetic studies, support the argument that meiotic recombination between homologous chromosomes in Drosophila females requires synapsis and SC formation. We have cloned the c(3)G gene and shown that it encodes a protein that is structurally similar to SC proteins from yeast and mammals. Immunolocalization of the C(3)G protein, as well as the analysis of a C(3)G-eGFP expression construct, reveals that C(3)G is present in a thread-like pattern along the lengths of chromosomes in meiotic prophase, consistent with a role as an SC protein present on meiotic bivalents. The availability of a marker for SC in Drosophila allowed the investigation of the extent of synapsis in exchange-defective mutants. These studies indicate that SC formation is impaired in certain meiotic mutants and that the synaptic defect correlates with the exchange defects. Moreover, the observation of interference among the residual exchanges in these mutant oocytes implies that complete SC formation is not required for crossover interference in Drosophila.
Keywords: Drosophila melanogaster, c(3)G, meiosis, synaptonemal complex, recombination
Meiotic prophase is marked by interactions between homologous chromosomes that culminate in their alignment with each other along a structure called the synaptonemal complex (SC) (von Wettstein et al. 1984; Zickler and Kleckner 1999; Walker and Hawley 2000). Synapsis and SC formation between homologs is associated with, or requisite for, the formation of exchanges between homologous sequences. These exchanges are later detectable physically as chiasmata, and genetically as recombination between loci on the chromosomes. In many meiotic systems, these genetic exchanges ensure the proper segregation of homologous chromosomes during anaphase I (Hawley 1988).
The SC is an almost universally conserved meiotic structure among eukaryotes (von Wettstein et al. 1984; Zickler and Kleckner 1999). After preliminary interactions align homologous chromosomes within ∼300 nm of each other, the chromosomal axes become juxtaposed at a distance of ∼100 nm, which is bridged by the SC. Electron microscopic (EM) studies show the SC as a lattice of transverse filaments (TFs) running between the homologs. The TFs connect the central element, located in the middle of the SC, with lateral elements along the axes of the chromosomes. The connections mediated by the SC are thought to provide a means for holding homologous chromosomes together during meiotic prophase (von Wettstein et al. 1984; Walker and Hawley 2000). In addition, the SC has been proposed to function in the regulation of meiotic recombination and the formation of chiasmata (von Wettstein et al. 1984).
Investigations in Saccharomyces cerevisiae, Drosophila melanogaster, and Caenorhabditis elegans have revealed a complex relationship between the SC and the initiation of recombination, which differs between species. In yeast, the SC is not necessary for meiotic recombination (Roeder 1997). However, the initiation of recombination and processing of recombination intermediates are required for the progression of synapsis. Time course studies in yeast have established that double strand break (DSB) formation precedes synapsis, and the processing of recombination intermediates occurs during the formation of the SC (Padmore et al. 1991; Schwacha and Kleckner 1994, 1995). In addition, recombination is initiated efficiently in mutants that eliminate or disrupt the SC (Sym and Roeder 1994; Storlazzi et al. 1996; Chua and Roeder 1998; Agarwal and Roeder 2000). SC formation is, however, abolished in mutants that fail to initiate recombination, and synapsis is defective or delayed in yeast mutants for the DSB processing and repair pathway (Roeder 1997). Similarly, other fungi, including Schizosaccharomyces pombe and Aspergillus nidulans, normally lack SC but still undergo meiotic recombination (Egel-Mitani et al. 1982; Bahler et al. 1993).
Conversely, the SC appears to be necessary for meiotic exchange in Drosophila females. In Drosophila, the mutant c(3)G (crossover suppressor on 3 of Gowen) essentially eliminates meiotic exchange (Gowen and Gowen 1922; Gowen 1933; Hall 1972), intragenic exchange, and gene conversion (Carlson 1972). High levels of meiotic nondisjunction result from the lack of exchange (Hall 1972). Although SC assembles along the length of each bivalent in wild-type Drosophila females (Carpenter 1975a, 1979b), EM studies of ovaries from c(3)G mutant females reveal the absence of SC (Meyer 1964; Smith and King 1968; Rasmussen 1975). Based on the lack of SC in c(3)G mutants, one hypothesis for the role of C(3)G was as a structural component of the SC (Smith and King 1968). Indeed, we present data here that c(3)G encodes a component of the SC, possibly the transverse filament (TF).
Components of the TF have been identified in the yeast S. cerevisiae (Zip1) and in several mammalian species (SCP1/Syn1) (Meuwissen et al. 1992; Sym et al. 1993; Dobson et al. 1994). Despite their apparently identical role within the structure of the SC, Zip1 and the SCP1 proteins bear little sequence similarity. However, these proteins share a similar structure, in that the central portions of the proteins are predicted to form coiled coils, which allow the proteins to dimerize to form the TFs.
Like TF proteins in other organisms (Zip1, SCP1), the c(3)G gene encodes a coiled-coil protein (Meuwissen et al. 1992; Sym et al. 1993). Antibodies raised against C(3)G stain prophase meiotic chromosomes in a thread-like pattern similar to Drosophila SC as analyzed by EM (Carpenter 1975a, 1979b). C(3)G localization in certain exchange-defective mutants reveals that problems in SC formation correlate with exchange defects, and further support the assertion that the SC is required for the completion of exchange in Drosophila females.
Results
Identification of the c(3)G gene
The c(3)G gene was mapped to a 17-kb interval in region 89A2-5 by P. Szauter (pers. comm.). Several transcripts from this region were identified by expressed sequence tags from the Berkeley Drosophila Genome Project (Rubin et al. 2000). A rescue construct, P{X203}, which contains 8 kb of genomic DNA from this interval (see Fig. 1 and Materials and Methods), was introduced into the Drosophila genome by P-element transformation. Although exchange in c(3)G68 homozygotes is nearly eliminated, in the presence of the P{X203} transgene, exchange is returned to a level slightly higher than that of wild type (Table 1). The ability of P{X203} to rescue the c(3)G phenotype shows that the c(3)G gene is contained within the 8-kb transgene construct.
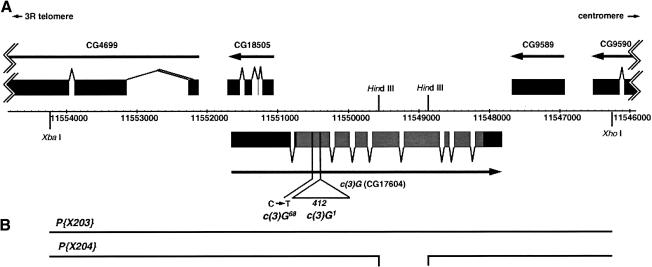
Figure 1.
The c(3)G (CG17604) locus. (A) The genomic structure of c(3)G (CG17604), based on the GM04379 and LD07655 cDNAs (Rubin et al. 2000); surrounding genes are shown as boxes representing exons. Genomic DNA coordinates in base pairs are from the Gadfly Drosophila genome annotation database (FlyBase 1999). The c(3)G (CG17604) open reading frame is shown in gray. c(3)G mutations are represented below the intron/exon structure of CG17604. CG18505 is encoded on the opposite strand as c(3)G, largely within the c(3)G 5′ untranslated region. (B) Rescue constructs P{X203} and P{X204} include genomic DNA from between the XbaI and XhoI sites indicated in A. P{X204} bears a deletion between the HindIII sites (shown in A) within the CG17604 gene.
Table 1.
Recombination and interference analysis
|
|
Maternal genotype
|
|||
|---|---|---|---|---|
| +a
|
c(3)G68
|
P{X203}/+; c(3)G68
|
P{X204}/+; c(3)G68
|
|
| n | 3915 | 3749 | 2657 | 1609 |
| Map length (standard error) | ||||
| 1 | 15.04 (0.57) | 0.027 (0.027) | 16.41 (0.72) | 1.43 (0.30) |
| 2 | 19.28 (0.63) | 0 | 22.36 (0.81) | 6.59 (0.62) |
| 3 | 19.82 (0.64) | 0 | 24.16 (0.83) | 15.10 (0.89) |
| 4 | 10.34 (0.49) | 0.027 (0.027) | 12.23 (0.64) | 16.16 (0.92) |
| Total | 64.48 | 0.053b | 75.16 | 39.28 |
| Interference (standard error) | ||||
| 1,2 | 0.798 (0.042) | n/a | 0.600 (0.064) | 1c |
| 2,3 | 0.519 (0.056) | n/a | 0.464 (0.061) | 0.500 (0.180) |
| 3,4 | 0.776 (0.052) | n/a | 0.452 (0.080) | 0.516 (0.111) |
| 1,3 | 0.024 (0.092) | n/a | −0.120 (0.105) | −1.015 (0.859) |
| 2,4 | 0.193 (0.097) | n/a | −0.032 (0.111) | 0.066 (0.243) |
Exchange was measured on the X chromosome for the maternal genotypes indicated; region 1, sc-cv; region 2, cv-v; region 3, v-f; region 4, f-y+.
A few recombinant chromosomes were recovered among the progeny of c(3)G68 homozygous females. This was also observed during previous studies (Hall 1972). These exchanges may either reflect a very low level of meiotic recombination in c(3)G68 females, or could be the result of mitotic recombination.
No double exchanges for regions 1 and 2 were observed in this experiment.
n/a, not applicable.
Based on sequence analysis (Adams et al. 2000), three complete and two partial transcription units were predicted to exist within the 8 kb covered by P{X203} (Fig. 1). The intact genes included a gene predicted to encode a uroporphyrin-III-C-methyltransferase protein (CG9589), a gene containing homology to acylphosphatase (CG18505), and a transcription unit predicted to encode a protein with a large coiled-coil region (CG17604). P{X203} also carries the 3′ end of a gene (CG9590) without its 5′ flanking promoter region and a gene that is truncated early in the transcript (CG4699; FlyBase 1999).
CG17604 was identified as a candidate for c(3)G because the 744-amino-acid conceptual translation product was predicted to be similar in structure to SC proteins Zip1 in yeast (Sym et al. 1993; Dong and Roeder 2000) and SCP1/Syn1 from mammals (Meuwissen et al. 1992; Dobson et al. 1994; Liu et al. 1996; Schmekel et al. 1996). The COILS v.2.1 secondary structure prediction program (Lupas et al. 1991) was used to predict the presence of coiled-coil segments within the CG17604 protein. The central portion (amino acids 158–646) of the protein was expected to form four stretches of coiled-coil structure, with the ends of the protein possibly assuming a random coil or globular structure. Although substantial amino acid sequence similarity did not exist between this gene and either Zip1 or SCP1, this predicted structure was very similar to these SC proteins. Amino acid similarity to other species would not necessarily be expected for SC proteins, because Zip1 and SCP1 show little sequence similarity to each other, even though they apparently play analogous roles in yeast and mammals, respectively (Liu et al. 1996; Schmekel et al. 1996; Dong and Roeder 2000). Interestingly, the total length of coiled coils in the CG17604 protein is predicted to be 67.86 nm, which is very similar to the total length of coiled coils predicted for Zip1 by the same methods, 68.16 nm. However, the length of coiled coils predicted for murine SCP1 is much higher, 87.76 nm. Therefore, although structurally similar to both Zip1 and SCP1, the CG17604 protein may be closer in size to Zip1.
To be certain that CG17604 corresponds to the c(3)G gene, the mutations in two c(3)G alleles, c(3)G1 and c(3)G68, were identified (Fig. 1). The original allele, c(3)G1 (also called c(3)G17 by Hall 1972), was discovered by Gowen as a spontaneous mutant in 1917 (Gowen and Gowen 1922). Southern blotting of digested c(3)G1 genomic DNA showed evidence for an insertional mutation (data not shown). Sequencing of the 5′ and 3′ ends of the insertion revealed 99% identity to the 412 retrotransposon long terminal repeat (Will et al. 1981). The insertion of the 412 element into codon 115 of the gene results in an amino acid change from glutamine to leucine followed by a stop codon, thus ending the open reading frame after 115 amino acids.
Sequencing of the CG17604 gene in DNA from c(3)G68 homozygotes revealed a C → T transition in codon 78. This changed a glutamine residue to a stop codon, which would truncate the protein after 77 amino acids. To ensure that this was not a polymorphism, this portion of the gene from seven independent c(3)G+ stocks was amplified and sequenced. Each c(3)G+ line matched the wild-type sequence, whereas the C → T mutation was found in three different c(3)G68 stocks. The identification of mutations in this gene in c(3)G mutant alleles confirms that the CG17604 gene is c(3)G.
Localization of C(3)G protein in female meiosis
A bacterially expressed fusion protein containing amino acids 565–743 of the predicted C(3)G protein was used to generate guinea pig antiserum. This antibody detected a protein of the expected size (85 kD) on Western blots of wild-type ovaries that was not present on Westerns of c(3)G68 ovaries (data not shown). Indirect immunofluorescence was performed on Drosophila ovaries to characterize the localization of C(3)G. The protein was detected in a subset of cells located within the germarium and early egg chambers (stages 1–6). The anti-C(3)G immunofluorescence was thread-like in appearance and colocalized with nuclear DNA (Fig. 2A–C). This pattern of staining is similar to the staining pattern reported by Huynh and St. Johnston (2000) for a cross-reacting antibody within antiserum raised against the inscuteable (insc) gene product. Although the antigen detected by the anti-Insc serum has not been identified, it is thought to be a component of the SC (Huynh and St. Johnston 2000).
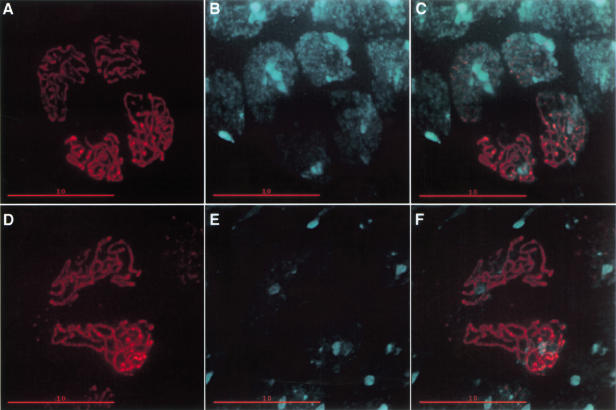
Figure 2.
C(3)G protein colocalizes in a thread-like pattern associated with nuclear DNA. (A) Anti-C(3)G immunofluorescence (red) in an early germ-line cyst (germarium region 2a), showing a thread-like pattern visible in four nuclei. The lower two adjacent nuclei are more intensely stained and are probably the two pro-oocytes. (B) Nuclear DNA in the cyst shown in A visualized by DAPI staining (cyan). (C) Merged image of A and B, showing colocalization of C(3)G (red) with the DNA (cyan). (D) C(3)G–eGFP (pseudocolored red) localizes in a thread-like pattern similar to the pattern visualized by anti-C(3)G immunofluorescence. Two brightly fluorescing nuclei are visible in the center of the frame, and two dimly fluorescing nuclei are positioned at the bottom and upper right edges of the frame. (E) Nuclear DNA in the cyst shown in D visualized by DAPI staining (cyan). (F) Merged image of D and E, showing colocalization of C(3)G–eGFP (red) with the DNA (cyan). Bars, 10 μm.
To confirm that the immunostaining pattern observed using the guinea pig antiserum corresponded to the c(3)G gene product, a construct for expression of C(3)G protein tagged with enhanced green fluorescent protein (eGFP) from the c(3)G promoter was introduced into Drosophila as a transgene. Microscopic analysis of ovaries expressing C(3)G–eGFP showed that the eGFP-tagged protein localized to nuclei in a thread-like pattern similar to that observed for anti-C(3)G immunofluorescence (Fig. 2D–F). Immunostaining of C(3)G–eGFP-expressing ovaries using anti-C(3)G showed that C(3)G–eGFP localization matched the anti-C(3)G immunofluorescence pattern (data not shown).
C(3)G is associated with paired chromosomes
Comparison with DAPI staining shows that chromatin is associated with the linear anti-C(3)G immunofluorescence pattern. Often, two masses of chromatin are observed along the thread-like anti-C(3)G signals (Fig. 3A–C). To further investigate the relationship between meiotic bivalents and C(3)G protein, fluorescence in situ hybridization (FISH) was used to identify a region of the X chromosome in ovarioles that were immunostained for C(3)G. The probe, a P1 clone from polytene region 5E, was visible as two paired spots in nuclei containing C(3)G. In these cases, the pair of FISH signals is associated with, and positioned on opposite sides of, a long segment of anti-C(3)G immunofluorescence (Fig. 3D,E). This result shows that the structure identified by anti-C(3)G lies between two paired homologous chromosomes.
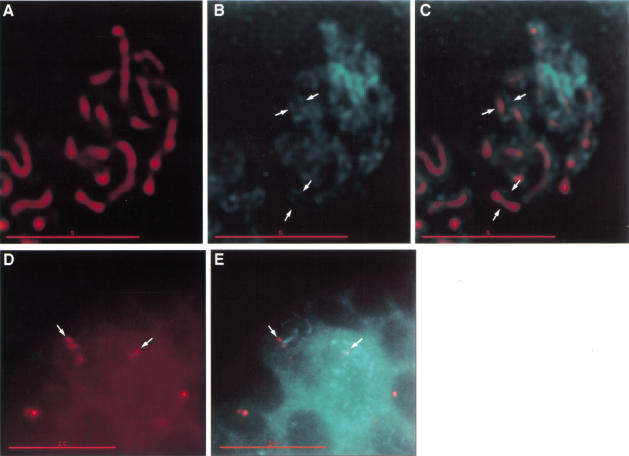
Figure 3.
C(3)G protein localizes between paired homologous chromosomes. (A) Deconvolved optical section through a pro-oocyte nucleus showing C(3)G localization (red). (B) DAPI image showing DNA (cyan) of the nucleus shown in A. Much of the chromatin is visible as paired linear tracks of staining (arrows). (C) Merged image of A and B, showing the association of the thread-like localization of C(3)G (red) to the paired linear tracks of DNA (cyan). In parts of this section, C(3)G can be observed between the tracks of DNA (arrows). Twisting of the chromosomes and/or position of the chromosomes within the section prevents the visualization of this arrangement of C(3)G and chromatin throughout the nucleus. A–C, bar, 5 μm. (D) Deconvolved optical section showing fluorescence in situ hybridization (FISH) of an X-chromosome probe (red) to an immunostained germarium. Pairs of FISH signals, representing paired homologous chromosomes, are visible in two cells (arrows). (E) Anti-C(3)G immunofluorescence (cyan) superimposed with the FISH signals (red) from D. The paired FISH signals are arranged on either side of the thread-like C(3)G immunofluorescence signal (arrows). D–E, bar, 10 μm.
Three-dimensional deconvolution microscopy (Agard et al. 1989) was used to reconstruct anti-C(3)G immunofluorescence images at high magnification (Fig. 4A). This allowed us to compare the thread-like immunofluorescence signals with reconstructions of Drosophila SC from serial sectioning EM (Carpenter 1975a,b, 1979a,b). The tracing of immunofluorescence signals in oocyte nuclei revealed five long continuous threads of signal. These match the five long stretches of SC observed by EM, and likely correspond to bivalents composed of the five major chromosome arms of Drosophila, XL, 2L, 2R, 3L, and 3R. One end of each of these putative chromosome arms was usually gathered near a DAPI-bright area of the nucleus. This area is almost certainly the heterochromatic chromocenter, described by Carpenter (1975a) for meiotic nuclei. In this area, the anti-C(3)G signals often converge, and determination of both arms of a single autosome is usually not possible. In addition, 1–3 shorter segments of anti-C(3)G signal were also observed in most pro-oocyte nuclei. One of these shorter segments is likely to correspond to chromosome 4. The somewhat variable number of shorter segments may have been derived from the presence of gaps or breaks in the thread-like signals, or the shorter segments could correspond to structures such as the right arm of the X chromosome, which may not always be recognizable in every nucleus.
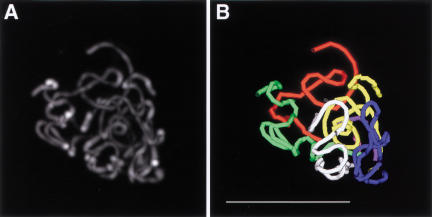
Figure 4.
Computer model of C(3)G localization. (A) Maximum intensity projection of all anti-C(3)G immunofluorescence signals from one nucleus (lower left nucleus from Fig. 2A). (B) Wire frame model of the anti-C(3)G immunofluorescence signals shown in A. The contiguous thread-like immunofluorescence signals were modeled in three dimensions using the deconvolved image stack used to create Figure 5A. Each contiguous segment of the model is shown in a different color. Bar, 5 μm.
The lengths of anti-C(3)G signals in five pro-oocytes are listed in Table 2. The total length of the anti-C(3)G signals varies between cells, as was observed by Carpenter (1979b) for SC length, but the relative lengths of the five putative bivalent arms are roughly similar and proportional to SC lengths measured previously. However, the lengths of anti-C(3)G signals are greater than the SC lengths observed previously by EM. The longest total length of SC described by Carpenter (1979b) was 73.5 μm, but the total lengths of anti-C(3)G signals exceed total SC lengths by >30 μm. The anti-C(3)G signal lengths were measured from three-dimensional computer models of the immunofluorescence patterns. This differs from the method of photographic measurement and calculation used to determine SC lengths from the EM data (Carpenter 1975a, 1979b), and therefore may account for the differences between SC and anti-C(3)G staining lengths. The striking similarity of the localization patterns of C(3)G and SC suggests that C(3)G is associated with the SC.
Table 2.
Measurements of linear segmentsa of C(3)G localization from three-dimensional models of anti-C(3)G immunofluorescence in w1118 pro-oocytes
| Pro-oocyte
|
Lengths (μm) of linear segments of C(3)G localization
|
Total
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 30.61 | 25.16 | 24.95 | 23.00 | 20.28 | 2.86 | 1.92 | 128.78 | |
| 2 | 28.20 | 24.48 | 24.11 | 23.32 | 21.96 | 3.31 | 125.36 | ||
| 3 | 22.02 | 21.93 | 21.34 | 20.83 | 19.68 | 3.61 | 2.21 | 0.94 | 112.56 |
| 4 | 24.41 | 22.26 | 19.57 | 19.56 | 17.65 | 4.44 | 1.04 | 1.02 | 109.95 |
| 5 | 25.76 | 23.82 | 19.29 | 17.84 | 14.86 | 1.68 | 1.51 | 0.64 | 105.40 |
| Mean | 116.41 | ||||||||
The identities of the chromosomes could not be determined.
Timing of C(3)G localization to chromosomes
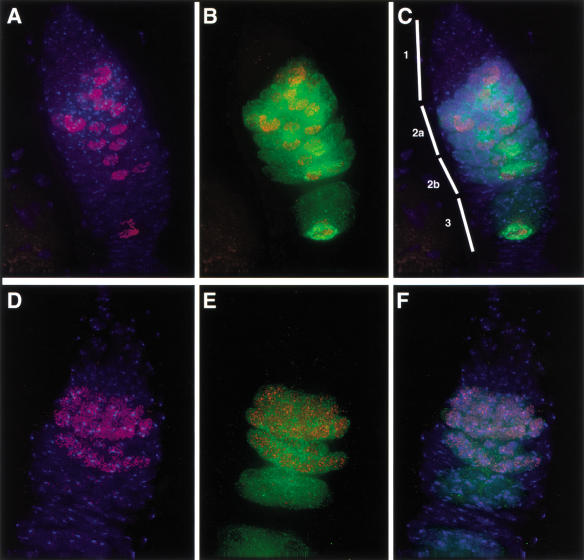
To further characterize the cells in which C(3)G appears, the localization of C(3)G was compared with that of the Drosophila Orb protein. Orb is an RNA-binding protein expressed in a complex pattern in germ-line cells during oogenesis. The protein is first detected in the cytoplasm of all cells in 16-cell cysts entering region 2a of the germarium, and accumulates in the cytoplasm of the oocyte in older cysts (Lantz et al. 1994). Coimmunostaining for C(3)G and Orb protein showed that C(3)G first appears at about the same time that Orb protein becomes visible in the cytoplasm of germ-line cyst cells, but before Orb is concentrated in the oocyte (Fig. 5A–C). Although Orb can be seen in the cytoplasm of all 16 cells in early 16-cell germ-line cysts (Lantz et al. 1994), C(3)G staining is usually present in ∼4 of the cells. In Drosophila, SC formation normally occurs in four germ-line cyst cells, the two four-ring canal cells and the two three-ring canal cells (Carpenter 1979b). Usually, two adjacent cells stain more brightly with anti-C(3)G than the other two cells (Fig. 2A). These less brightly staining cells are always adjacent to the brightly staining cells, but not necessarily adjacent to each other. The brightly staining nuclei probably belong to the two pro-oocytes, which share a ring canal, whereas the other two cells are most likely the two cells that possess three ring canals. C(3)G therefore marks the pro-oocytes earlier than Orb, in region 2a.
Figure 5.
Comparison of C(3)G and Orb localization in wild-type and egl germaria. (A) Maximum intensity projection of a deconvolved image stack containing an entire w1118 germarium. Anti-C(3)G immunofluorescence (red) is visible in a subset of nuclei (visualized by DAPI staining, blue). (B) Comparison of C(3)G (red) and Orb localization (green) in the germarium shown in A. C(3)G is expressed in germ-line cells beginning at the time when Orb is first detected in the cytoplasm of region 2a cysts. In early cysts, C(3)G is detected in ∼4 cells per cyst. By region 2b, only two cells retain C(3)G, one of which is the oocyte, as determined by the accumulation of Orb protein. C(3)G is then lost from the other pro-oocyte by the time cysts leave the germarium. (C) Germarium from A, showing C(3)G (red), Orb (green), and DNA (blue). Regions 1, 2a, 2b, and 3 of the germarium are indicated at left. (D) Maximum intensity projection of a deconvolved image stack containing an entire germarium from a female homozygous for egl1. Anti-C(3)G immunofluorescence (red) is visible in numerous nuclei (visualized by DAPI staining, blue) in region 2a and early region 2b, in contrast to wild type. (E) Comparison of C(3)G (red) and Orb localization (green) in the germarium shown in D. Orb protein is expressed and is present in the cytoplasm of germ-line cells starting in region 2a, but Orb fails to accumulate in the oocyte cytoplasm, because no oocyte is determined in egl mutant ovaries. C(3)G is present in the nuclei of all 16 cells of the germ-line cyst starting in region 2a, at the same time that Orb becomes detectable. By late region 2b, all 16 cells lose anti-C(3)G staining. (F) Germarium from D, showing C(3)G (red), Orb (green), and DNA (blue).
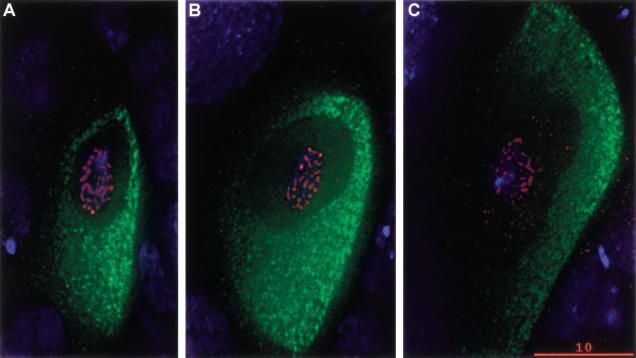
By region 2b, C(3)G staining is restricted to two adjacent nuclei in each cyst. One of these can be identified as the oocyte on the basis of cytoplasmic Orb accumulation (Fig. 5C). The C(3)G protein is lost from the second pro-oocyte as the cyst enters the vitellarium. C(3)G association with chromatin in the oocyte nucleus persists in the early vitellarium stages. As the egg chambers mature, the thread-like C(3)G staining pattern gradually breaks down (Fig. 6). Much of the chromosomal localization of C(3)G is lost by stage 6 of oogenesis. While this occurs, hazy extrachromosomal anti-C(3)G immunofluorescence signal becomes visible within the oocyte nucleus and grows in intensity as the chambers age. This may represent C(3)G protein that has disassembled from the chromosomes. These data indicate that the C(3)G protein localizes to oocyte chromosomes during early meiotic prophase and is removed during karyosome formation, when meiotic recombination is thought to be complete.
Figure 6.
Localization of C(3)G in vitellarial cysts from w1118 females. (A) Deconvolved optical section showing C(3)G (red) in a stage 3 oocyte nucleus. The Orb protein (green) is accumulated in the oocyte cytoplasm. C(3)G remains localized in thread-like patterns associated with DNA (blue) at this stage. (B) Deconvolved optical section showing C(3)G (red) in a stage 4 oocyte nucleus. Orb protein and DNA are stained as in A. Thread-like C(3)G colocalization with DNA is visible, but is beginning to dissipate. (C) Deconvolved optical section showing C(3)G (red) in a stage 6 oocyte nucleus. Orb protein and DNA are stained as in A. C(3)G localization to chromatin is decreased, and hazy extrachromosomal anti-C(3)G immunofluorescence is starting to become visible in the nucleus. In A–C, the posterior of each egg chamber is to the right. Bar, 10 μm.
Chromosomal localization of C(3)G is not observed in ovaries from homozygous c(3)G1 or c(3)G68 females. Although Orb protein is expressed and localized normally, no nuclear C(3)G is detected (data not shown). This would be expected, for both alleles are expected to encode truncated proteins. Because the antibody was raised against a portion of the C-terminal end of C(3)G, it is possible that a small truncated protein is produced, but was undetectable using this antibody. The lack of C(3)G in meiotic nuclei correlates with the lack of SC in c(3)G mutants (Meyer 1964; Smith and King 1968; Rasmussen 1975), and is consistent with a role for C(3)G in SC structure.
C(3)G transiently assembles in all 16 cells in egalitarian germ-line cysts
If C(3)G forms a component of the SC, localization of this protein should be altered in mutants that affect the assembly of SC. In females homozygous for the egalitarian (egl) mutant, all 16 cells of the germ-line cyst enter meiosis and form SC, as determined by EM (Carpenter 1994). However, an oocyte is not selected, and all 16 cells eventually take on a nurse cell fate. Anti-C(3)G staining of egl1 mutant ovaries reveals nuclear C(3)G staining in all 16 cells of 1–3 cysts per germarium (Fig. 5D–F). The staining is thread-like and essentially identical to the staining observed in wild-type pro-oocyte nuclei. In region 2b of the germarium, the staining disappears from all 16 cells simultaneously, and by region 3 no cells contain C(3)G. These results match the observations of SC assembly in egl mutants made by Carpenter (1994), who reported that the SC disassembles from all 16 cells and no oocyte is selected, and are comparable to the results of immunostaining egl ovaries using the anti-Insc serum, which apparently detects an SC component (Huynh and St. Johnston 2000). Similarly, Orb protein is expressed in this mutant background, but fails to accumulate in a single cell.
Synapsis defects caused by a mutant C(3)G protein
To rule out rescue by the other genes on P{X203}, a second construct P{X204} was made by deleting a HindIII fragment from the P{X203} construct, thus creating a mutated version of the CG17604 gene (Fig. 1B). This mutant is expected to produce a protein in which amino acids 340–552 are removed in-frame from within the coiled-coil region. This internally deleted C(3)G protein is analogous to proteins encoded by the mutant zip1 constructs used by Tung and Roeder (1998) to analyze Zip1 function. In that study, yeast expressing mutant Zip1 proteins that bear deletions in the central coiled-coil region either underwent synapsis with a narrowing of the SC (Zip1-M2p), or failed to synapse even though the mutant protein localized to the chromosomal axes (Zip1-M1p).
As shown in Table 1, the exchange-deficient phenotype of c(3)G was only partially rescued by the P{X204} construct. The X-chromosome map distance was reduced to 39.28, or ∼60% of wild type. This result suggests that the CG17604 coiled-coil gene is responsible for the c(3)G phenotype, because the remaining transcription units contained on the transgene were unaffected by the deletion mutation. Had one of the other genes been responsible for c(3)G, P{X204} should have fully rescued the phenotype, as did P{X203}.
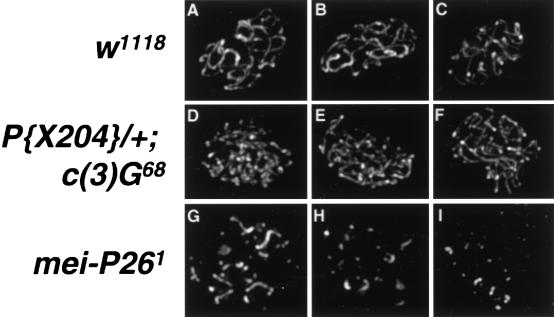
Localization of the mutant C(3)G protein was assessed by anti-C(3)G immunofluorescence in P{X204}/+; c(3)G68 females. In ovaries of this genotype, the mutant C(3)G protein encoded by P{X204} should be the only protein detected by anti-C(3)G. The mutant C(3)G protein was found to be localized in linear arrays, similar to wild type (Fig. 7A–C). However, the number of long linear arrays is increased, and an increased number of shorter lines is visible (Fig. 7D–F). These results may be explained by the localization of the protein to unsynapsed or partially synapsed chromosomes, similar to the Zip1-M1p protein in yeast (Tung and Roeder 1998), or noncontiguous localization of C(3)G along chromosomes. Most significantly, this indicates that disruption of the SC using a mutant SC protein can lead to a meiotic exchange defect.
Figure 7.
Localization of C(3)G in recombination-defective mutants. (A–C) Maximum intensity projections of anti-C(3)G immunofluorescence in three pro-oocyte nuclei from w1118 females. (D–F) Maximum intensity projections of anti-C(3)G immunofluorescence in three pro-oocyte nuclei from P{X204}/+; c(3)G68/c(3)G68 females. The mutant form of C(3)G encoded by the P{X204} construct localizes to chromosomes (data not shown), but forms an increased number of stained segments suggestive of incomplete synapsis. (G–I) Maximum intensity projections of anti-C(3)G immunofluorescence in three pro-oocyte nuclei from mei-P261 homozygous females. Pro-oocytes in mei-P261 germaria were identified by cytoplasmic accumulation of Orb protein (data not shown). C(3)G forms spots and short segments of linear localization, indicating an extensive lack of normal SC.
Localization of C(3)G in a second meiotic mutant mei-P26
The correlation of disruptions in SC formation with abnormal exchange frequencies suggested that other meiotic mutants might also have defects in SC formation. A large number of mutants in Drosophila form a class of meiotic mutants that have come to be known as precondition mutants. These are defective in specifying the wild-type frequency and distribution of meiotic exchanges (Baker and Hall 1976). It was postulated that certain preconditions, such as pairing and synapsis, must occur in order for any given chromosomal interval to undergo exchange. Because these mutants affect the positions of exchanges in addition to the exchange frequency, they were thought to disrupt these preconditions, rather than affect the recombination machinery itself (Carpenter and Sandler 1974). The precondition mutants generally share a phenotype that results in a decrease in total exchange frequency, with the distinction of having the frequency of exchange in chromosomal intervals distal to the centromere most greatly decreased, whereas exchange in proximal intervals remains at wild-type or even increased levels. Interestingly, the exchange observed for the P{X204} mutant c(3)G construct has this altered distribution, which is known as a polar exchange distribution. Thus, the mutant C(3)G protein encoded by P{X204} appears to disrupt synapsis, which is thought to be a precondition for exchange, and produces a polar exchange defect.
To investigate the hypothesis that at least some precondition mutants disrupt synapsis, we examined C(3)G localization in mei-P261, a mutant that produces this phenotype. Severe mutations in mei-P26 cause defects in gametogenesis in both sexes that lead to sterility (Page et al. 2000). Females homozygous for the hypomorphic allele mei-P261 are fertile but show a polar meiotic exchange phenotype (Sekelsky et al. 1999; Page et al. 2000). In ovaries of this genotype, C(3)G fails to localize normally on meiotic chromosomes (Fig. 7G–I). Wild-type germarial cysts in which Orb protein has begun accumulating in the oocyte cytoplasm show a thread-like pattern of C(3)G localization. In contrast, C(3)G localization in mei-P261 oocytes that are accumulating Orb ranges from weak, dotty C(3)G staining to short, linear tracks of C(3)G. This indicates that the failure of mei-P261 ovaries to properly assemble the SC may underlie the defects in exchange frequency.
Complete SC is not required for interference in Drosophila
Exchanges are not distributed along the chromosomes at random. Instead, exchanges usually occur on each chromosome, and two exchanges are rarely seen in proximity to each other. The phenomenon known as interference is thought to represent the mechanism by which this is achieved (Muller 1916). Zip1 appears to be essential for crossover interference in yeast (Sym and Roeder 1994; Storlazzi et al. 1996). In zip1 null mutants, exchange decreases to 60%–70% of wild type, but the residual exchanges no longer show interference. The connection between Zip1 and interference was also shown by a series of mutant Zip1 proteins, which showed that mutants that allow extensive synapsis generally showed interference, but those that failed to undergo or complete synapsis showed little or no interference (Tung and Roeder 1998).
In females that express a mutant version of C(3)G, interference is slightly decreased but not eliminated (Table 1). Similarly, calculations from previously published data for exchange on chromosome 2 in mei-P261 females (Table 3 in Page et al. 2000) indicate only slight decreases in interference. In mei-P261 females, interference is reduced to 0.589 for the adjacent net–dp and dp–b intervals, compared with 0.672 for wild type, and reduced to 0.350 for dp–b and b–pr, compared with 0.568 in wild type. Based on anti-C(3)G immunofluorescence, SC does not form normally in these mutants. Therefore, full-length SC does not appear to be required for mediating interference. SC is not sufficient for establishing interference either, because meiotic mutants mei-41 and mei-218 build apparently normal SC, but have alterations in interference (Baker and Carpenter 1972; Carpenter 1979a).
Discussion
C(3)G is a coiled-coil SC protein similar to Zip1 and SCP1
The lack of known SC components has until now precluded the detailed investigation of meiotic synapsis in Drosophila. Visualization of Drosophila SC has been possible by EM analysis of serially sectioned germaria (Carpenter 1975a,b, 1979a,b). The identification of an SC protein allows the visualization of the SC at the level of the light microscope and the comparison of SC formation with localization of specific proteins involved in synapsis and recombination. We have shown by phenotypic rescue, mutation analysis, and by protein localization that the c(3)G gene encodes a Drosophila SC protein. C(3)G is a 744-amino-acid protein predicted to contain a central domain rich in coiled coils, flanked by globular domains at the N and C termini. This protein structure is characteristic of proteins comprising the transverse filaments (TFs) of the SC (Meuwissen et al. 1992; Sym et al. 1993; Dobson et al. 1994). In both Zip1 and SCP1/Syn1, the proteins appear to form parallel dimers oriented with the N terminus in the center of the SC, and the C terminus adjacent to the lateral element (Dobson et al. 1994; Liu et al. 1996; Schmekel et al. 1996; Dong and Roeder 2000). The N termini of the dimers interdigitate across the width of the SC, forming the TFs.
We hypothesize that C(3)G forms a similar structure in the SC for several reasons. First, the phenotype of a c(3)G mutant includes the absence of SC (Meyer 1964; Smith and King 1968; Rasmussen 1975). Second, the C(3)G protein appears to localize along synapsed bivalents in wild type, specifically in the cells in which SC is observed using EM (Carpenter 1975a, 1979b). Third, thread-like C(3)G localization is observed in all 16 cells per cyst in egl ovaries, matching previous observations by EM (Carpenter 1994). Fourth, a mutated version of the protein containing an in-frame deletion fails to localize normally, suggesting defects in synapsis similar to those observed for a deleted version of Zip1 (Tung and Roeder 1998).
The relationship between synapsis and recombination in Drosophila females
The lack of both exchange and the SC in c(3)G mutants has long bolstered the classical view of meiosis, in which homolog pairing and synapsis precede, and are required for, recombination (Hawley and Arbel 1993). Our results suggest that c(3)G is an essential component of the SC and that it is required for both synapsis and exchange in Drosophila females. Several lines of evidence suggest that c(3)G is necessary for recombination: (1) Essentially no meiotic exchange is observed in c(3)G mutants (Gowen and Gowen 1922; Gowen 1933; Hall 1972; this study). (2) c(3)G homozygotes do not undergo intragenic exchange or gene conversion (Carlson 1972). (3) Homozygosity for c(3)G rescues the egg polarity defects in the okra mutant, which suggests that double strand breaks are not made in c(3)G, and thus do not persist in okra; c(3)G double mutants (Ghabrial and Schüpbach 1999). (4) C(3)G protein localizes to chromosomes prior to Mei-P22 localization (H. Liu, J.K. Jang, N. Kato, and K.S. McKim, pers. comm.). (5) Mei-P22 fails to localize to chromosomes in the absence of C(3)G (H. Liu, J.K. Jang, N. Kato, and K.S. McKim, pers. comm.).
Detailed studies of meiosis in yeast have shown that DSB formation occurs before synapsis and is necessary for synapsis to occur (Roeder 1997). Yeast may use the initiation of recombination as a strategy to align homologous chromosomes, with the SC a means to stabilize the alignment. This process may be reordered in Drosophila, such that the pairing and intimate synapsis of homologs are needed before recombination can initiate. Evidence from both flies and C. elegans suggests that synapsis occurs in the absence of DSB formation (Dernburg et al. 1998; McKim et al. 1998).
The precondition meiotic exchange phenotype can result from defective synapsis
Three types of exchange-defective meiotic mutants have been characterized in Drosophila females: (1) those that lack SC and completely lack exchange, such as c(3)G (Gowen and Gowen 1922); (2) those that form SC but lack exchange, such as mei-W68 and mei-P22 (McKim et al. 1998); and (3) those that form SC but have an altered frequency or distribution of exchanges, such as mei-9 and mei-218 (Sekelsky et al. 1995; McKim et al. 1996). This third class of mutants can be further subdivided into three subclasses: those that only affect the frequency of exchange, but not the distribution of exchange events (e.g., mei-9); those that only affect the distribution of exchange, and not the frequency (e.g., mei-352); and those that affect both the frequency and distribution of exchanges (e.g., mei-218, mei-P26). This last subclass has often been called the precondition class of mutants. The residual exchange in these mutants takes on a polar distribution, such that exchange is most severely depressed in the distal intervals of a chromosome arm, but proximal intervals are less severely affected (Carpenter and Sandler 1974).
Using a transgenic construct encoding a mutant C(3)G protein, we have shown that defects in SC formation can result in an exchange-defective phenotype. Moreover, analysis of exchange in females carrying P{X204} reveals an altered distribution of exchanges similar to that of precondition meiotic mutants. The mutant protein encoded by this construct failed to localize normally, suggesting that synapsis is defective, and that these defects result in the precondition phenotype. This appears to be the case for at least one such mutant, mei-P261, which acts as a classical precondition mutant (Page et al. 2000). The abnormal localization of C(3)G in mei-P261 suggests that the exchange defects may be due to a lack of synapsis.
The precondition phenotype may not always occur because of synapsis defects, however. Studies of SC formation by EM in the mutant mei-218 showed normal SC formation but abnormalities in the numbers of recombination nodules (Carpenter 1979a). Exchange in hypomorphic mutants of mei-W68 and mei-P22 also follows a polar distribution (Baker et al. 1980; H. Liu, J.K. Jang, N. Kato, and K.S. McKim, pers. comm.). Therefore, the precondition phenotype may also result from deficiencies in recombination machinery components. Evidence for the polar exchange phenotype in apparent weak mutants of c(3)G (McKinley et al. 1979; this study), suggests that the SC is necessary for recombination in Drosophila.
Complete SC is not required for interference in Drosophila
Null mutations in the yeast Zip1 gene appear to eliminate interference, which can be measured in yeast because zip1 mutants have only a moderate effect on exchange frequencies (Sym and Roeder 1994; Storlazzi et al. 1996). These residual exchanges do not show interference in the absence of Zip1. A role for SC in mediating interference is further suggested by the lack of interference in S. pombe and Aspergillus, which do not make SC (Egel-Mitani et al. 1982; Bahler et al. 1993; Kohli and Bahler 1994; Munz 1994). The lack of any exchange in c(3)G homozygotes precludes the analysis of interference in c(3)G null mutants. However, in mutant genotypes in which the localization of C(3)G is abnormal, and exchange is reduced, the remaining exchanges show interference. This result differs from data presented by Tung and Roeder (1998) for Zip1 mutants, which showed a loss of interference in mutants that disrupted synapsis.
Interference might be controlled otherwise in Drosophila than in yeast. Recombination initiates efficiently in yeast mutants that lack SC (Sym and Roeder 1994; Storlazzi et al. 1996; Chua and Roeder 1998; Agarwal and Roeder 2000), and SC appears to assemble in the presence of recombination intermediates (Padmore et al. 1991; Schwacha and Kleckner 1994, 1995). Interference in yeast may require the presence of SC during the processing of recombination intermediates, or during their resolution. In contrast, SC formation in Drosophila may be necessary only for the initiation of recombination, and interference may be established through other means. Other factors unrelated to the SC must be involved in promoting interference, because full-length SC is not sufficient for interference to occur (Baker and Carpenter 1972; Carpenter 1979a). Alternatively, interference in Drosophila may also require the presence of SC, and the SC merely has an additional function required for recombination initiation.
Materials and methods
Genetic analyses
The genetic markers and chromosomes used in this study are described in Lindsley and Zimm (1992) and FlyBase (1999). Flies were reared on standard cornmeal–molasses–dextrose medium at 25°C. Exchange along the X chromosome was scored among the progeny of females of the genotype y sc cv v f · y+/X; 2/2; 3/3 crossed to y sc cv v f car/BSY males, where X, 2, and 3 represent chromosomes X, 2, and 3 with the genotypes indicated in Table 1. For most crosses, only female progeny were scored (the presence of w on the X chromosome in some experiments would obscure the scoring of v in male progeny). For any two chromosomal intervals A and B, interference (I) is calculated using the formula I = 1 − (d/ab), where a and b are the total frequencies of exchange in intervals A and B, respectively, and d is the observed frequency of double exchanges involving intervals A and B.
Coiled-coil analysis
The COILS v.2.1 secondary structure prediction program (Lupas et al. 1991), located at http://www.ch.embnet.org/software/COILS_form.html, was used to predict the presence of coiled-coil segments within the C(3)G (CG17604), yeast Zip1, and mouse SCP1 proteins. The COILS program was run using the MTIDK matrix and a 21-residue window. Amino acids with a score of 0.5 or higher were predicted to form a coiled-coil structure. Estimates of the physical length of coiled-coil segments were made by multiplying the number of amino acids in each segment predicted to assume a coiled-coil conformation by 0.1485 nm/residue, the mean axial rise per residue in a coiled coil (Steinert et al. 1993).
Germ-line transformation
The rescue construct P{X203} was built by inserting a 7963-bp genomic XhoI–XbaI restriction fragment into the vector pUAST (P. Szauter, pers. comm.). The genomic fragment encompasses the DNA from the XhoI site located at 11546254 to the XbaI site 11554217 on chromosome 3R, as described in the Gadfly Drosophila genome annotation database (FlyBase 1999). The P{X204} construct was made by removing a 702-bp HindIII fragment (corresponding to base pairs 11548874–11549575) from within the genomic fragment carried by P{X203} and re-ligating the plasmid. P{c(3)G–eGFP} contains a construct for the expression of a C(3)G–eGFP fusion protein from the c(3)G promoter. An NheI–NcoI fragment of genomic DNA (corresponding to base pairs 11552577–11550307) containing the c(3)G promoter, first exon, and part of the second exon was fused to an NcoI–XhoI restriction fragment derived from the c(3)G (CG17604) cDNA LD07655 and inserted in the transformation vector pCasPeR4In (obtained from J.J. Sekelsky, University of North Carolina, Chapel Hill). The eGFP coding region was amplified from the peGFP-N1 vector (Clontech) and inserted into an EcoRI site in the second to last codon of the c(3)G open reading frame, allowing a fusion of eGFP to the C-terminal end of the C(3)G protein.
The transformation constructs P{X203}, P{X204}, and P{c(3)G–eGFP}were introduced into Drosophila as described previously (Page et al. 2000). An insertion of P{X203} on the third chromosome was recovered. The construct was then jumped to the X chromosome using P transposase. The ability of this construct to rescue the c(3)G phenotype was tested by crossing y w P{X203}/y sc cv v f · y+; c(3)G68 females with y sc cv v f car/BSY males and scoring for exchange on the X chromosome. Similarly, an insertion of P{X204} on the second chromosome was used to test for rescue of the c(3)G phenotype. The P{c(3)G–eGFP} insertion used in this study is on chromosome 2.
Production of anti-C(3)G antibodies
The Escherichia coli expression vector pHEX was generated by inserting an XhoI–HindIII linker fragment encoding six histidine residues between the XhoI and HindIII sites of pGEX-KG. Nucleotides 4007–4547 of the c(3)G (CG17604) cDNA LD07655 (Rubin et al. 2000) were amplified using primers that added an XbaI site to the 5′ end and an XhoI site to the 3′ end. This fragment was digested with XbaI and XhoI and ligated between the XbaI and XhoI sites of pHEX to form pHEX–c(3)G. pHEX–c(3)G encodes a GST–C(3)G–6xHis fusion protein containing amino acids 565–743 of C(3)G tagged with GST at the N terminus and a 6xHis tag at the C terminus. The GST–C(3)G–6xHis fusion protein was expressed in E. coli BL21 cells and purified to be used as antigen to produce guinea pig polyclonal anti-C(3)G antibodies.
Ovary fixation and immunofluorescence
Females were aged 2–3 d in vials containing males and yeast paste. Ovaries were dissected in PBS and immediately fixed for 20 min in 200 μL of PBS containing 2% EM-grade formaldehyde (Ted Pella, Redding, CA) and 0.5% Nonidet P-40, plus 600 μL of heptane. Fixed ovaries were then rinsed three times in PBST (PBS plus 0.2% Tween-20), and washed three times for 5 min in PBST. Prior to immunostaining, the ovarioles were teased apart and blocked by incubating in PBST plus 1% BSA at room temperature for 1 h. Primary antibody incubation was at room temperature for 1 h, followed by three 20-min washes in PBST. This procedure was repeated for incubation with secondary antibodies, except that during one wash, PBS containing 0.5 μg/mL DAPI was substituted for the PBST.
Guinea pig anti-C(3)G serum was used at a dilution of 1:500. Hybridoma culture supernatants for mouse monoclonal anti-Orb antibodies 4H8 and 6H4 (Lantz et al. 1994) were used together at a dilution of 1:30. Secondary antibodies Alexa 488-conjugated anti-mouse IgG (Molecular Probes), Alexa 488-conjugated anti-guinea pig IgG (Molecular Probes), and Cy3-conjugated anti-guinea pig IgG (Jackson Immunoresearch) were each used at a dilution of 1:500.
In situ hybridization on immunostained tissue was performed essentially as described by Hunt et al. (1995), except that the tissue and probe were denatured together at 85°C for 5 min, and all the steps were performed with the tissue suspended in solution in microfuge tubes. The probe used was P1 clone DS00715 containing Drosophila genomic DNA from region 5E, which was directly labeled with Cy3-dCTP (Amersham) by nick translation.
Prior to viewing, immunostained tissue was embedded in polyacrylamide gel using a procedure based on methods published previously (Urata et al. 1995; Bass et al. 1997). Briefly, the immunostained ovarioles in PBST were placed on a coverslip within an area enclosed by dried nail polish or Scotch invisible tape, and as much PBST as possible was aspirated. Then ∼15–20 μL of polyacrylamide solution (10% polyacrylamide from a 30% 29:1 acrylamide:bis acrylamide solution, 0.68% ammonium persulfate, and 0.08% sodium sulfite in PBS) was added to the ovarioles and covered with a silanized coverslip. After 30 min, the silanized coverslip was removed, and the resulting polyacrylamide film was equilibrated with glycerol containing 2.5% 1,4-diazabicyclo[2.2.2]octane (DABCO) and mounted on a glass slide.
Microscopy and image processing
Images were collected using a DeltaVision reconstruction microscopy system (Applied Precision), consisting of an Olympus IX70 inverted fluorescence microscope equipped with an Olympus 60× oil, 1.4 NA PlanApo objective lens and high-resolution CCD camera. Image data were corrected and deconvolved using the softWoRx v. 2.5 software package (Applied Precision). Computer models of anti-C(3)G immunofluorescence were constructed from deconvolved image stacks by placing points connected by lines through contiguous regions of immunofluorescence using the SoftWoRx program 3D Model. Lengths of the linear objects thus created were calculated by the 3D Model computer program.
Acknowledgments
We thank Paul Szauter for generously providing vital mapping information, fly stocks, and clones. We are also grateful to Michael Paddy for the FISH images in Figure 3 and to both Michael Paddy and Heiner Matthies for helpful advice. We thank Adelaide Carpenter, Kim McKim, and members of the McKim and Hawley laboratories for helpful discussions. Thanks also go to Ed van Veen, Amy Tong, and Annette Lentz for technical assistance. The monoclonal antibodies 4H8 and 6H4 developed by Paul Schedl were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa Department of Biological Sciences. S.L.P. was supported by a fellowship from the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation (DRG-1476). This work was supported by a grant to R.S.H. from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rsh@stowers-institute.org; FAX (816) 926–2060.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.935001.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Agard DA, Hiraoka Y, Shaw P, Sedat JW. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. doi: 10.1016/s0091-679x(08)60986-3. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Roeder GS. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–255. doi: 10.1016/s0092-8674(00)00029-5. [DOI] [PubMed] [Google Scholar]
- Bahler J, Wyler T, Loidl J, Kohli J. Unusual nuclear structures in meiotic prophase of fission yeast: A cytological analysis. J Cell Biol. 1993;121:241–256. doi: 10.1083/jcb.121.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BS, Carpenter ATC. Genetic analysis of sex chromosomal meiotic mutants in Drosophila melanogaster. Genetics. 1972;71:255–286. doi: 10.1093/genetics/71.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BS, Hall JC. Meiotic mutants: Genetic control of meiotic recombination and chromosome segregation. In: Ashburner M, Novitski E, editors. The genetics and biology of Drosophila. 1a. New York, NY: Academic Press; 1976. pp. 351–434. [Google Scholar]
- Baker BS, Gatti M, Carpenter ATC, Pimpinelli S, Smith DA. Effects of recombination-deficient and repair-deficient loci on meiotic and mitotic chromosome behavior in Drosophila melanogaster. In: Generoso WM, Shelby MD, de Serres FJ, editors. DNA repair and mutagenesis in eukaryotes. New York, NY: Plenum Press; 1980. pp. 189–208. [DOI] [PubMed] [Google Scholar]
- Bass HW, Marshall WF, Sedat JW, Agard DA, Cande WZ. Telomeres cluster de novo before the initiation of synapsis: A three- dimensional spatial analysis of telomere positions before and during meiotic prophase. J Cell Biol. 1997;137:5–18. doi: 10.1083/jcb.137.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson PS. The effects of inversions and c(3)G mutation on intragenic recombination in Drosophila. Genet Res. 1972;19:129–132. doi: 10.1017/s001667230001435x. [DOI] [PubMed] [Google Scholar]
- Carpenter ATC. Electron microscopy of meiosis in Drosophila melanogaster females. I. Structure, arrangement, and temporal change of the synaptonemal complex in wild-type. Chromosoma. 1975a;51:157–182. doi: 10.1007/BF00319833. [DOI] [PubMed] [Google Scholar]
- ————— Electron microscopy of meiosis in Drosophila melanogaster females. II. The recombination nodule—A recombination-associated structure at pachytene? Proc Natl Acad Sci. 1975b;72:3186–3189. doi: 10.1073/pnas.72.8.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Recombination nodules and synaptonemal complex in recombination-defective females of Drosophila melanogaster. Chromosoma. 1979a;75:259–292. doi: 10.1007/BF00293472. [DOI] [PubMed] [Google Scholar]
- ————— Synaptonemal complex and recombination nodules in wild-type Drosophila melanogaster females. Genetics. 1979b;92:511–541. doi: 10.1093/genetics/92.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Egalitarian and the choice of cell fates in Drosophila melanogaster oogenesis. Ciba Found Symp. 1994;182:223–246. [PubMed] [Google Scholar]
- Carpenter ATC, Sandler L. On recombination-defective meiotic mutants in Drosophila melanogaster. Genetics. 1974;76:453–475. doi: 10.1093/genetics/76.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua PR, Roeder GS. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell. 1998;93:349–359. doi: 10.1016/s0092-8674(00)81164-2. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Dobson MJ, Pearlman RE, Karaiskakis A, Spyropoulos B, Moens PB. Synaptonemal complex proteins: Occurrence, epitope mapping and chromosome disjunction. J Cell Sci. 1994;107:2749–2760. doi: 10.1242/jcs.107.10.2749. [DOI] [PubMed] [Google Scholar]
- Dong H, Roeder GS. Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. J Cell Biol. 2000;148:417–426. doi: 10.1083/jcb.148.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel-Mitani M, Olson LW, Egel R. Meiosis in Aspergillus nidulans: Another example for lacking synaptonemal complexes in the absence of crossover interference. Hereditas. 1982;97:179–187. doi: 10.1111/j.1601-5223.1982.tb00761.x. [DOI] [PubMed] [Google Scholar]
- FlyBase. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 1999;27:85–88. doi: 10.1093/nar/27.1.85. http://flybase.bio.indiana.edu/ . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial A, Schüpbach T. Activation of a meiotic checkpoint regulates translation of Gurken during Drosophila oogenesis. Nat Cell Biol. 1999;1:354–357. doi: 10.1038/14046. [DOI] [PubMed] [Google Scholar]
- Gowen JW. Meiosis as a genetic character in Drosophila melanogaster. J Exp Zool. 1933;65:83–106. [Google Scholar]
- Gowen MS, Gowen JW. Complete linkage in Drosophila melanogaster. Amer Naturalist. 1922;61:286–288. [Google Scholar]
- Hall JC. Chromosome segregation influenced by two alleles of the meiotic mutant c(3)G in Drosophila melanogaster. Genetics. 1972;71:367–400. doi: 10.1093/genetics/71.3.367. [DOI] [PubMed] [Google Scholar]
- Hawley RS. Exchange and chromosomal segregation in eucaryotes. In: Kucherlapati R, Smith G, editors. Genetic recombination. Washington, DC.: American Society of Microbiology; 1988. pp. 497–527. [Google Scholar]
- Hawley RS, Arbel T. Yeast genetics and the fall of the classical view of meiosis. Cell. 1993;72:301–303. doi: 10.1016/0092-8674(93)90108-3. [DOI] [PubMed] [Google Scholar]
- Hunt P, LeMaire R, Embury P, Sheean L, Mroz K. Analysis of chromosome behavior in intact mammalian oocytes: Monitoring the segregation of a univalent chromosome during female meiosis. Hum Mol Genet. 1995;4:2007–2012. doi: 10.1093/hmg/4.11.2007. [DOI] [PubMed] [Google Scholar]
- Huynh J, St Johnston D. The role of BicD, egl, orb and the microtubules in the restriction of meiosis to the Drosophila oocyte. Development. 2000;127:2785–2794. doi: 10.1242/dev.127.13.2785. [DOI] [PubMed] [Google Scholar]
- Kohli J, Bahler J. Homologous recombination in fission yeast: Absence of crossover interference and synaptonemal complex. Experientia. 1994;50:295–306. doi: 10.1007/BF01924013. [DOI] [PubMed] [Google Scholar]
- Lantz V, Chang JS, Horabin JI, Bopp D, Schedl P. The Drosophila orb RNA binding protein is required for the formation of the egg chamber and establishment of polarity. Genes & Dev. 1994;8:598–613. doi: 10.1101/gad.8.5.598. [DOI] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG. The genome of Drosophila melanogaster. San Diego, CA: Academic Press; 1992. [Google Scholar]
- Liu JG, Yuan L, Brundell E, Bjorkroth B, Daneholt B, Hoog C. Localization of the N-terminus of SCP1 to the central element of the synaptonemal complex and evidence for direct interactions between the N-termini of SCP1 molecules organized head-to-head. Exp Cell Res. 1996;226:11–19. doi: 10.1006/excr.1996.0197. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- McKim KS, Dahmus JB, Hawley RS. Cloning of the Drosophila melanogaster meiotic recombination gene mei-218: A genetic and molecular analysis of interval 15E. Genetics. 1996;144:215–228. doi: 10.1093/genetics/144.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Green-Marroquin BL, Sekelsky JJ, Chin G, Steinberg C, Hawley RS. Meiotic synapsis in the absence of recombination. Science. 1998;279:876–878. doi: 10.1126/science.279.5352.876. [DOI] [PubMed] [Google Scholar]
- McKinley CM, Generoso EE, Grell RF. A “leaky” c(3)G mutant. Genetics. 1979;91 Suppl.:s79. [Google Scholar]
- Meuwissen RL, Offenberg HH, Dietrich AJ, Riesewijk A, van Iersel M, Heyting C. A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J. 1992;11:5091–5100. doi: 10.1002/j.1460-2075.1992.tb05616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer GF. A possible correlation between the submicroscopic structure of meiotic chromosomes and crossing over. In: Titlbach M, editor. Proceedings of the 3rd European regional conference on electron microscopy. Prague, Czechoslovakia: Publishing House of the Czechoslovak Academy of Sciences; 1964. pp. 461–462. [Google Scholar]
- Muller HJ. The mechanism of crossing-over II. Amer Naturalist. 1916;50:284–305. [Google Scholar]
- Munz P. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics. 1994;137:701–707. doi: 10.1093/genetics/137.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore R, Cao L, Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- Page SL, McKim KS, Deneen B, Van Hook TL, Hawley RS. Genetic studies of mei-P26 reveal a link between the processes that control germ cell proliferation in both sexes and those that control meiotic exchange in Drosophila. Genetics. 2000;155:1757–1772. doi: 10.1093/genetics/155.4.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SW. Ultrastructural studies of meiosis in males and females of the c(3)G17 mutant of Drosphila melanogaster Meigen. Compt Rend Trav Lab Carlsberg. 1975;40:163–173. [Google Scholar]
- Roeder GS. Meiotic chromosomes: It takes two to tango. Genes & Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmekel K, Meuwissen RL, Dietrich AJ, Vink AC, van Marle J, van Veen H, Heyting C. Organization of SCP1 protein molecules within synaptonemal complexes of the rat. Exp Cell Res. 1996;226:20–30. doi: 10.1006/excr.1996.0198. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- ————— Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- Sekelsky JJ, McKim KS, Chin GM, Hawley RS. The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics. 1995;141:619–627. doi: 10.1093/genetics/141.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky JJ, McKim KS, Messina L, French RL, Hurley WD, Arbel T, Chin GM, Deneen B, Force SJ, Hari KL, et al. Identification of novel Drosophila meiotic genes recovered in a P-element screen. Genetics. 1999;152:529–542. doi: 10.1093/genetics/152.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PA, King RC. Genetic control of synaptonemal complexes in Drosophila melanogaster. Genetics. 1968;60:335–351. doi: 10.1093/genetics/60.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert PM, Marekov LN, Fraser RD, Parry DA. Keratin intermediate filament structure. Crosslinking studies yield quantitative information on molecular dimensions and mechanism of assembly. J Mol Biol. 1993;230:436–452. doi: 10.1006/jmbi.1993.1161. [DOI] [PubMed] [Google Scholar]
- Storlazzi A, Xu L, Schwacha A, Kleckner N. Synaptonemal complex (SC) component Zip1 plays a role in meiotic recombination independent of SC polymerization along the chromosomes. Proc Natl Acad Sci. 1996;93:9043–9048. doi: 10.1073/pnas.93.17.9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M, Roeder GS. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 1994;79:283–292. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- Sym M, Engebrecht J, Roeder GS. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Tung KS, Roeder GS. Meiotic chromosome morphology and behavior in zip1 mutants of Saccharomyces cerevisiae. Genetics. 1998;149:817–832. doi: 10.1093/genetics/149.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urata Y, Parmelee SJ, Agard DA, Sedat JW. A three-dimensional structural dissection of Drosophila polytene chromosomes. J Cell Biol. 1995;131:279–295. doi: 10.1083/jcb.131.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein D, Rasmussen SW, Holm PB. The synaptonemal complex in genetic segregation. Annu Rev Genet. 1984;18:331–413. doi: 10.1146/annurev.ge.18.120184.001555. [DOI] [PubMed] [Google Scholar]
- Walker MY, Hawley RS. Hanging on to your homolog: The roles of pairing, synapsis and recombination in the maintenance of homolog adhesion. Chromosoma. 2000;109:3–9. doi: 10.1007/s004120050407. [DOI] [PubMed] [Google Scholar]
- Will BM, Bayev AA, Finnegan DJ. Nucleotide sequence of terminal repeats of 412 transposable elements of Drosophila melanogaster. A similarity to proviral long terminal repeats and its implications for the mechanism of transposition. J Mol Biol. 1981;153:897–915. doi: 10.1016/0022-2836(81)90458-7. [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]