Abstract
Nearly a decade since the completion of the first draft of the human genome, the biomedical community is positioned to usher in a new era of scientific inquiry that links fundamental biological insights with clinical knowledge. Accordingly, holistic approaches are needed to develop and assess hypotheses that incorporate genotypic, phenotypic, and environmental knowledge. This perspective presents translational bioinformatics as a discipline that builds on the successes of bioinformatics and health informatics for the study of complex diseases. The early successes of translational bioinformatics are indicative of the potential to achieve the promise of the Human Genome Project for gaining deeper insights to the genetic underpinnings of disease and progress toward the development of a new generation of therapies.
Keywords: Translational bioinformatics, systems medicine, systems biology, bioinformatics, biomedical informatics, knowledge representation, information retrieval, phylogenetics, modeling physiologic and disease processes, linking the genotype and phenotype, identifying genome and protein structure and function, visualization of data and knowledge, simulation of complex systems (at all levels: molecules to work groups to organizations), knowledge representations, uncertain reasoning and decision theory, languages, computational methods, statistical analysis of large datasets, advanced algorithms, discovery, text and data mining methods, natural-language processing, automated learning, ontologies
Introduction
The study of complex diseases requires the effective integration and analysis of disparate features that originate from genotypic, phenotypic, and environmental sources. In contrast to microscopic approaches that focus on detailed analyses of a single data type, a macroscopic approach offers a holistic view for exploring systems of relationships.1 Meaningful insights from a systems theory approach require the coalescence of many, often intractable, heterogeneous data types.2 Traditionally, biomedical informatics innovations have focused (‘microscopically’) on innovations constrained to particular domains3 (eg, clinical innovations in health informatics; biological innovations in bioinformatics). This has led to a perceived gulf between bioinformatics and health informatics, thus decreasing the potential impact of a ‘macroscopic’ approach. Recent years have seen recognition of the growing need to bridge these domains through the development of trans-disciplinary training programs and curricula4 as well as venues specifically designed to share innovations that span the laboratory and clinical spaces (eg, the AMIA Summit on Translational Bioinformatics). Translational bioinformatics (TBI) has thus emerged as a systems theory approach to bridge the biological and clinical divide through a combination of innovations and resources across the entire spectrum of biomedical informatics.5 Along with complementary areas of emphasis, such as those focused on developing systems and approaches within clinical research contexts,6 insights from TBI may enable a new paradigm for the study and treatment of disease.
The rapid escalation of activity in TBI can be attributed to parallel advancements in the biological and clinical realms. In biology, we have seen unprecedented advances in technology, such as those associated with generation of molecular sequences.7 In healthcare, we are observing a new era of clinical data acquisition and decision support that is driven by Federal legislation fostering adoption of electronic health records and enablement of seamless exchange of health information.8 9 The challenges have been paralleled in the biological and clinical realms, where there are common challenges in heterogeneous data integration, missing data, and semantic mapping. Nonetheless, opportunities to develop linkages between genetic and clinical information are also increasing as a result of participatory initiatives, such as those promoted by some direct-to-consumer genetic test vendors.10 Furthermore, there is great opportunity to leverage complementary approaches to address these common challenges (eg, some of the tools developed by clinical research informatics researchers6).
The promise of the $2.7 billion Human Genome Project was to enable scientists to understand the genetic basis of human disease.11 However, nearly a decade since the completion of the first draft of the human genome,12 there is still much to be elucidated. Through technological and computational advances, the $1000 genome is becoming a very real possibility.13 The availability of a large number of complete human genomes with clinical, phenotype, and environmental information may enable a new paradigm for the development of new sets of hypotheses pertaining to complex diseases, such as those that involve multiple genes and environmental parameters.14 A major goal of TBI is thus to develop informatics approaches for linking across traditionally disparate data and knowledge sources enabling both the generation and testing of new hypotheses.15 As large volumes of linked biological and clinical data become available, the complexity of disease may be dissected using novel TBI approaches designed in silico, but validated in traditional in vitro or even in vivo interventions.
Building on previous successes
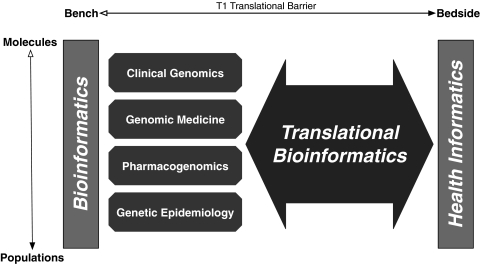
TBI is built on the successes of research that have evolved in the 30 years since the first use16 of the term ‘bioinformatics.’ Four notable areas germane to the present discourse are clinical genomics, genomic medicine, pharmacogenomics, and genetic epidemiology (figure 1). The acceptance of clinical genomics (which has the purpose of identifying clinically relevant molecular biomarkers) by the clinical community can be measured by the growing number of clinically relevant genetic tests.17 Genomic medicine, or ‘personalized medicine,’ (which aims to identify genotype–phenotype correlations relevant to individuals, or haplotype variation) is positioned to uncover large-scale genotype–phenotype associations as a result of genome-wide testing and increased resolution of representation of clinical data. Pharmacogenomics may also benefit from ascertaining correlations with data captured for clinical purposes (eg, such as captured in electronic health records). For example, it may enable correlation of genomic measurements with clinical phenotypes observed relative to pharmacological substances (eg, as listed in the Pharmacogenomics Knowledge Base (PharmGKB)18). It may also potentially provide patient-specific prescribing advice through decision support systems. Finally, genetic epidemiology is rising to new levels with the aggregation of genome-based data alongside public health and environmental registries (eg, such as cataloged in HuGENet19). Collectively, these sub-disciplines of bioinformatics have been suggested as core to the integration of biological and health data.20 However, the mere availability of observations or statistically significant associations is of little practical value without explanations of potential clinical utility. This challenge of finding true biomedical explanations has been reflected before in medicine, for example, when improved methods for acquiring physiological data were developed.21
Figure 1.
Bridging biological and clinical knowledge using translational bioinformatics. Bioinformatics approaches, focused on areas from molecules to populations (eg, clinical genomics, genomic medicine (‘personalized medicine’), pharmacogenomics, and genetic epidemiology), form the foundation of approaches that are used by translational bioinformatics (TBI; large bidirectional arrow). TBI thus bridges knowledge acquired from both the biological (using bioinformatics) and health (using health informatics) domains. Accordingly, the success of TBI will result in the crossing of the T1 translational barrier, and thus link innovations from bench to bedside.
The ability to sequence a patient's genome as routinely as other routine clinical laboratory tests is no longer a far-fetched possibility.13 Accordingly, the sheer volume of potentially available data poses significant challenges for their integration in a form that can be used to either test current hypotheses or develop new ones. The heterogeneity of data suggests the need for new multi-dimensional paradigms for knowledge integration, requiring a deeper understanding of biology than previously required by informatics practitioners. Should one only consider single nucleotide polymorphic markers, or also include intronic (non-coding DNA) regions that have been shown to participate in gene regulation? Can gene expression measurements capture the effects of the environment? How do we then integrate relevant biological data, such as from proteomic studies, and correlate them with fidelity to phenotype data to track subtle, but essential, environmental phenomena? Parallel to the difficulty in addressing these queries there will be significant ethical, legal, and social implication issues to consider.22
At the core of TBI is the development of new hypotheses originating from the integration of genomic and clinical data. TBI reflects a new era of trans-disciplinary science, and reflects the needed unification of multi-scale biological and clinical information for enabling the formal postulation of a deeper understanding of disease such as originally proposed by Blois23 and more recently by Kalet.24 Understanding the genomic influences on the complex evolution of disease, the impact of therapeutic approaches as can be measured by molecular biomarkers, and the overall consistency of genotype–phenotype–environmental correlations across populations forms the basis of focus for the TBI community.
Challenges in studying complex diseases
Understanding complex diseases toward the development and assessment of putative therapies requires traversing between the bench and bedside, often referred to as the ‘T1 translational barrier.’25 26 As a goal, the objective is uncomplicated—to ascertain how basic science observations can be applied to clinical contexts, either in the form of prognostic, diagnostic, or therapeutic approaches to disease. As an endeavor, it represents a grand challenge in modern medicine and also a potential paradigm shift for how to integrate a broad set of data points.
The high dimensionality of potential data types when considering the full array of biological and clinical data that can be generated dwarfs any previous attempt at heterogeneous data integration. There is therefore a need to develop the next generation of clinical decision support systems that can incorporate data from massive biological datasets that will need to be combined with relevant disease phenotype information and computable knowledge bases to offer clinically useful suggestions. Perhaps more mundane, but of equal significance, is the need to develop approaches that can accommodate a dizzying set of file formats and representation standards. These are not, by themselves, completely new challenges to the biomedical informatics community. Nonetheless, they reflect a core area of emphasis where energy is needed to integrate knowledge across clinical genomics, genomic medicine, pharmacogenomics, and genetic epidemiology in light of the avalanche of additional genomic and clinical data and the corresponding knowledge of inter-relationships.
Amidst the challenges of knowledge integration and handling unprecedented volumes of data, TBI is greatly challenged with developing approaches that can bridge biological knowledge and place it into a meaningful clinical context. The volume of data can lead to spurious correlations that may be an artifact of the data and neither biologically nor clinically insightful. For example, if a physician had access to a patient's entire genome, how could it be leveraged to provide clinically insightful knowledge that would not have been possible using solely data already in a medical chart (eg, family history of a disease)? As shown for the genomic era's ‘Patient 0,’ it is plausible to integrate genomic data with relevant clinical data to develop prognostic approaches.27 The potential to provide appropriate care with respect to predicted disease outcome or efficacy of therapeutics offers great incentive for developing TBI approaches that integrate the full complement of biological, clinical, and environmental data. For this reason, phenotypic annotation of samples whose gene expression or single nucleotide polymorphic information is available in genomic data repositories such as GEO28 and dbGAP29 is underway in different laboratories,30 31 involving methodologies that are widely used in health informatics (eg, natural language processing, ontology mapping). Finally, approaches such as those implemented by the Crimson system32 hold promise for capitalizing on the clinical data that are captured as an artifact of standard clinical care. The extent to which this type of relatively noisy data can be used for research is still the object of active research by the TBI community.
Projects that involve TBI approaches to integrate biological and clinical data are already underway. The NIH-funded eMERGE (Electronic Medical Records and Genomics) project is a multi-site endeavor exploring issues involved with linking genomic information (from genome-wide association studies) with clinical data for individuals with specific conditions.33 Other efforts such as the Personal Genome Project,34 the Exome Project,35 the Million Veteran Program,36 and the 1000 Genomes Project37 reflect the increasing interest of the biomedical research and clinical communities in studying the complexity of genotype–phenotype relationships as well as postulating hypotheses for disease that incorporate genomic data. In addition to human-based genome projects, there are also initiatives such as the Human Microbiome Project (HMP38) and Metagenomics of the Human Intestinal Tract (MetaHIT39) that strive to provide a census of commensal microbial flora potentially related to disease.40
The emerging TBI toolbox
The relationship between bioinformatics and health informatics, while conceptually related under the umbrella of biomedical informatics,26 has not always been very clear. The TBI community is specifically motivated with the development of approaches to identify linkages between fundamental biological and clinical information. As technological advances continue to produce data that enhance our ability to further understand the biological underpinnings of complex diseases,41 the clinical community will depend on the development of approaches to interpret these data such that they can be clinically actionable.
TBI approaches are emerging as a melding of a complementary suite of techniques that strive to meet this need. Network approaches42 have led to the development of new techniques to study drug–target43 and gene–disease relationships44 as well as to provide a deeper understanding of the human metabolism.45 Techniques have also been developed to combine genomic and public datasets for studying allelic variation at the population level.46 Systems biology approaches have been used to identify genomic signatures that correlate with the potential efficacy of vaccines.47 Finally, high-throughput sequence based approaches are showing promise for the identification of prognostic genetic markers for increasing numbers of rare diseases.48–50 As the results of these early successes suggest, the TBI community is beginning to work closely with biomedical scientists to develop a new cadre of approaches to study the complex relationships between genotypic, phenotypic, and environmental data. Building on these endeavors will bring us closer than ever before to an entirely new generation of prognostic tests and highly effective and personalized clinical interventions.
Conclusion
The decade following the completion of the first draft of the human genome has witnessed unprecedented technological advancements that have led to the increasing prominence and importance of bioinformatics and health informatics for biology and healthcare, respectively. The exponential growth of genomic data, along with parallel achievements in acquiring and analyzing clinical data position the biomedical research enterprise to deliver on the promise of the Human Genome Project. TBI is accordingly positioned to enable a systems view of complex disease.
Acknowledgments
The authors wish to acknowledge Casey Overby, PhD and Elizabeth Chen, PhD for valuable discussion.
Footnotes
Funding: INS is funded in part by a grant from the National Institutes of Health (R01LM009725). AJB is funded in part by grants from the Lucile Packard Foundation for Children's Health, Hewlett Packard Foundation, and the National Institutes of Health (R01LM009719). YAL is funded in part by a grant from the National Institutes of Health (UL1RR024999). LOM is funded in part by grants from the National Institutes of Health (R01LM009520, U54HL108460, and UL1RR031980), the Komen Foundation, and the Agency for Healthcare Research and Quality (R01HS019913). PTH is funded in part by grants from the Washington Life Sciences Discovery Fund (‘Institute for Genomic Medicine’) and National Institutes of Health (T15LM07442, UL1RR025014, P41LM007242, R01HG02288).
Competing interests: INS, YAL, PTH, and LOM declare they have no competing interests. AJB COI has been submitted in accordance to the ICMJE COI form.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Rosnay JD. The Macroscope: A New World Scientific System. 1st edn New York: Harper & Row, 1979 [Google Scholar]
- 2.Von Bertalanffy L. General System Theory: Foundations, Development, Applications. New York: George Braziller, Inc, 1968 [Google Scholar]
- 3.Louie B, Mork P, Martin-Sanchez F, et al. Data integration and genomic medicine. J Biomed Inform 2007;40:5–16 [DOI] [PubMed] [Google Scholar]
- 4.Tarczy-Hornoch P, Markey M, Smith J, et al. Bio*Medical Informatics and Genomic Medicine: Research and Training. J Biomed Inform 2007;40:1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butte AJ. Translational bioinformatics: coming of age. J Am Med Inform Assoc 2008;15:709–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Embi PJ, Payne PR. Clinical research informatics: challenges, opportunities and definition for an emerging domain. J Am Med Inform Assoc 2009;16:316–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schadt EE, Turner S, Kasarskis A. A window into third-generation sequencing. Hum Mol Genet 2010;19:R227–40 [DOI] [PubMed] [Google Scholar]
- 8.Blumenthal D, DesRoches C, Donelan K, et al. Health Information Technology in the United States: The Information Base for Progress, 2006 [Google Scholar]
- 9.Blumenthal D. Launching HITECH. N Engl J Med 2010;362:382–5 [DOI] [PubMed] [Google Scholar]
- 10.Ginsburg GS, Willard HF. Genomic and personalized medicine: foundations and applications. Transl Res 2009;154:277–87 [DOI] [PubMed] [Google Scholar]
- 11.Watson JD. The human genome project: past, present, and future. Science 1990;248:44–9 [DOI] [PubMed] [Google Scholar]
- 12.Varmus H. Ten years on—the human genome and medicine. N Engl J Med 2010;362:2028–9 [DOI] [PubMed] [Google Scholar]
- 13.Wolinsky H. The thousand-dollar genome. Genetic brinkmanship or personalized medicine? EMBO Rep 2007;8:900–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson G. It Takes a Genome: How a Clash Between Our Genes and Modern Life is Making Us Sick. Upper Saddle River, N.J: Pearson Education, 2009 [Google Scholar]
- 15.Butte AJ. Translational bioinformatics applications in genome medicine. Genome Med 2009;1:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogeweg P, Hesper B. Interactive instruction on population interactions. Comput Biol Med 1978;8:319–27 [DOI] [PubMed] [Google Scholar]
- 17.Pagon RA, Tarczy-Hornoch P, Baskin PK, et al. GeneTests-GeneClinics: genetic testing information for a growing audience. Hum Mutat 2002;19:501–9 [DOI] [PubMed] [Google Scholar]
- 18.Thorn CF, Klein TE, Altman RB. PharmGKB: the pharmacogenetics and pharmacogenomics knowledge base. Methods Mol Biol 2005;311:179–91 [DOI] [PubMed] [Google Scholar]
- 19.Little J, Hawken S. On track? Using the human genome epidemiology roadmap. Public Health Genomics 2010;13:256–66 [DOI] [PubMed] [Google Scholar]
- 20.Martin-Sanchez F, Iakovidis I, Norager S, et al. Synergy between medical informatics and bioinformatics: facilitating genomic medicine for future health care. J Biomed Inform 2004;37:30–42 [DOI] [PubMed] [Google Scholar]
- 21.Walshe FM. The integration of medicine. BMJ 1945;1:723–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malin BA. An evaluation of the current state of genomic data privacy protection technology and a roadmap for the future. J Am Med Inform Assoc 2005;12:28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blois MS. Information holds medicine together. MD Computing 1987;4:42–6 [PubMed] [Google Scholar]
- 24.Kalet I. Principles of Biomedical Informatics. 1st edn Amsterdam, Boston: Academic Press/Elsevier, 2009 [Google Scholar]
- 25.Westfall JM, Mold J, Fagnan L. Practice-based research—“Blue Highways” on the NIH roadmap. JAMA 2007;297:403–6 [DOI] [PubMed] [Google Scholar]
- 26.Sarkar IN. Biomedical informatics and translational medicine. J Transl Med 2010;8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashley EA, Butte AJ, Wheeler MT, et al. Clinical assessment incorporating a personal genome. Lancet 2010;375:1525–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res 2011;39(Database issue):D1005–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet 2007;39:1181–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lacson R, Pitzer E, Kim J, et al. DSGeo: software tools for cross-platform analysis of gene expression data in GEO. J Biomed Inform 2010;43:709–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah NH, Rubin DL, Espinosa I, et al. Annotation and query of tissue microarray data using the NCI thesaurus. BMC Bioinformatics 2007;8:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy S, Churchill S, Bry L, et al. Instrumenting the health care enterprise for discovery research in the genomic era. Genome Res 2009;19:1675–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Church GM. The personal genome project. Mol Syst Biol 2005;1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The Exome Project. http://exome.gs.washington.edu/
- 36.U.S. Department of Veterans Affairs VA Boston Healthcare System, http://www.boston.va.gov/News_and_Media/million_veteran_program.asp
- 37.Durbin RM, Abecasis GR, Altshuler DL, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature 2010;467:1061–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.http://www.commonfund.nih.gov/hmp/overview.aspx
- 39.MetaHIT. http://www.metahit.eu/
- 40.Friedrich MJ. Microbiome project seeks to understand human body's microscopic residents. JAMA 2008;300:777–8 [DOI] [PubMed] [Google Scholar]
- 41.Feero WG, Guttmacher AE, Collins FS. Genomic medicine—an updated primer. N Engl J Med 2010;362:2001–11 [DOI] [PubMed] [Google Scholar]
- 42.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet 2011;12:56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yildirim MA, Goh KI, Cusick ME, et al. Drug-target network. Nat Biotechnol 2007;25:1119–26 [DOI] [PubMed] [Google Scholar]
- 44.Goh KI, Cusick ME, Valle D, et al. The human disease network. Proc Natl Acad Sci U S A 2007;104:8685–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duarte NC, Becker SA, Jamshidi N, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci U S A 2007;104:1777–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang MH, Lindegren ML, Butler MA, et al. Prevalence in the United States of selected candidate gene variants: Third National Health and Nutrition Examination Survey, 1991–1994. Am J Epidemiol 2009;169:54–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Querec TD, Akondy RS, Lee EK, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol 2009;10:116–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lupski JR, Reid JG, Gonzaga-Jauregui C, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med 2010;362:1181–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roach JC, Glusman G, Smit AF, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 2010;328:636–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bilguvar K, Oztürk AK, Louvi A, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 2010;467:207–10 [DOI] [PMC free article] [PubMed] [Google Scholar]