Abstract
Although MDM2 plays a major role in regulating the stability of the p53 tumor suppressor protein, other poorly understood MDM2-independent pathways also exist. Human adenoviruses have evolved strategies to regulate p53 function and stability to permit efficient viral replication. One mechanism involves adenovirus E1B55K and E4orf6 proteins, which collaborate to target p53 for degradation. To determine the mechanism of this process, a multiprotein E4orf6-associated complex was purified and shown to contain a novel Cullin-containing E3 ubiquitin ligase that is (1) composed of Cullin family member Cul5, Elongins B and C, and the RING-H2 finger protein Rbx1(ROC1); (2) remarkably similar to the von Hippel-Lindau tumor suppressor and SCF (Skp1–Cul1/Cdc53–F-box) E3 ubiquitin ligase complexes; and (3) capable of stimulating ubiquitination of p53 in vitro in the presence of E1/E2 ubiquitin-activating and -conjugating enzymes. Cullins are activated by NEDD8 modification; therefore, to determine whether Cullin complexes are required for adenovirus-induced p53 degradation, studies were conducted in ts41 Chinese hamster ovary cells that are temperature sensitive for the NEDD8 pathway. E4orf6/E1B55K failed to induce the degradation of p53 at the nonpermissive temperature. Thus, our results identify a novel role for the Cullin-based machinery in regulation of p53.
Keywords: p53, adenovirus, protein degradation, Cullin complexes, ubiquitination, Elongin B, Elongin C, NEDD8
The p53 cellular tumor suppressor induces cell cycle arrest and apoptosis by functioning as a transcriptional regulator (for review, see Woods and Vousden 2001). Intracellular levels of activated p53 are known to be regulated by a variety of mechanisms, including phosphorylation, acetylation, and targeted degradation. In most cells p53 is degraded, at least in part, through ubiquitin-dependent proteolysis involving MDM2 (for review, see Woods and Vousden 2001), which binds near the N terminus and functions as an E3 ubiquitin ligase for p53 (Honda et al. 1997). Nuclear export appears to be required for MDM2-dependent p53 degradation, which is blocked by both the drug leptomycin B (Roth et al. 1998; Lain et al. 1999) and the p53-binding protein ARF (for review, see Zhang and Xiong 2001). ARF binds MDM2, thus blocking MDM2-mediated ubiquitination and degradation. Although MDM2 is clearly a major regulator of p53 stability, MDM2-independent cellular pathways for p53 degradation exist, including the c-Jun N-terminal kinase (JNK), which binds to and destabilizes p53 (Fuchs et al. 1998), and calpain proteases (Gonen et al. 1997; Kubbutat and Vousden 1997; Zhang et al. 1997).
Infection of cells by many viruses induces the stabilization and activation of p53 (for review, see Roulston et al. 1999). In the case of human adenoviruses, binding of the viral early region 1A (E1A) protein with either the retinoblastoma tumor suppressor family or p300/CBP/p400 histone acetyl transferases induces the activation, stabilization, and accumulation of p53 (Chiou and White 1997; Querido et al. 1997b; Samuelson and Lowe 1997). Increased levels of p53 could therefore result in the induction of cell cycle arrest and early apoptotic cell death, effects that would severely limit the yield of viral progeny. Adenoviruses and other small DNA tumor viruses have evolved various types of mechanisms to prevent the deleterious effects of p53. The large T antigen of simian virus 40 binds to the DNA-binding domain of p53 and prevents p53-mediated transcriptional effects (Sarnow et al. 1982; Bargonetti et al. 1992). This process leads to a large accumulation of p53, but it is nonfunctional. The E6 products of human papillomavirus types 16 and 18 bind p53 (Werness et al. 1990) and target it for degradation by the ubiquitin pathway. Targeting is mediated by a cellular E6-associated protein, E6AP-100K, that functions as a ubiquitin ligase, with E6 functioning as a bridge between the ligase and the p53 substrate (Scheffner et al. 1993).
Although adenovirus E1A can induce a large accumulation of p53 when expressed alone, such increased p53 levels are not usually apparent in wild-type adenovirus-infected cells. Two products of early regions 1B and 4, E1B55K and E4orf6, bind p53 near the N and C termini, respectively, and inhibit transcriptional activation by the tumor suppressor (Sarnow et al. 1982; Yew and Berk 1992; Yew et al. 1994; Dobner et al. 1996; Teodoro and Branton 1997; E. Querido, S. Gurd, and P.E. Branton, unpubl.). In addition, several studies, including our own, have shown that E4orf6 and E1B55K function together to reduce the half-life of p53 and induce efficient p53 degradation (Moore et al. 1996; Querido et al. 1997a; Steegenga et al. 1998; Cathomen and Weitzman 2000; Nevels et al. 2000; Shen et al. 2001). Furthermore, the E1B55K/E4orf6 complex (Sarnow et al. 1984) can induce p53 turnover in cells lacking either MDM2 or ARF, indicating that neither is required for this process (Querido et al. 2001). E4orf6 is a nuclear protein containing both nuclear localization and nuclear retention signals that are believed to be responsible for targeting E4orf6 and E4orf6/E1B55K complexes to the nucleus (Goodrum et al. 1996; Orlando and Ornelles 1999; Nevels et al. 2000). In addition, both E4orf6 and E1B55K have nuclear export signals, and several groups have shown they can shuttle between the nucleus and the cytoplasm (Dobbelstein et al. 1997; Weigel and Dobbelstein 2000; Rabino et al. 2000; Dosch et al. 2001).
In the present studies, we have determined the mechanism of adenovirus-induced p53 degradation through the identification of E4orf6-associated cellular proteins. Such proteins were found to be members of a novel Cullin-containing complex that is required for E4orf6–E1B55K-mediated p53 degradation and capable of inducing ubiquitination of p53 in vitro. These findings suggest that similar complexes could also function in the regulation of p53 levels in normal cells.
Results
Identification of cellular E4orf6-binding proteins.
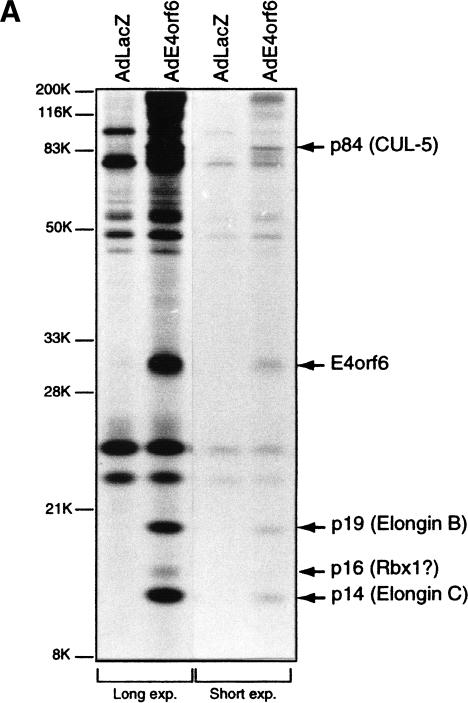
To investigate the roles of the E1B55K and E4orf6 proteins in regulating p53 destruction in adenovirus-infected cells, we affinity purified an E4orf6-containing complex from whole cell extracts of [35S]methionine/[35S]cysteine-labeled p53-null human H1299 cells infected with either an adenovirus vector expressing Ad5 E4orf6 (AdE4orf6) or the control vector AdLacZ. Using this procedure, we observed that approximately stoichiometric amounts of 35S-labeled cellular proteins of ∼84 kD, 19 kD, and 14 kD, as well as lower levels of a 16-kD protein, were reproducibly and specifically immunoprecipitated with the monoclonal antibody 1D5 raised against E4orf6 (Fig. 1A; Boivin et al. 1999). Ion trap mass spectrometry of the E4orf6-associated proteins revealed that the ∼84-kD protein was the Cullin family member Cul5 (Kipreos et al. 1996; Byrd et al. 1997), also known as the vasopressin-activated calcium mobilization receptor or VACM-1, and that the ∼19-kD and 14-kD proteins were Elongins B and C, respectively.
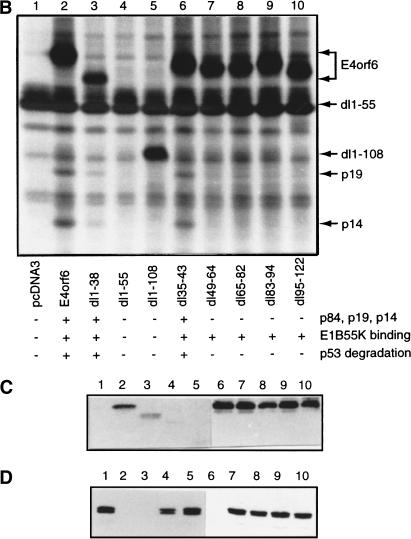
Figure 1.
Analysis of E4orf6 complex formation. (A) Detection of E4orf6 complexes. Extracts from H1299 cells infected with AdLacZ or AdE4orf6 and labeled with 35S were immunoprecipitated with 1D5 anti-E4orf6 antibody followed by SDS-PAGE and autoradiography. Two exposures have been included: positions of migration of prestained standard size markers (left) and those of E4orf6 and coimmunoprecipitating proteins (right). Identities for these species were derived from mass spectroscopy analysis of tryptic peptides. (B) Analysis of complex formation using E4orf6 deletion mutants. H1299 cells were transfected with plasmid DNAs encoding wild-type or mutant E4orf6, and extracts were immunoprecipitated and analyzed as in A. Coimmunoprecipitation of p19 and p14 cellular proteins with E4orf6 has been summarized below as + and −. Binding of p84 showed a similar pattern, as determined from other exposures of the gel (data not shown). Binding of E4orf6 to E1B55K (shown in C) and p53 turnover experiments with E4orf6 and E1B55K (D) have also been indicated as + and −. (C) Complex formation between E4orf6 mutants and E1B55K. Extracts from H1299 cells transfected with plasmid DNAs expressing E1B55K (pCA14 HH55K) and wild-type or mutant E4orf6 proteins (pcDNA3E4orf6) were immunoprecipitated with anti-E1B55K antibody (2A6), and membranes were immunoblotted with anti-E4orf6 antibody (1807). (D) E4orf6/E1B55K-induced turnover of p53. H1299 cells were transfected with plasmid DNAs pcDNA3 p53wt (p53), pCA14 HH55K (E1B-55K), and pcDNA3E4orf6 (wild-type or mutant E4orf6), and extracts were analyzed by Western blotting using anti-p53 antibodies 1801 and 421.
We have previously constructed and partially characterized a series of E4orf6 in-frame deletion mutants (Querido et al. 2001). Consistent with the hypothesis that the coimmunoprecipitating E4orf6/Cul5/ElonginBC complex is involved in the destabilization of p53, we observed a correlation between the ability of E4orf6 mutants to promote p53 degradation and to bind not only E1B55K but also Elongin B and Elongin C (Fig. 1B–D). To determine the ability of E4orf6 mutants to participate in p53 degradation, p53-null H1299 cells were cotransfected with cDNAs expressing human p53, histidine-tagged E1B55K (HH55Kwt), and wild-type or mutant E4orf6. As shown in Figure 1D, p53 levels were reduced in whole cell extracts from cells expressing E1B55K and wild-type E4orf6 or E4orf6 mutants dl1–38 and dl35–43 (lanes 2,3,6), but not from cells expressing E4orf6 mutants dl1–55, dl1–108, dl49–64, dl65–83, dl83–94, or dl95–122. Similarly, endogenous Elongin BC could be immunoprecipitated with wild-type E4orf6 and mutants dl1–38 and dl35–43, but not with any of the other mutants tested, suggesting that a region required for the interaction may reside between residues 44 and 108 of E4orf6. Interaction of E4orf6 with E1B55K is not sufficient to promote degradation of p53, as several of the E4orf6 mutant proteins that could still bind E1B55K but failed to bind Elongin BC also failed to support p53 degradation (Fig. 1C).
The mammalian Elongin BC complex was shown to function as a regulator of RNA polymerase II elongation factor Elongin A (for review, see Reines et al. 1999). Elongin BC is also a component of the large von Hippel-Lindau (VHL) tumor suppressor complex (Duan et al. 1995; Kibel et al. 1995). Binding of Elongin BC with both Elongin A and VHL appears to involve the interaction of Elongin C with a short region termed the BC-box (consensus sequence [A,P,S,T]LXXXCXXX[A,I,L,V]), which is found in VHL and Elongin A (Duan et al. 1995; Kibel et al. 1995; Aso et al. 1996; Stebbins et al. 1999; Ohh et al. 2000). In addition, Elongin BC binds to many additional proteins, including members of the so-called SOCS-box protein family, each of which contains an Elongin BC-binding site (Kamura et al. 1998; Zhang et al. 1999). The VHL complex was shown to function as an E3 ubiquitin ligase (Iwai et al. 1999; Lisztwan et al. 1999) that targets the α-subunits of the hypoxia-inducible transcription factors HIF1 and HIF2 for ubiquitination (Maxwell et al. 1999; Cockman et al. 2000; Kamura et al. 2000; Ohh et al. 2000). VHL functions as a substrate recognition subunit (Cockman et al. 2000; Kamura et al. 2000; Ohh et al. 2000), whereas Elongins B and C serve to link these proteins to a module containing Cul2 and Rbx1. The Cul2/Rbx1 module activates an E2 ubiquitin-conjugating enzyme to ubiquitinate HIF-α (Kamura et al. 2000). The VHL complex is remarkably similar to the yeast and mammalian SCF ubiquitin ligase complexes (for review, see Patton et al. 1998), each of which contain an F-box protein substrate recognition subunit linked to a Cul1(Cdc53)/Rbx1 module by the Skp1 protein (Kamura et al. 1999; Ohta et al. 1999; Seol et al. 1999; Skowyra et al. 1999; Tan et al. 1999). Thus, the association of a Cullin family member and Elongin BC with E4orf6 could suggest that the adenovirus E4orf6 and E1B55K proteins target p53 for degradation by forming a Cullin E3 ligase and recruiting it to p53.
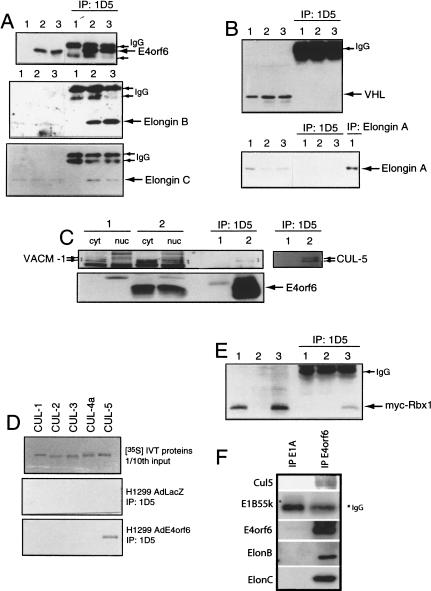
To confirm the interaction of Elongin BC with E4orf6, H1299 cells were infected with either the control adenovirus vector AdLacZ (Fig. 2A, lane 1) or with vectors encoding E4orf6 (lane 2) or a combination of vectors expressing E4orf6, His-tagged E1B55K, and p53 (lane 3). Western blotting with the anti-E4orf6 antibody 1807, with anti-Elongin B antiserum, or with an anti-Elongin C antibody was then performed on whole cell extracts (left) or on proteins immunoprecipitated with anti-E4orf6 1D5 monoclonal antibody (right). As expected, E4orf6 was detected only in whole cell extracts from E4orf6-expressing cells (top panel). Elongin B (middle panel) and Elongin C (bottom panel) were specifically precipitated with E4orf6 whether or not E1B55K and p53 were present. Figure 2B shows that neither VHL nor Elongin A coimmunoprecipitated with E4orf6. Confirming that VHL is not required for E4orf6/E1B55K-mediated p53 degradation, both human VHL-positive and VHL-null tumor cell lines supported p53 turnover by these viral proteins at similar levels (data not shown).
Figure 2.
Identification of E4orf6 binding proteins. (A) H1299 cells were infected with AdLacZ (lane 1), AdE4orf6 (lane 2), or AdE4orf6 plus AdHH55K plus Adp53wt (lane 3). Whole cell extracts (left three lanes) and 1D5 immunoprecipitates (right three lanes) were analyzed by Western blotting using anti-E4orf6 antibody 1807 (top), anti-Elongin B antibody (middle), and anti-Elongin C antibody (bottom). (B) Same as Figure 1A except that immunoblotting was performed with anti-VHL Ig32 antibody (top) or anti-Elongin A antibody (bottom). At the right (lane 1) is shown an immunoprecipitate prepared with anti-Elongin A and blotted with the same antibody. (C) H1299 cells were infected with AdLacZ (lane 1) or AdE4orf6 (lane 2), and cytoplasmic and nuclear extracts as well as whole cell extracts were prepared. Whole cell extracts immunoprecipitated with 1D5 anti-E4orf6 antibody were analyzed by Western blotting using either a polyclonal antibody to rabbit VACM-1 (left six lanes) or a polyclonal antibody against human Cul5 (right two lanes). E4orf6 (bottom) was detected using 1807 antibody. (D) Human Cul1, Cul2, Cul3, and Cul5 and mouse Cul4A were synthesized (top) by in vitro transcription/translation, and aliquots of these 35S-labeled proteins were combined with 1D5 immunoprecipitates from AdLacZ-infected (middle) or AdE4orf6-infected (bottom) H1299 cells. (E) H1299 cells were transfected with plasmid DNAs expressing Myc–Rbx1 (lane 1), E4orf6 (lane 2), and Myc–Rbx1 plus E4orf6 (lane 3). Whole cell extracts (left lanes) or 1D5 immunoprecipitates (right lanes) were analyzed by Western blotting with anti-Myc 9E10 antibody. (F) Human 293 cells were infected with wild-type Ad5 at a MOI of 40, and at 15 h postinfection, cell extracts were prepared and immunoprecipitated with either anti-E4orf6 (1809) or anti-E1A (M73) antibodies. The pattern of associated proteins determined by Western blotting as above.
To verify the interaction of E4orf6 and Cul5, nuclear and cytoplasmic extracts were prepared from cells infected with AdLacZ or AdE4orf6 and immunoprecipitated with anti-E4orf6 antibody 1D5. Following Western blot analysis, an antibody raised against rabbit Cul5 (VACM-1; Burnatowska-Hledin et al. 1995) detected two closely migrating Cul5 species in cytoplasmic fractions from AdLacZ-infected (Fig. 2C, lane 1) and AdE4orf6-infected cells (Fig. 2C, lane 2); however, in 1D5 immunoprecipitates and in nuclear fractions, Cul5 was only detected when E4orf6 was present (Fig. 2C, cf. lanes 1 and 2). To confirm that this species represented Cul5, an antipeptide polyclonal antiserum was raised against a synthetic peptide corresponding to the C terminus of human Cul5. Figure 2C shows that similar results were obtained with this antiserum (see lanes 1 and 2 at right). When this work commenced, Cul5 had not been shown to play a role in ubiquitination; however, in parallel studies we have found that it appears to be a component of a larger family of Elongin BC-based E3 ubiquitin ligases (Kamura et al. 2001). It was also suggested recently that Cul5 is able to form a complex that ubiquitinates E2F1 in vitro (Ohta and Xiong 2001). To determine whether E4orf6 can associate with other members of the Cullin family, in vitro translated human Cul1, Cul2, Cul3, and Cul5 or mouse Cul4a were tested for the ability to bind 1D5 immunoprecipitates from AdLacZ- or AdE4orf6-infected H1299 cells. As shown in Figure 2D, all Cullins were synthesized at similar levels (top); however, only Cul5 was specifically precipitated with E4orf6 (bottom panels).
The 16-kD Rbx1 (ROC1) protein is a component of both the SCF and VHL tumor suppressor complexes (Kamura et al. 1999; Ohta et al. 1999; Seol et al. 1999; Skowyra et al. 1999; Tan et al. 1999). As shown in Figure 2E, myc-tagged Rbx1 specifically co-immunoprecipitated with E4orf6 from cells transfected with cDNAs expressing these two proteins. Thus, Rbx1 associates with E4orf6 and probably represents the 16-kD species detected at low levels in Figure 1A.
To determine if the Cul5/ElonginB/C complex also associates with E4orf6/E1B55K during productive infection, human 293 cells were infected with wild-type Ad5 and cell extracts prepared at 15 h post-infection were immunoprecipitated using either anti-E4orf6 antibodies (1807) or, as a control, anti-E1A M73 mouse monoclonal antibodies (Harlow et al. 1985). Analysis by Western blotting using appropriate antibodies indicated that Cul 5 (appearing again as a doublet), Elongin B, and Elongin C were detected in association with E4orf6 but not with E1A products (Fig. 2F). E1B55K also associated with E4orf6, as shown many times previously (Querido et al. 2001). The material present in the position of migration of E1B55K in the M73 immunoprecipitate was undoubtedly antibody heavy chain, as this protein has not been found previously to associate with E1A products.
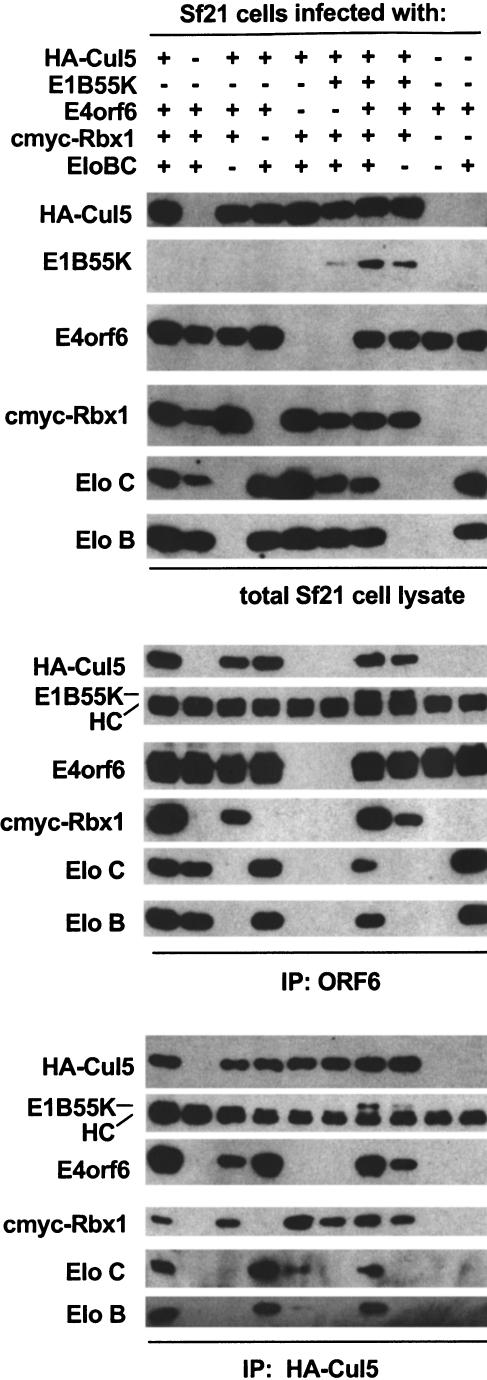
As a final verification of the interaction of E4orf6 and E1B55K with Cul5, Elongin BC, and Rbx1, Sf21 insect cells were infected with various combinations of baculoviruses encoding E4orf6, E1B55K, Elongin BC, HA-tagged Cul5 (HA-Cul5), and c-myc-tagged Rbx1 (cmyc–Rbx1), and complexes were immunoprecipitated from cell lysates with either 1D5 antibodies against E4orf6 or antibodies against the HA-epitope and subjected to immunoblotting. As shown in Figure 3, complexes containing E4orf6, E1B55K, Cul5, Elongin BC, and Rbx1 could be isolated from insect cells. Consistent with the results obtained in mammalian cells, the interaction of E4orf6 with Cul5, Elongin BC, and Rbx1 did not require E1B55K. In addition, Elongin BC and Cul5 were able to interact with E4orf6 in lysates from cells not overexpressing other complex components. This observation does not, however, necessarily imply that Cul5/Rbx1 binds directly to E4orf6, as previous results indicate that insect Elongins B and C can assemble into functional complexes with mammalian Elongin BC-binding proteins and Cullins (Brower et al. 1999; Kamura et al. 2000). Taken together, the findings presented thus far indicate that E4orf6 can assemble with Elongin BC, Cul5, and Rbx1 in a complex that is distinct from the Elongin BC-containing complexes formed with VHL and Elongin A.
Figure 3.
Reconstitution of E4orf6/E1B55K/Cul5/Rbx1/Elongin B and C complexes in Sf21 cells. Sf21 cells were infected with baculoviruses encoding the proteins indicated. The lysates were immunoprecipitated using either anti-HA (12CA5) or anti-E4orf6 (1D5) antibody. Crude lysates (top) and washed immune complexes (middle, bottom) were separated by SDS-PAGE and immunoblotted with the indicated antibodies.
E4orf6 relocates Cul5 but not Cul2 to the nucleus
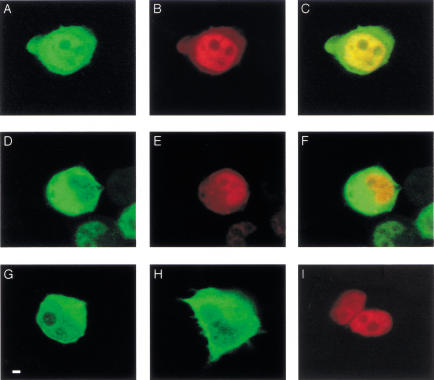
Given the remarkable architectural similarity between Cul1/Cdc53-containing SCF and Cul2-containing VHL ubiquitin ligases and the E4orf6 complex, these results raise the possibility that the complex of E4orf6, Cul5, Elongin BC, and Rbx1 may also function as a ubiquitin ligase. Because E4orf6 is found largely in the nucleus (Goodrum et al. 1996; Dobbelstein et al. 1997) and Cul5 had been reported previously as largely cytoplasmic (Burnatowska-Hledin et al. 1999), it was important to determine whether Cul5 and E4orf6 colocalize in cells. H1299 cells were transfected with cDNAs expressing human HA-Cul5 or HA-Cul2 and/or E4orf6, and cells were examined by laser confocal microscopy. E4orf6 was detected using a rabbit polyclonal antibody (1807) directed toward the E4orf6 C terminus (Boivin et al. 1999) and visualized using Alexa 594 anti-rabbit antibody (red color). The Cullins were detected using mouse monoclonal anti-HA antibodies and visualized using Alexa 488 anti-mouse antibody (green color). Figure 4 shows examples of the staining patterns obtained and are representative of the majority of cells in the preparations. Figure 4, G and H, shows that when expressed independently, Cul2 (Fig. 4G) and Cul5 (Fig. 4H) were localized primarily to the cytoplasm, whereas E4orf6 (Fig. 4I) was largely nuclear. Figure 4, A–C and D–E, shows cells that coexpressed E4orf6 and either Cul5 or Cul2, respectively. In the presence of E4orf6, Cul5 (Fig. 4A), but not Cul2 (Fig. 4D), was present at high levels in the nucleus. The appearance of yellow nuclear fluorescence following superimposition of Figure 4, panels A and B (Fig. 4C), indicated that Cul5 co-localized in the nucleus with E4orf6. This was not the case with Cul2, as the nuclear fluorescence remained largely unchanged following superimposition of Figure 4, D and E (Fig. 4F). These data are consistent with our observation that E4orf6 binds preferentially to Cul5, and they suggest that E4orf6 may transport Cul5 to the nucleus, as it does E1B55K (Goodrum et al. 1996). We also tested the ability of E4orf6 mutants dl1–38, dl1–55, and R240E/R241E to direct nuclear localization of Cul5. The E4orf6 mutant dl1–38 was wild-type in its ability to direct Cul5 to the nucleus, whereas the other two mutants were deficient (data not shown). These observations correlate with the ability of these mutants to cooperate with E1B55K in p53 turnover (Fig. 1; Querido et al. 2001).
Figure 4.
Intracellular colocalization of E4orf6 and Cul5. H1299 cells were transfected with plasmid DNAs expressing HA–Cul2, HA–Cul5, or E4orf6, and cells were fixed and examined by laser confocal microscopy. E4orf6 was detected using rabbit polyclonal antibody (1807) and Alexa 594 (red) anti-rabbit IgG antibody. HA-tagged human Cul2 and Cul5 were detected using mouse monoclonal anti-HA antibody (HA.11) and Alexa 488 (green) anti-mouse IgG antibody. (A–C) HA–Cul5 and E4orf6. (D–F) HA–Cul2 and E4orf6. C and F were generated by scanning for both the red and the green signal to detect any colocalization as bright yellow. (G) HA–Cul2 alone. Bar, 20 μm; cells shown are representative. (H) HA–Cul5 alone. (I) E4orf6 alone.
E4orf6 complex ubiquitinates p53 in vitro
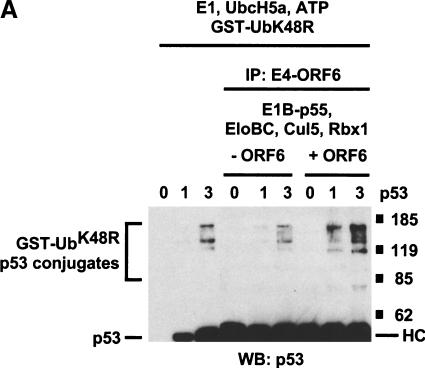
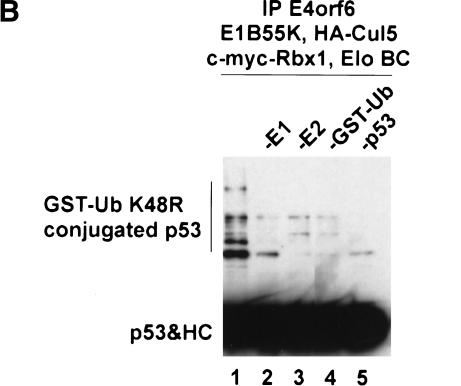
To test directly the possibility that the E4orf6 complex can function as an E3 ubiquitin ligase that targets p53, we examined the ability of immunopurified E4orf6 complexes to support the ubiquitination of recombinant p53 in vitro. Conjugation of ubiquitin to target proteins requires, in addition to an E3 ubiquitin ligase, an E1 ubiquitin-activating enzyme that transfers ubiquitin to the active site cysteine of an E2 ubiquitin-conjugating enzyme. E2 and E3 then act together to catalyze the formation of an isopeptide bond between lysine residue(s) on the target protein and the C terminus of ubiquitin. The E3 binds both the target protein and E2 and accordingly plays a central role in determining the substrate specificity of the reaction. Cul5, E1B55K, Rbx1, and Elongins B and C were expressed in insect cells, with or without E4orf6, and insect cell lysates were immunoprecipitated with anti-E4orf6 1D5 antibodies. Immunoprecipitated complexes were supplemented with various concentrations of recombinant p53 and with recombinant yeast ubiquitin-activating enzyme (yUba1), the E2 ubiquitin-conjugating enzyme hUbc5A, ATP, and GST–ubiquitinK48R. Ubiquitination of p53 was assessed by the appearance of slower-migrating forms of p53, as detected by Western blotting using an anti-p53 monoclonal antibody. The ∼35-kD GST–ubiquitin fusion protein GST–ubiquitinK48R was used in these reactions because it causes a larger and more easily visualized shift in the migration of p53 than does the ∼7-kD ubiquitin molecule. In addition, GST–ubiquitinK48R does not efficiently form polyubiquitinated products, resulting in a simpler pattern of GST–ubiquitin–p53 conjugates. Figure 5A (left three lanes) shows that in the absence of 1D5 immunoprecipitate, low levels of ubiquitinated p53 could be detected in the presence of 65 ng of recombinant p53, but not in the absence of p53 or with 20 ng of p53. Identical background levels of p53 ubiquitination were observed in reactions containing no immunoprecipitates (left three lanes) or 1D5 immunoprecipitates from cells not expressing E4orf6 (middle three lanes). In the presence of immunoprecipitates from cells expressing E4orf6 (right three lanes), significantly higher levels of ubiquitinated p53 were formed in reactions containing either 20 or 65 ng of p53. As expected, formation of p53-ubiquitin conjugates depends on the presence of E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme, and GST–ubiquitin (Fig. 5B). Only background levels of ubiquitinated p53 were formed in reactions performed with immunoprecipitates from cells expressing E4orf6 alone, in the absence of other proteins of the E4orf6 complex (data not shown). Taken together, these results argue that the E4orf6 complex can function with a ubiquitin-activating enzyme and a ubiquitin-conjugating enzyme to facilitate p53 ubiquitination in vitro, and they are consistent with the idea the E4orf6/E1B55K complex targets p53 for ubiquitination and subsequent degradation via this Cul5/Elongin BC/Rbx1 complex in cells.
Figure 5.
In vitro ubiquitination of p53 by E4orf6 complexes. (A) Lysates from Sf21 cells infected with baculoviruses encoding E1B55K, Elongin B, Elongin C, HA–Cul5, and Myc–Rbx1, with or without baculovirus encoding E4orf6, were immunoprecipitated with anti-E4orf6 antibody (1D5). Increasing amounts of purified p53 protein were incubated with the immunoprecipitates and with yUba1, hUbc5a, and GST–UbK48R in ubiquitination reaction buffer. Reaction mixtures were incubated for 30 min at 25°C and analyzed by Western blotting using anti-p53 antibody (421). (B) p53 was incubated with immunoprecipitates and with various combinations of yUba1, hUbc5a, and GST–UbK48R. The indicated components were omitted from reactions: E1, yUba1; E2, hUBC5a; GST-Ub, GST-UbK48R. HC indicates heavy chain.
Degradation of p53 by E4orf6–E1B55K requires functional Cullins
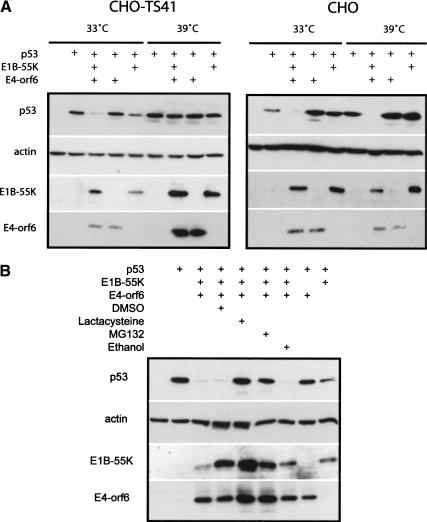
Although we have shown that E4orf6 binds to a Cul5/Elongin BC/Rbx1 complex that is capable of the in vitro ubiquitination of p53 and that this interaction correlates with the ability of E4orf6/E1B55K to degrade p53, we wished to show that Cullins are required for adenovirus-mediated p53 degradation. Some time ago, a Chinese hamster ovary (CHO) cell line, ts41, was isolated (Hirschberg and Marcus 1982). ts41 cells are temperature sensitive for progression into G2/M and thus undergo multiple rounds of DNA synthesis without cell division (Handeli and Weintraub 1992). Recently, ts41 cells have been shown to be defective in the NEDD8 activation pathway. The ts41 mutation can be complemented by overexpression of amyloid precursor protein-binding protein 1 (APP-BP1), which together with hUba3 forms the NEDD8-specific E1-activating enzyme (Gong and Yeh 1999; Chen et al. 2000). NEDD8, and its yeast counterpart Rub1, are highly conserved ubiquitin-like proteins that, following cleavage, are conjugated with a number of proteins (Kamitani et al. 1997). Like ubiquitination, conjugation of NEDD8 to target proteins requires E1-3 activating, conjugating, and ligating enzymes, which are distinct from enzymes of the ubiquitin conjugation pathway (Osaka et al. 1998; Gong and Yeh 1999; for review, see Yeh et al. 2000). A number of reports now indicate that Cullins require modification by NEDD8 to participate in the degradation of target proteins (Freed et al. 1999; Hori et al. 1999; Liakopoulos et al. 1999; Wada et al. 1999). Recently, it has been shown using ts41 cells that ongoing neddylation is required by the SCF and VHL–Elongin C–Cullin 2 complexes for the degradation of targets in mammalian cells, including HIF1α, p27, and IκBα (M. Ohh, J.J. Moslehi, Y. Chen, V. Chau, M.A. Read, and W.G. Kaelin, in prep.). Thus, it appeared possible to use ts41 cells to confirm that a Cullin-mediated pathway is involved in the degradation of p53 by the adenovirus E4orf6/E1B55K complex. Wild-type CHO and ts41 cells were incubated at 33°C, and following infection with adenovirus vectors expressing human p53 and E1B55K or E4orf6, either singly or in combination, some cultures were shifted to the nonpermissive temperature of 39°C. Cell extracts were prepared and analyzed by Western blotting using antibodies against p53, E4orf6, or E1B55K. Figure 6A (right panel) shows that in wild-type CHO cells, expression of both E4orf6 and E1B55K resulted in the degradation of p53 at both temperatures. Little degradation was apparent with E4orf6 alone; however, as observed previously (Querido et al. 2001), some degradation was seen in both cell types using the E1B55K vector alone, as low levels of E4orf6 are produced from the vector backbone, which contains the E4 region. In the case of ts41 cells (Fig. 6A, left panel), p53 degradation by E4orf6/E1B55K was also apparent at 33°C, but little degradation occurred at the nonpermissive temperature of 39°C. Although these results leave open the possibility that the defect in ts41 cells also affects other targets that are indirectly involved, they do suggest that the mechanism of degradation of p53 by E4orf6-E1B55 is dependent on neddylation and is likely to require functional Cullins, which are the best known targets of this form of modification. As we found in Figure 2 that Cul5 was the only Cullin to associate with E4orf6, it is likely that Cul5 is the Cullin responsible for this process.
Figure 6.
E4orf6/E1B55K require NEDDylated Cullins to degrade p53. (A) Wild-type and ts41 Chinese hamster ovary (CHO) cells maintained at 33°C were infected with various combinations of adenovirus vectors for 48 h (see + at top). Vectors included AdrtTA (vector control), Adp53wt, AdE4orf6, and AdHH55K. Some cultures were shifted to the 39°C for the last 15 h of incubation. Whole cell extracts analyzed by Western blotting using to detect p53 (1801), E4orf6 (1807), E1B55K (2A6), and actin. (B) CHO cells infected with various combinations of adenovirus vectors (as in A). Proteasome inhibitors MG132, lactacystin, or their solvents were added for the last 6 h of infection. Cell extracts were analyzed as in A.
Treatment of CHO cells with the proteasome inhibitors lactacystin or MG132 inhibited the degradation of p53 by the E4orf6/E1B55K complex (Fig. 6B), confirming that p53 turnover in these cells required functional proteasomes.
Discussion
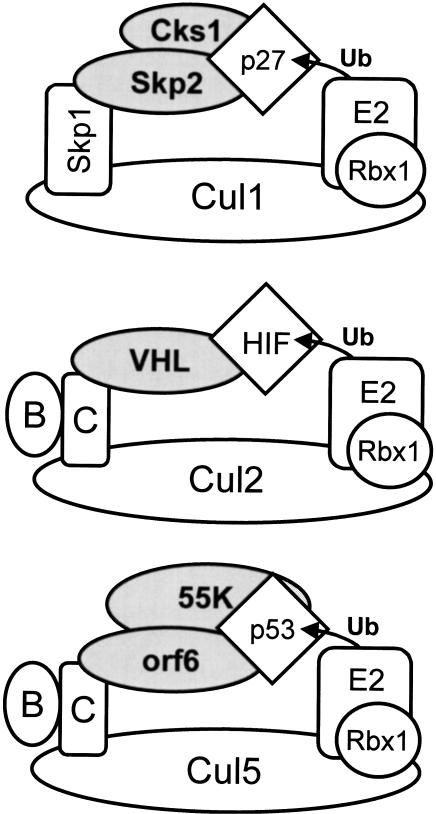
We and others had shown previously that the adenovirus proteins E4orf6 and E1B55K function together to promote degradation of p53; however, little was known about the mechanism of this process. Our previous work using the inhibitor MG132 suggested that such degradation requires functional proteasomes (Querido et al. 2001). In this report, we have presented evidence that assembly of E4orf6 into a multiprotein complex containing E1B55K, Elongins B and C, Cul5, and Rbx1 is required for E4orf6/E1B55K-dependent p53 turnover in cells. In addition, we have shown that this E4orf6-containing complex functions as an E3 ubiquitin ligase that promotes ubiquitination of p53 in vitro. As modeled in Figure 7, the E4orf6 complex is strikingly similar to VHL and SCF E3 ubiquitin ligase complexes, in that each contains a substrate recognition module linked to a Cullin–Rbx1 subcomplex via either Elongin BC or the Elongin C-like protein Skp1. The Cks1 cyclin-dependent kinase-binding protein, recently shown to be required for SCF-mediated degradation of p27Kip1 in vivo, is shown in the SCF complex (Bartek and Lukas 2001; Spruck et al. 2001). Figure 7 also shows the positions of the substrate (diamond), the E2-conjugating enzyme, and ubiquitin proteins based on computer modeling and the recent elucidation of the X-ray crystallographic structure of the Skp1/Skp2/Cul1/Rbx1 complex (N. Zheng and N.P. Pavletich, pers. comm.). It should be noted that the interaction interface between E4orf6 and the Cul5/Elongin BC/Rbx1 complex has not yet been determined. E4orf6 contains two putative BC-box sequences; however, the interaction between E4orf6 and Elongins B and C was unaffected by mutation of either BC-box sequence individually (Q. Yin, J.W. Conaway, and R.C. Conaway, unpubl.). Thus, it is possible that the interaction occurs by a novel mechanism, and further studies will be required to establish the precise nature of this interaction.
Figure 7.
Parallels between the SCF, VHL, and E4orf6 ubiquitin ligase complexes. The SCF (top), VHL (middle), and E4orf6 (bottom) ubiquitin ligase complexes have been illustrated. (B) Elongin B; (C) Elongin C; (p27) p27Kip1; (Ub) ubiquitin; (orf6) Ad5 E4orf6; (55K) Ad5 E1B55K; (Cul1) the mammalian homolog of Cdc53; and (Rbx1) Rbx1/ROC1. The architecture of the SCF complex shown is based on the X-ray structure of a Skp1/Skp2/Cul1/Rbx1 crystal determined recently (N. Zheng and N.P. Pavletich, pers. comm.). Models of the VHL and E4orf6 complexes are theoretical only, and the interaction interface between E4orf6 and the Cul5/Elongin BC/Rbx1 complex has not yet been determined.
In addition, we are still unsure of the identity of the ubiquitin-conjugating enzyme used by this complex. Our in vitro studies used hUbc5; however, in vivo assays detected HA-E2Cdc34 in association with E4orf6 complexes (data not shown). Further studies will be required to identify the conjugating enzyme(s) involved.
Several other questions also exist. Ubiquitination of p53 in vitro was observed to occur at similar levels in the presence or absence of E1B55K (data not shown). It is possible that E4orf6 present in Cul5 complexes is no longer able to bind p53 efficiently, and thus, E1B55K is required to introduce high levels of the p53 substrate. Additionaly, the role of E1B55K in vivo may be to position p53 in the complex for optimal ubiquitination. In support of this hypothesis, previous studies have shown that E1B55K and E4orf6 must bind each other for p53 destabilization to occur (Cathomen and Weitzman 2000; Querido et al. 2001). Furthermore, a recent study of E1B55K single amino-acid substitution mutants found that E1B55K R240A, although still able to bind E4orf6 and support viral late gene expression, was defective in binding p53 (Shen et al. 2001). Significantly, an adenovirus encoding this mutant as well as wild-type E4orf6, ONYX-051, lost the ability to target p53 for degradation. It is also possible that E1B55K is required to supply a nuclear export signal (Kratzer et al. 2000; Dosch et al. 2001). Previous studies indicated that deletion of the E4orf6 NES had little effect on p53 degradation by E4orf6/E1B55K (Querido et al. 2001); however, it is still unclear if degradation occurs via cytoplasmic proteasomes following shuttling to the cytoplasm, as is the case with MDM2-mediated p53 degradation (Lain et al. 1999), or if it occurs via nuclear proteasomes. One final question arises because of our observation that the efficiency of p53 ubiquitination in our reconstituted system is relatively inefficient. Many SCF ubiquitin ligases specifically recognize phosphorylated substrates (Patton et al. 1998). Similarly, recognition of HIF1α by the VHL ubiquitin ligase depends on prior hydroxylation of a proline residue within the HIF1α oxygen-dependent degradation domain (Ivan et al. 2001; Jaakkola et al. 2001). By analogy, it is possible that the majority of the recombinant p53 used in our reactions is not appropriately modified and accordingly is not recognized by the E4orf6 complex. An interesting recent finding suggests that p53 must be phosphorylated by the COP9 signalosome before being recognized by the HPV E6/E6AP and MDM2 E3 ligases for ubiquitin-mediated degradation to take place (Bech-Otschir et al. 2001). This or a related process may represent a general requirement for degradation of p53 by ubiquitin-dependent pathways and may explain the low levels of p53 ubiquitination observed in vitro. Alternatively, efficient ubiquitination of p53 may require additional activities not present in our reconstituted reactions.
Information on the mechanism of action of viral proteins has often led to insights into previously unrecognized normal cellular pathways. The present studies indicate that E4orf6 may have evolved functions that parallel those of unidentified cellular regulators of p53 degradation. It is tempting to speculate that cells may contain cellular E4orf6 homologs that function with Cullin-containing complexes to regulate the degradation of p53.
Materials and methods
Cells
Human small cell carcinoma H1299 cells carrying a homologous deletion of the p53 gene (Mitsudomi et al. 1992), CHO cells, and derivative ts41 cells (Hirshberg and Marcus 1982) were maintained in Dulbecco's modified Eagle's medium (GIBCO BRL) supplemented with 10% fetal calf serum (CanSera). Sf21 insect cells were cultured as described previously (Kamura et al. 1998).
Vectors
The adenovirus vectors used in this study are derivatives of vectors described by Bacchetti and Graham (1993). They are deleted in the E1 and E3B regions, and the gene of interest is inserted in the E1 region under the cytomegalovirus (CMV) promoter. Because the E1A coding region is absent, the resident Ad5 vector genes are expressed at almost undetectable levels. AdLacZ expresses the Escherichia coli β-galactosidase gene, and Adp53wt expresses human wild-type p53 (Bacchetti and Graham 1993). The adenovirus vectors AdHH55K, expressing HMK and histidine-tagged Ad5 E1B55K, and AdE4orf6, expressing Ad5 E4orf6, were described previously (Querido et al. 1997a). AdrtTA, which was used as a control vector in some cases, expresses reverse tetracycline transactivator protein (Gossen et al. 1995) and was produced by standard methods (Bett et al. 1994). The HPC4–Elongin B, HSV–Elongin C, and MYC–Rbx1 baculovirus vectors are described in Kamura et al. (1999), and the HA–Cul5 baculovirus vector is described in Kamura et al. (2001). E4orf6 and E1B55K were subcloned into BacPAK8, and recombinant baculoviruses were generated with the BacPAK baculovirus expression system (Clontech). HA–Cul2 and HA–Cul5 were expressed in mammalian cells using pcDNA3 and pCI–neo vectors, respectively. The following plasmids were also used: MycRbx1 encodes myc-tagged murine Rbx1 (Kamura et al. 1999), HA–E2cdc34 expresses HA-tagged human E2cdc34 (Lisztwan et al. 1998), and Flag-VHL encodes human Flag-tagged VHL (Kamura et al. 1999). Plasmids encoding human Cul1, Cul2, Cul3, and Cul5 and mouse Cul4A, are described in Michel and Xiong (1998). The plasmid pcDNA3 p53 wt encodes human wild-type p53, and pCA14 HH55K encodes HMK and histidine-tagged Ad5 E1B55K. The pcDNA3 E4orf6 wild-type plasmid, as well as all in-frame deletion mutants generated using PCR-based protocols, are described in Querido et al. 2001.
Antisera
Anti-E4orf6 mouse monoclonal antibody 1D5 was described in Querido et al. (2001), and E4orf6-specific rabbit polyclonal antibody 1807 was described in Boivin et al. (1999). Anti-p53 pAb421 and pAb1801 hybridoma supernatants were prepared as described in Querido et al. (2001). E1B55K was detected with the 2A6 monoclonal antibody (Sarnow et al. 1982). Anti-Elongin A, anti-Elongin B, and anti-Elongin C goat polyclonal antibodies were purchased from Santa Cruz Biotechnology, and Ig32 anti-VHL mouse monoclonal antibody was from Pharmingen. The rabbit polyclonal antibody generated against the C terminus of rabbit VACM-1 was a generous gift from Maria Burnatowska-Hledin (Hope College, Holland, MI). A rabbit polyclonal antibody against human Cul5 was made for us by Genemed Synthesis Inc. using a synthetic peptide (EHKIRRDESDINTFIYMA) corresponding to the C terminus of human Cul5. Anti-Myc 9E10 antibody (Santa Cruz), anti-HA mouse monoclonal HA.11 (BAbCO) and mouse monoclonal 12CA5 (Boehringer-Mannheim), and anti-Flag M2 antibody (Sigma), were used to detect the tagged epitopes. The anti-γ-actin mouse monoclonal antibody was a generous gift from Gordon Shore (McGill University, Quebec, Canada).
Identification of E4orf6-binding proteins
H1299 cells growing on 100-mm-diameter dishes (Corning Glass Works, NY) were infected at a multiplicity of infection (MOI) of 30 plaque-forming units (pfu) per cell with the indicated adenovirus vectors. Cell cultures were labeled from 18 to 22 h postinfection with 200 μCi per plate of [35S]methionine/[35S]cysteine EasyTag Express protein labeling mix (>1000 Ci/mmole; DuPont NEN) in methionine/cysteine-free medium. Whole cell extracts were then prepared in nonionic detergent lysis buffer X (50 mM Tris-HCl at pH 8.0, containing 250 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM EGTA, and 2 μg/mL each of aprotinin, leupeptin, and pepstatin), immunoprecipitated using 1D5 anti-E4orf6 antibody, and analyzed by SDS-PAGE and autoradiography, as described previously (Boivin et al. 1999). Prestained standard size markers (Bio-Rad) were used, and their positions are indicated in Figure 1A. The potential identities shown for the p84, p19, and p14 species were derived from mass spectroscopy analysis of tryptic peptides derived from gel-purified material (Borealis Biosciences, Toronto, Canada). For the analysis of complex formation using E4orf6 deletion mutants, a study similar to that of Figure 1A was performed in H1299 cells, except that cells in 100-mm-diameter plates were transfected according to the GIBCO BRL Lipofectin protocol with 10 μg of pcDNA3 plasmid DNA encoding wild-type or mutant E4orf6.
E4orf6 protein-binding assays
In some cases, H1299 cells growing in 100-mm-diameter dishes were infected with adenovirus vectors, whereas in other cases, H1299 cells were seeded in 60-mm plates and lipofected according to the GIBCO BRL Lipofection protocol with 2 μg total of plasmid DNA. After 24 h, whole cell extracts were prepared in buffer X, immunoprecipitations were performed with the indicated antibodies, and samples were separated by SDS-PAGE. Following transfer to nitrocellulose (Schleicher & Schuell/Optitran), Western blotting was performed, and species were visualized by enhanced chemiluminescence (NEN LifeScience). For the binding assays of E4orf6 with the various Cullins, immunoprecipitates were prepared from AdLacZ- or AdE4orf6-infected H1299 cells using 1D5 anti-E4orf6 antibody. Following the final wash, the precipitates were divided into five equal portions, resuspended in 100 μL of buffer X and combined with a 10 μL aliquot of samples containing various Cullins, human Cul1, Cul2, Cul3, and Cul5 and mouse Cul4A (Michel and Xiong 1998), which were synthesized using a standard in vitro transcription/translation kit (TnT, Promega) in the presence of 40 μCi of translation grade [35S]methionine (Dupont-NEN). The mixtures were incubated at room temperature for 1 h, and after five washes with buffer X, the precipitates were separated by SDS-PAGE. The presence of labeled Cullins was assessed by autoradiography.
In vivo p53 degradation assays
The E4orf6/E1B55K-induced p53 turnover was studied using either plasmids or adenovirus vectors to introduce the viral proteins and p53 into cells, as described more fully in Querido et al. (2001). H1299 cells were grown on 60-mm dishes and transfected using 12 μL of Lipofectin reagent (GIBCO BRL) with the following plasmid DNAs: 1 μg of pcDNA3 p53wt, 4 μg of pCA14 HH55K, and 4 μg of pcDNA3 E4orf6 (expressing wild-type or mutant E4orf6 proteins). Total plasmid DNA was kept equal by adding empty pcDNA3 plasmid DNA. Expression from all plasmid DNAs was under the CMV promoter. After 24 h, cells were harvested and lysed in Western blot lysis buffer (50 mM HEPES at pH 7.9, containing 400 mM KCl, 0.1% NP-40, 4 mM NaF, 4 mM NaVO4, 0.2 mM EDTA, 0.2 mM EGTA, 2 mg/mL aprotinin, leupeptin, and pepstatin, and 1 mM dithiothreitol). Protein concentrations were measured by the Bradford assay (Bio-Rad), and 40 μg of each extracts was separated by SDS-PAGE and immunoblotted with anti-p53, anti-E1B55K, or anti-E4orf6 antibody. The E4orf6 immunoblots were checked for the equivalent expression of each E4orf6 mutant, and the E1B55K expression, which did not vary, allowed us to verify equal transfection efficiency. Equal sample loading was verified by staining the membranes with Ponceau S red (Sigma) after transfer. Wild-type CHO cells and ts41 cells (Hirschberg and Marcus 1982) were infected with various combinations of adenovirus vectors at an MOI of 90 pfu per cell for each virus. Vectors included AdrTTA, Adp53wt, AdE4orf6, and AdHH55K. The total amount of virus was kept constant at 270 pfu per cell by adding AdrTTA (vector control) virus as needed. Cells were harvested 48 h after infection, and cell extracts were prepared and analysed as above. Western blotting was performed with anti-p53, anti-E1B55K, anti-E4orf6, or anti-actin antibody. In some cases, wild-type CHO cells were also treated with proteasome inhibitors MG132 (20 μM, Calbiochem), lactacysteine (20 μM, Calbiochem) or their solvents, ethanol (0.2%) and DMSO (0.2%) for the last 6 h of infection.
Reconstitution of E4orf6 complexes in Sf21 cells
Plates containing 1 × 106 Sf21 cells were infected with baculoviruses encoding various proteins. Cells were lysed in 1 mL of 40 mM HEPES-NaOH (pH 7.9), containing 150 mM NaCl, 1 mM DTT, 0.5% (v/v) Triton X-100, 10% (v/v) glycerol, 5 μg/mL leupeptin, 5 μg/mL antipain, 5 μg/mL pepstatin A, and 5 μg/mL aprotinin. Approximately 100 μg of Sf21 cell lysate was immunoprecipitated with either 2 μg of anti-HA (12CA5) or 2 μg of anti-E4orf6 (1D5) antibody and 10 μL of protein A Sepharose. Crude lysates and washed immune complexes were separated by SDS-PAGE and immunoblotted with the indicated antibodies.
In vitro p53 ubiquitination assay
Lysates from Sf21 cells infected with baculoviruses encoding E1B55K, Elongin B, Elongin C, HA–Cul5, and Rbx1, with or without E4orf6 baculovirus, were immunoprecipitated with 1D5 antibody. To the precipitates was added 0, 20, or 65 ng of recombinant histidine-tagged p53 protein (Wang et al. 1993) that had been expressed in insect cells and purified by nickel chromatography, as described previously (Kamura et al. 1999) and the mixtures were incubated with 50 ng of yUba1, 100 ng of hUbc5a, or 500 ng of GST–UbK48R in 10 μL of ubiquitination reaction buffer (40 mM HEPES-NaOH at pH 7.9, containing 60 mM potassium acetate, 2 mM DTT, 6 mM MgCl2, and 0.5 mM EDTA at pH 7.9, 10% [v/v] glycerol, and 1.5 mM ATP). Reaction mixtures were incubated for 30 min at 25°C, separated by SDS-PAGE, and analyzed by Western blotting with anti-p53 antibody (pAB421).
Confocal microscopy
H1299 cells were seeded in four-chamber culture slides (Falcon) and transfected with 0.4 μg of plasmid DNA according to the GIBCO BRL Lipofection protocol. After 24 h, cells were fixed with 4% paraformaldehyde and permeabilized with PBS 0.1% Triton X-100. A solution of PBS with 3% bovine serum albumin was used for blocking and antibody incubation. E4orf6 was detected using rabbit polyclonal 1807 antibody and Alexa 594 (red) anti-rabbit IgG antibody (Molecular Probes). Recombinant Cullin proteins tagged with the HA epitope were detected using mouse monoclonal anti-HA.11 antibody (BAbCO) and Alexa 488 (green) anti-mouse antibody (Molecular Probes). The fluorophores were visualized using a laser confocal Zeiss microscope.
Acknowledgments
We wish to thank Rachael Neve for ts41 cells; Maria Burnatowska-Hledin for the rabbit VACM-1 antibodies; Yue Xiong for plasmids expressing human Cul1, Cul2, Cul3, and Cul5 and murine Cul4A; Peter Tegtmeyer for the p53 baculovirus; Wilhelm Krek for the plasmid expressing HA–E2Cdc34; and Dennis Takayesu for technical assistance. We also thank Ning Zheng and Nikola Pavletich for information about their structure of the SCF complex before publication. This work was supported by grants from the Canadian Institutes of Health Research and the National Cancer Institute of Canada (P.E.B.) and the National Institute of General Medicine (GM41628, R.C.C.). This work was performed while J.W.C. was an associate investigator of the Howard Hughes Medical Institute. E.Q. was supported by a studentship from the Fonds FRSQ-FCAR-Santé du Québec; and P.B., a postdoctoral fellowship from Défi Corporatif Canderel.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL branton@med.mcgill.ca; FAX (514) 398-8845.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.926401.
References
- Aso T, Haque D, Barstead RJ, Conaway RC, Conaway JW. The inducible elongin A elongation activation domain: Structure, function, and interaction with the elongin BC complex. EMBO J. 1996;15:5557–5566. [PMC free article] [PubMed] [Google Scholar]
- Bacchetti S, Graham FL. Inhibition of cell proliferation by an adenovirus vector expressing the human wild type p53 protein. Int J Oncol. 1993;3:781–788. doi: 10.3892/ijo.3.5.781. [DOI] [PubMed] [Google Scholar]
- Bargonetti J, Reynisdottir I, Friedman PN, Prives C. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes & Dev. 1992;6:1886–1898. doi: 10.1101/gad.6.10.1886. [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas J. p27 destruction: Cks1 pulls the trigger. Nat Cell Biol. 2001;3:E95–E98. doi: 10.1038/35070160. [DOI] [PubMed] [Google Scholar]
- Bech-Otschir D, Kraft R, Huang X, Henklein P, Kapelari B, Pollmann C, Dubiel W. COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J. 2001;20:1630–1639. doi: 10.1093/emboj/20.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bett AJ, Haddara W, Prevec L, Graham FL. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc Natl Acad Sci. 1994;91:8802–8806. doi: 10.1073/pnas.91.19.8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin D, Morrison MR, Marcellus RC, Querido E, Branton PE. Analysis of synthesis, stability, phosphorylation, and interacting polypeptides of the 34-kilodalton product of open reading frame 6 of the early region 4 protein of human adenovirus type 5. J Virol. 1999;73:1245–1253. doi: 10.1128/jvi.73.2.1245-1253.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower CS, Shilatifard A, Mather T, Kamura T, Takagi Y, Haque D, Treharne A, Foundling SI, Conaway JW, Conaway RC. The elongin B ubiquitin homology domain: Identification of Elongin B sequences important for interaction with Elongin C. J Biol Chem. 1999;274:13629–13636. doi: 10.1074/jbc.274.19.13629. [DOI] [PubMed] [Google Scholar]
- Burnatowska-Hledin M, Spielman WS, Smith WL, Shi P, Meyer JM, Dewitt DL. Expression cloning of an AVP-activated calcium-mobilizing receptor from rabbit kidney medulla. Am J Physiol. 1995;268:F1198–F1210. doi: 10.1152/ajprenal.1995.268.6.F1198. [DOI] [PubMed] [Google Scholar]
- Burnatowska-Hledin M, Lazdins IB, Listenberger L, Zhao P, Sharangpani A, Folta V, Card B. VACM-1 receptor is specifically expressed in rabbit vascular endothelium and renal collecting tubule. Am J Physiol. 1999;276:F199–F209. doi: 10.1152/ajprenal.1999.276.2.F199. [DOI] [PubMed] [Google Scholar]
- Byrd PJ, Stankovic T, McConville CM, Smith AD, Cooper PR, Taylor AM. Identification and analysis of expression of human VACM-1, a cullin gene family member located on chromosome 11q22-23. Genome Res. 1997;7:71–75. doi: 10.1101/gr.7.1.71. [DOI] [PubMed] [Google Scholar]
- Cathomen T, Weitzman MD. A functional complex of adenovirus proteins E1B-55kDa and E4orf6 is necessary to modulate the expression level of p53 but not its transcriptional activity. J Virol. 2000;74:11407–11412. doi: 10.1128/jvi.74.23.11407-11412.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, McPhie DL, Hirschberg J, Neve RL. The amyloid precursor protein-binding protein APP-BP1 drives the cell cycle through the S-M checkpoint and causes apoptosis in neurons. J Biol Chem. 2000;275:8929–8935. doi: 10.1074/jbc.275.12.8929. [DOI] [PubMed] [Google Scholar]
- Chiou S-K, White E. p300 binding by E1A cosegregates with p53 induction but is dispensable for apoptosis. J Virol. 1997;71:3515–3525. doi: 10.1128/jvi.71.5.3515-3525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockman ME, Masson N, Mole DR, Jaakkola P, Chang G-W, Clifford SC, Maher ER, Pugh CW, Ratcliffe PJ, Maxwell PH. Hypoxia inducible factor-α binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein J. Biol Chem. 2000;275:25733–25741. doi: 10.1074/jbc.M002740200. [DOI] [PubMed] [Google Scholar]
- Dobbelstein M, Roth J, Kimberly WT, Levine AJ, Shenk T. Nuclear export of the E1B-55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;16:4276–4282. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- Dosch T, Horn F, Schneider G, Kratzer F, Dobner T, Hauber J, Stauber RH. The adenovirus type 5 E1B-55K oncoprotein actively shuttles in virus-infected cells, whereas transport of E4orf6 is mediated by a CRM1-independent mechanism. J Virol. 2001;75:5677–5683. doi: 10.1128/JVI.75.12.5677-5683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan DR, Pause A, Burgess WH, Aso T, Chen DY, Garrett KP, Conaway RC, Conaway JW, Linehan WM, Klausner RD. Inhibition of transcriptional elongation by the VHL tumor suppressor protein. Science. 1995;269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- Freed E, Lacey KR, Huie P, Lyapina SA, Deshaies RJ, Stearns T, Jackson PK. Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle. Genes & Dev. 1999;13:2242–2257. doi: 10.1101/gad.13.17.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SY, Adler V, Buschmann T, Yin Z, Wu X, Jones SN, Ronai Z. JNK targets p53 ubiquitination and degradation in nonstressed cells. Genes & Dev. 1998;12:2658–2663. doi: 10.1101/gad.12.17.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen H, Shkedy D, Barnov S, Kosower NS, Ciechanover A. On the involvement of calpains in the degradation of the tumor suppressor protein p53. FEBS Lett. 1997;406:17–22. doi: 10.1016/s0014-5793(97)00225-1. [DOI] [PubMed] [Google Scholar]
- Gong L, Yeh ETH. Identification of the activating and conjugating enzyme of the NEDD conjugation pathway. J Biol Chem. 1999;274:12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- Goodrum FD, Shenk T, Ornelles DA. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J Virol. 1996;70:6323–6335. doi: 10.1128/jvi.70.9.6323-6335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- Handeli S, Weintraub H. The ts41 mutation in Chinese hamster cells leads to successive phases in the absence of intervening G2, M, and G1. Cell. 1992;71:599–611. doi: 10.1016/0092-8674(92)90594-3. [DOI] [PubMed] [Google Scholar]
- Harlow E, Franza BR, Jr, Schley C. Monoclonal antibodies specific for adenovirus early region 1A proteins: Extensive heterogeneity in early region E1A products. J Virol. 1985;55:533–546. doi: 10.1128/jvi.55.3.533-546.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg J, Marcus M. Isolation by a replica-plating technique of Chinese hamster temperature-sensitive cell cycle mutants. J Cell Physiol. 1982;113:159–166. doi: 10.1002/jcp.1041130125. [DOI] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Hori T, Osaka F, Chiba T, Miyamoto C, Okabayashi K, Shimbara N, Kato S, Tanaka K. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene. 1999;18:6829–6834. doi: 10.1038/sj.onc.1203093. [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFα targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Iwai K, Yamanaka K, Kamura T, Minato N, Conaway RC, Conaway JW, Klausner RD, Pause A. Identification of the von Hippel-Lindau tumor suppressor protein as the substrate recognition subunit of an E3 ubiquitin ligase complex. Proc Natl Acad Sci. 1999;96:12436–12441. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- Kamura T, Sato S, Haque D, Liu L, Kaelin WG, Conaway RC, Conaway JW. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes & Dev. 1998;12:3872–3881. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Elledge SJ, Conaway RC, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- Kamura T, Sato S, Iwai K, Czyzyk-Krzeska M, Conaway RC, Conaway JW. Activation of HIF1α ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc Natl Acad Sci. 2000;97:10430–10435. doi: 10.1073/pnas.190332597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Burian D, Yan Q, Schmidt SL, Lane WS, Querido E, Branton PE, Shilatifard A, Conaway RC, Conaway JW. MUF1, a novel elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J Biol Chem. 2001;276:29748–29753. doi: 10.1074/jbc.M103093200. [DOI] [PubMed] [Google Scholar]
- Kibel A, Iliopoulos O, DeCaprio JA, Kaelin WG. Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- Kratzer F, Rosorius O, Heger P, Hirschmann N, Dobner T, Hauber J, Stauber RH. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene. 2000;19:850–857. doi: 10.1038/sj.onc.1203395. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Vousden KH. Proteolytic cleavage of human p53 by calpain: A potential regulator of protein stability. Mol Cell Biol. 1997;17:460–468. doi: 10.1128/mcb.17.1.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lain S, Midgley C, Sparks A, Lane EB, Lane DP. An inhibitor of nuclear export activates the p53 response and induces the localization of HDM2 and p53 to U1A-positive nuclear bodies associated with the PODs. Exp Cell Res. 1999;248:457–472. doi: 10.1006/excr.1999.4433. [DOI] [PubMed] [Google Scholar]
- Liakopoulos D, Busgen T, Brychzy A, Jentsch S, Pause A. Conjugation of the ubiquitin-like NEDD8 to cullin-2 is linked to von Hippel-Lindau tumor suppressor function. Proc Natl Acad Sci. 1999;96:5510–5515. doi: 10.1073/pnas.96.10.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisztwan J, Marti A, Sutterluty H, Gstaiger M, Wirbelauer C, Krek W. Association of human CUL-1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein p45(SKP2): Evidence for evolutionary conservation in the subunit composition of the CDC34–SCF pathway. EMBO J. 1998;17:368–383. doi: 10.1093/emboj/17.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisztwan J, Imbert G, Wirbelauer C, Gstaiger M, Krek W. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin–protein ligase activity. Genes & Dev. 1999;13:1822–1833. doi: 10.1101/gad.13.14.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang G-W, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Michel JJ, Xiong Y. Human CUL-1, but not other cullin family members, selectively interacts with SKP1 to form a complex with SKP2 and cyclin A. Cell Growth Differ. 1998;9:435–449. [PubMed] [Google Scholar]
- Mitsudomi T, Steinberg SM, Nau MM, Carbone D, D'Amico D, Bodner S, Oie HK, Linnoila RI, Mulshine JL, Minna JD, et al. p53 gene mutations in non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene. 1992;7:171–180. [PubMed] [Google Scholar]
- Moore M, Horikoshi N, Shenk T. Oncogenic potential of the adenovirus E4orf6 protein. Proc Natl Acad Sci. 1996;93:11295–11301. doi: 10.1073/pnas.93.21.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevels M, Rubenwolf S, Spruss T, Wolf H, Dobner T. Two distinct activities contribute to the oncogenic potential of the adenovirus type 5 E4orf6 protein. J Virol. 2000;74:5168–5181. doi: 10.1128/jvi.74.11.5168-5181.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohh M, Park CW, Ivan M, Hoffman MA, Kim T-Y, Huang LE, Pavletich N, Chau V, Kaelin WG. Ubiquitination of hypoxia-inducible factor requires direct binding to the β-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- Ohta T, Xiong Y. Phosphorylation- and SKP1-independent in vitro ubiquitination of E2F1 by multiple ROC-cullin ligases. Cancer Res. 2001;61:1347–1353. [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- Orlando JS, Ornelles DA. An arginine-faced amphipathic α-helix is required for adenovirus type 5 E4orf6 protein function. J Virol. 1999;73:4600–4610. doi: 10.1128/jvi.73.6.4600-4610.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka F, Kawasaki H, Aida N, Saeki M, Chiab T, Kawashima S, Tanaka K, Kato S. A new NEDD8-ligating system for cullin-4a. Genes & Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: Don't Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- Querido E, Marcellus RC, Lai A, Charbonneau R, Teodoro JG, Ketner G, Branton PE. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J Virol. 1997a;71:3788–3798. doi: 10.1128/jvi.71.5.3788-3798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querido E, Teodoro JG, Branton PE. Accumulation of p53 induced by the adenovirus E1A protein requires regions involved in the stimulation of DNA synthesis. J Virol. 1997b;71:3526–3533. doi: 10.1128/jvi.71.5.3526-3533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querido E, Morisson MR, Chu-Pham-Dang H, Thirlwell SW, Boivin D, Branton PE. Identification of three functions of the adenovirus E4orf6 protein that mediate p53 degradation by the E4orf–E1B55K complex. J Virol. 2001;75:699–709. doi: 10.1128/JVI.75.2.699-709.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabino C, Aspegren A, Corbin-Lickfett K, Bridge E. Adenovirus late gene expression does not require a Rev-like nuclear RNA export pathway. J Virol. 2000;74:6684–6688. doi: 10.1128/jvi.74.14.6684-6688.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D, Conaway RC, Conaway JW. Mechanism and regulation of transcriptional elongation by RNA polymerase II. Curr Opin Cell Biol. 1999;11:342–346. doi: 10.1016/S0955-0674(99)80047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J, Dobbelstein M, Freedman DA, Shenk T, Levine AJ. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulston A, Marcellus RC, Branton PE. Viruses and apoptosis. Annu Rev Microbiol. 1999;53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]
- Samuelson AV, Lowe SW. Selective induction of p53 and chemosensitivity in RB-deficient cells by E1A mutants unable to bind the RB-related proteins. Proc Natl Acad Sci. 1997;94:12094–12099. doi: 10.1073/pnas.94.22.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnow P, Ho YS, Williams J, Levine AJ. Adenovirus E1B 58kD tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kD cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- Sarnow P, Hearing P, Anderson CW, Halbert DN, Shenk T, Levine AJ. Adenovirus early region 1B 58,000-dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J Virol. 1984;49:692–700. doi: 10.1128/jvi.49.3.692-700.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6–AP complex functions as a ubiquitin–protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes & Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Kitzes G, Nye JA, Fattaey A, Hermiston T. Analyses of single-amino-acid substitution mutants of adenovirus type 5 E1B-55K protein. J Virol. 2001;75:4297–4307. doi: 10.1128/JVI.75.9.4297-4307.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstruction of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- Spruck C, Strohmaier H, Watson M, Smith APL, Ryan A, Krek W, Reed SI. A CDK-independent function of mammalian Cks1: Targeting of SCFSkp2 to the CDK inhibitor p27Kip1. Mol Cell. 2001;7:639–650. doi: 10.1016/s1097-2765(01)00210-6. [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Kaelin WG, Pavletich NP. Structure of the VHL–Elongin C–Elongin B complex: Implications for VHL tumor suppressor function. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- Steegenga WT, Riteco N, Jochemsen AG, Fallaux FJ. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene. 1998;16:349–357. doi: 10.1038/sj.onc.1201540. [DOI] [PubMed] [Google Scholar]
- Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan Z-Q. Recruitment of a ROC1–CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- Teodoro JG, Branton PE. Regulation of p53-dependent apoptosis, transcriptional repression, and cell transformation by phosphorylation of the 55-kilodalton E1B protein of human adenovirus type 5. J Virol. 1997;71:3620–3627. doi: 10.1128/jvi.71.5.3620-3627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Yeh ET, Kamitani T. Identification of NEDD8-conjugation site in human cullin-2. Biochem Biophys Res Commun. 1999;257:100–105. doi: 10.1006/bbrc.1999.0339. [DOI] [PubMed] [Google Scholar]
- Wang Y, Reed M, Wang P, Stenger JE, Mayr G, Anderson ME, Schwedes JF, Tegtmeyer P. p53 domains: Identification and characterization of two autonomous DNA-binding regions. Genes & Dev. 1993;7:2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- Weigel S, Dobbelstein M. The nuclear export signal within the E4orf6 protein of adenovirus type 5 supports virus replication and cytoplasmic accumulation of viral mRNA. J Virol. 2000;74:764–772. doi: 10.1128/jvi.74.2.764-772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- Woods DB, Vousden KH. Regulation of p53 function. Exp Cell Res. 2001;264:56–66. doi: 10.1006/excr.2000.5141. [DOI] [PubMed] [Google Scholar]
- Yeh ET, Gong L, Katani T. Ubiquitin-like proteins: New wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- Yew PR, Berk AJ. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- Yew PR, Liu X, Berk AJ. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes & Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- Zhang JG, Farley A, Nicholson SE, Willson TA, Zugaro LM, Simpson RJ, Moritz RL, Cary D, Richardson R, Hausmann G, et al. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc Natl Acad Sci. 1999;96:2071–2076. doi: 10.1073/pnas.96.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Lu Q, Xie ZJ, Mellgren RL. Inhibition of the growth of WI-38 fibroblasts by benzyloxycarbonyl-Leu-Leu-Tyr-diazomethyl ketone: Evidence that cleavage of p53 by a calpain-like protease is necessary for G1 to S-phase transition. Oncogene. 1997;14:255–263. doi: 10.1038/sj.onc.1200841. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y. Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell Growth Differ. 2001;12:175–186. [PubMed] [Google Scholar]