Abstract
Cells defective in any of the RAD51 paralogs (RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3) are sensitive to DNA cross-linking agents and to ionizing radiation. Because the paralogs are required for the assembly of DNA damage-induced RAD51 foci, and mutant cell lines are defective in homologous recombination and show genomic instability, their defect is thought to be caused by an inability to promote efficient recombinational repair. Here, we show that the five paralogs exist in two distinct complexes in human cells: one contains RAD51B, RAD51C, RAD51D, and XRCC2 (defined as BCDX2), whereas the other consists of RAD51C with XRCC3. Both protein complexes have been purified to homogeneity and their biochemical properties investigated. BCDX2 binds single-stranded DNA and single-stranded gaps in duplex DNA, in accord with the proposal that the paralogs play an early (pre-RAD51) role in recombinational repair. Moreover, BCDX2 complex binds specifically to nicks in duplex DNA. We suggest that the extreme sensitivity of paralog-defective cell lines to cross-linking agents is owing to defects in the processing of incised cross links and the consequential failure to initiate recombinational repair at these sites.
Keywords: Recombination, DNA repair, genomic instability
Genome stability requires the efficient repair of double-strand breaks (DSBs) that arise during DNA replication or are caused by DNA-damaging agents. Vertebrate cells use two main mechanisms of repair, known as nonhomologous end joining (NHEJ) and homologous recombination (HR). NHEJ is a relatively simple process that promotes religation of the broken DNA, whereas homologous recombination is rather more complex because it involves interactions with the sister chromatid, which acts as a template for repair (van Gent et al. 2001). This requirement means that HR is particularly effective in S-phase as the DNA undergoes replication (Liang et al. 1998; Takata et al. 1998; Essers et al. 2000), and that replication-induced breaks are likely to be an important initiator of HR (Cox et al. 2000).
In the late 1980s, two cell lines (irs1 and irs1SF) were identified that were sensitive to ionizing radiation, UV, and cross-linking agents (Jones et al. 1987; Fuller and Painter 1988). The genes responsible for the defects were cloned by complementation and are known as XRCC2 and XRCC3, respectively (Tebbs et al. 1995; Tambini et al. 1997). Mutant cell lines are deficient in homologous recombination (Johnson et al. 1999; Pierce et al. 1999), mildly sensitive (twofold) to ionizing radiation, and show high levels of chromosomal aberrations both in control and irradiated cultures (Liu et al. 1998; Cui et al. 1999; Deans et al. 2000), properties that are suggestive of the accumulation of unrepaired replication-induced DSBs (Brenneman et al. 2000). The XRCC2 and XRCC3 mutants are unusually sensitive (60-fold to 100-fold) to damaging agents like cisplatin, nitrogen mustard, or mitomycin C that introduce cross links in DNA.
Sequence analyses of the XRCC2 and XRCC3 genes revealed that the protein products share significant sequence homology with the cellular recombinase RAD51, and are likely to have arisen through gene duplication and divergent evolution (Cartwright et al. 1998b; Liu et al. 1998). It was therefore rather surprising when three more RAD51 paralogs were identified by database analyses (Albala et al. 1997; Rice et al. 1997; Cartwright et al. 1998a; Dosanjh et al. 1998; Pittman et al. 1998). These genes are known as RAD51B (also known as RAD51L1, hREC2, R51H2), RAD51C (RAD51L2), and RAD51D (RAD51L3 or R51H3). All five RAD51 paralogs have been disrupted in chicken DT40 cells and mutants shown to exhibit similar recombination/repair defective phenotypes (Takata et al. 2000, 2001). All show reduced growth rates, exhibit chromosomal instability, and accumulate spontaneous breaks presumably attributable to an inability to repair broken replication forks. In the mouse, disruption of RAD51B, RAD51D, or XRCC2 confers embryonic lethality (Shu et al. 1999; Deans et al. 2000; Pittman and Schimenti 2000).
Although no direct homologs of RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3 exist in simple eukaryotes, significant homologies exist between the RAD51 paralogs and the yeast RAD55 and RAD57 proteins. XRCC2 is closest to RAD55, and RAD51D and XRCC3 are closest to RAD57 (Thacker 1999; Tsutsui et al. 2000). In yeast, defects in RAD55 or RAD57 result in radiation sensitivity, a phenotype that can be partially complemented by overexpression of RAD51 protein (Hays et al. 1995). Biochemical studies of RAD55 and RAD57 show that the two proteins form a heterodimer that interacts with RAD51 and stimulates RAD51-mediated pairing and strand-exchange reactions (Sung 1997b). The heterodimer is thought to facilitate assembly of the RAD51 nucleoprotein filament that initiates recombination. The vertebrate RAD51 paralogs may play a similar role, because the formation of DNA-damage-induced subnuclear RAD51 foci (sites where repair is thought to occur) is dependent on the functions of the RAD51 paralogs (Bishop et al. 1998; Takata et al. 2000, 2001; Gasior et al. 2001; Sonoda et al. 2001). Moreover, one of the paralogs, XRCC3, interacts with RAD51 (Liu et al. 1998; Schild et al. 2000), and the DNA repair defects associated with mutations in the RAD51 paralogs can be partially overcome by RAD51 overexpression (Takata et al. 2001).
At the present time, very little is known about the cellular functions of the RAD51 paralogs. A number of protein–protein interactions have been shown, suggesting that the five proteins might function as a complex (Schild et al. 2000). Biochemical studies indicate that the individual proteins suffer insolubility problems in isolation, but it has been possible to purify RAD51D (Braybrooke et al. 2000) and a complex containing RAD51C–XRCC3 (Kurumizaka et al. 2001; Masson et al. 2001). RAD51D was shown to bind single-stranded DNA (ssDNA) and have a DNA-stimulated ATPase activity. Similarly, the RAD51C–XRCC3 complex bound ssDNA and was shown to promote DNA–DNA interactions resulting in DNA annealing. At the present time, however, it is not known which RAD51 paralog complexes represent the biologically relevant forms of the protein that are found in vivo.
In this work, we show that the RAD51 paralogs are found in two distinct complexes in human cells. One complex contains RAD51B, RAD51C, RAD51D, and XRCC2 (here called the BCDX2 complex), whereas the second contains RAD51C with XRCC3. The nature of these complexes has been confirmed by overexpression of all five proteins using baculovirus systems, and both complexes have been purified to homogeneity. We describe the properties of the BCDX2 complex and show that it binds specifically to single-stranded DNA, and to single-stranded regions in duplex DNA, suggestive of a presynaptic role in recombination. Most importantly, BCDX2 binds to nicks in duplex DNA, leading us to propose that BCDX2 is capable of the specific recognition of incised cross links, thereby providing the necessary link between incision by nucleotide excision repair enzymes and recombinational repair.
Results
The RAD51 paralogs form two distinct protein complexes
In previous studies, yeast two-hybrid systems were used to show a series of interactions between the human RAD51 paralogs, including (1) RAD51B with RAD51C, (2) RAD51C with XRCC3, (3) RAD51C with RAD51D, (4) RAD51D with XRCC2, and (5) XRCC3 with RAD51 (Dosanjh et al. 1998; Liu et al. 1998; Braybrooke et al. 2000; Schild et al. 2000). To determine whether these interactions are representative of those that take place in human cells, all five RAD51 paralog genes were cloned from a human cDNA library into baculovirus vectors, and monoclonal (mAbs) and polyclonal (pAbs) antibodies were raised against the purified denatured proteins. The pAbs were then used to immunoprecipitate potential RAD51 paralog complexes from HeLa cell-free extracts, and the interacting proteins were identified by Western blotting using mAbs.
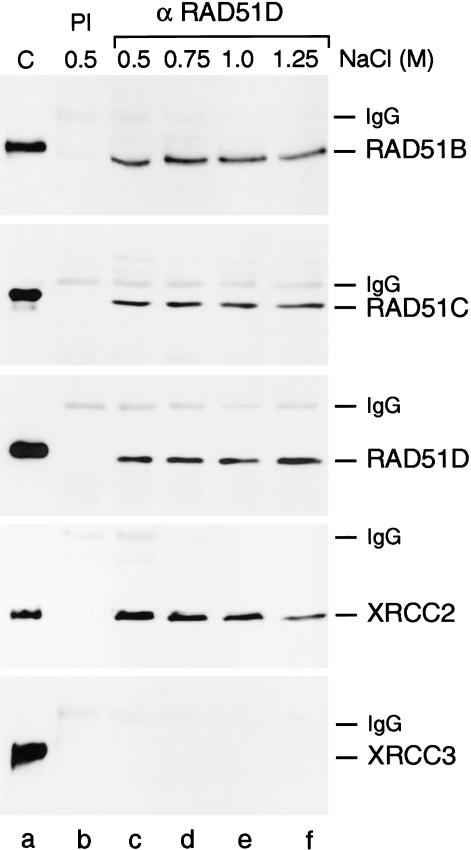
In the most striking experiment, shown in Figure 1, we observed that anti-RAD51D pAbs pulled down endogenous RAD51D together with RAD51B, RAD51C, and XRCC2. The interactions were strong and complex formation was resistant to 1.25 M NaCl (Fig. 1, lanes c–f). There was, however, no evidence that the fifth paralog, XRCC3, was part of this complex. The failure to detect XRCC3 in this complex was not simply due to a problem of detection (e.g., low amounts of protein or antibody affinity), because anti-RAD51C pAbs pulled down endogenous RAD51C and XRCC3 under identical conditions (Masson et al. 2001). Preimmune sera failed to pull down any of the RAD51 paralogs (Fig. 1, lane b).
Figure 1.
Interactions between the RAD51 paralogs in HeLa cell-free extracts. Coimmunoprecipitation of endogenous RAD51B, RAD51C, RAD51D, and XRCC2 from HeLa cell-free extracts. Protein complexes were precipitated using preimmune serum (PI; lane b) or anti-RAD51D pAbs (lanes c–f). The complexes were washed in buffer containing NaCl, as indicated, and visualized by Western blotting using anti-RAD51B, anti-RAD51C, anti-RAD51D, anti-XRCC2, or anti-XRCC3 mAbs, as indicated. (Lane a) Marker proteins (RAD51Bhis6, RAD51Chis10, RAD51Dhis6, XRCC2, XRCC3his6). The His-tagged controls migrate more slowly than the endogenous human proteins.
These results lead us to suggest that there are two distinct complexes containing the RAD51 paralogs in HeLa cells: one complex containing RAD51B, RAD51C, RAD51D, and XRCC2 (the BCDX2 complex), and a second complex containing RAD51C and XRCC3. This finding was supported by pull-down experiments using RAD51B or XRCC2 pAbs that again brought down RAD51B, RAD51C, RAD51D, and XRCC2, but not XRCC3 (data not shown). As expected, RAD51C pAbs pulled down all five paralogs. Using anti-RAD51 pAbs, we failed to pull down any of the RAD51 paralogs from HeLa extracts, and RAD51 was not found in association with the BCDX2 and RAD51C–XRCC3 complexes, as determined in RAD51C pAb pull downs (data not shown).
To provide further evidence for the specific nature of the RAD51C–XRCC3 interaction, Sf9 insect cells were multiply infected with baculovirus encoding all five paralog proteins, one of which (XRCC3) carried a histidine tag at the N terminus. Protein extracts were loaded onto a Talon affinity column, which retained XRCC3-His6 and associated proteins, while allowing nonassociated proteins to flow through. We found that the column bound XRCC3-his6 and RAD51C (Fig. 2, lanes h–j), but not RAD51B, RAD51D, and XRCC2, all of which were present in the flow through (Fig. 2, lane a). A significant portion of the RAD51C, however, appeared with RAD51B, RAD51D, and XRCC2 in the column flow through, again supporting the notion that RAD51C is involved in two complexes: one containing BCDX2 and the other containing RAD51C and XRCC3.
Figure 2.
Specific interactions between XRCC3 and RAD51C. Sf9 cells were coinfected with pBAC51B, pBAC51C, pBAC51D, pBACX2, and pBACX3his6. Following protein expression, extracts were loaded onto a Talon column to bind XRCC3-His6 and associated proteins, which were subsequently eluted and identified by Western blotting using mAbs. (Lane a) Flow through; (lane b) 30 mM imidazole wash; (lanes c–j) fractions 4–18.
To provide more direct evidence for the two complexes, extracts from Sf9 cells multiply infected with pBAC51B, pBAC51Chis10, pBAC51D, pBACX2, and pBACX3his6 were passed through a Superdex 200 gel filtration column. The five paralog proteins eluted with a broad profile, with the bulk of the RAD51B, RAD51D, and XRCC2 present in fractions 22/23 (Fig. 3A). A considerable amount of the RAD51C also eluted in these fractions, and in addition RAD51C was found in fractions 24/25, where it coeluted with XRCC3. These results are consistent with the presence of two overlapping peaks, confirming the presence of the two complexes indicated schematically in Figure 3B.
Figure 3.
Complex formation by the RAD51 paralogs. (A) Gel filtration analysis of RAD51 paralog complexes. Extracts from Sf9 cells infected with pBAC51B, pBAC51Chis10, pBAC51D, pBACX2, and pBACX3his6 were analyzed by gel filtration as described in Materials and Methods. (B) Diagrammatic representation of two distinct complexes as indicated by the data presented in Figures 1 and 2 and part A.
The protein peak containing the BCDX2 complex (fractions 22/23) showed a molecular mass corresponding to ∼180 kD. Because the calculated molecular mass of BCDX2 is 146 kD, and we have not observed self-associations between any of the RAD51 paralogs, these results most likely indicate a 1:1:1:1 stoichiometry of the four subunits. The gel filtration profile of the RAD51Chis10–XRCC3his6 complex (calculated molecular mass 85 kD) was indicative of a heterodimer between the proteins. In the experiment shown in Figure 3A very little monomeric protein was detected, except for some RAD51D, which eluted in fraction 29. The elution profile of the complexes was essentially the same in buffer containing 1.0 M NaCl, indicative of stable interactions between the RAD51 paralogs (data not shown).
Dissection of interactions within the BCDX2 complex
To analyze protein–protein interactions within the BCDX2 complex we used a similar approach to that shown in Figure 2. Sf9 cells were infected with various baculovirus combinations, and in each case one of the expressed proteins was histidine-tagged for absorption to Talon. Proteins that interacted with the single tagged paralog were then identified by Western blotting. We found that RAD51B (Fig. 4A) and RAD51D (Fig. 4B) were individually bound by RAD51Chis10, and that RAD51B and RAD51D could be bound simultaneously by RAD51Chis10 (Fig. 4C). In the absence of RAD51C, however, interactions between RAD51Bhis6 and RAD51D were not observed (Fig. 4D). Similarly, we failed to detect interactions between XRCC2his6 and RAD51B or RAD51C in the absence of RAD51D (Fig. 4E), whereas RAD51Chis10, RAD51B, RAD51D, and XRCC2 all interacted together and were bound by Talon (data not shown).
Figure 4.
Dissection of the interactions between RAD51C, RAD51B, RAD51D, and XRCC2. Sf9 cells were infected with the indicated baculovirus and extracts were loaded onto a Talon column to bind the single His-tagged protein and associated proteins. The interacting proteins were eluted and identified by Western blotting using mAbs. The column flow through (FT), wash (W), and eluting fractions (4–20) are indicated. A summary of interactions is shown diagrammatically at the right. (A) Sf9 infected with pBAC51Chis10 and pBAC51B. (B) pBAC51Chis10 with pBAC51D. (C) pBAC51Chis10 expressed with both pBAC51B and pBAC51D. Blot probed with RAD51C mAbs (top panel), and with RAD51B and RAD51D mAbs (lower panel). (D) pBAC51Bhis6 with pBAC51D. (E) pBACX2his6 with both pBAC51B and pBAC51C. Blot probed with RAD51B and XRCC2 mAbs (upper panel), and with RAD51C and XRCC2 mAbs (lower panel).
Together, these results indicate that the architecture of the BCDX2 complex is as follows: RAD51B interacts specifically with RAD51C, which in turn interacts with RAD51D, and RAD51D interacts with XRCC2 (as indicated in the diagram of Fig. 3B). When RAD51 was expressed in the presence of RAD51B, RAD51Chis10, RAD51D, XRCC2, and XRCC3, and extracts were applied to a Talon column, the RAD51 was found in the column flow through (data not shown). The lack of direct interaction between RAD51 and BCDX2 confirmed our earlier immunoprecipitation analyses using HeLa extracts.
Purification of the two RAD51 paralog complexes
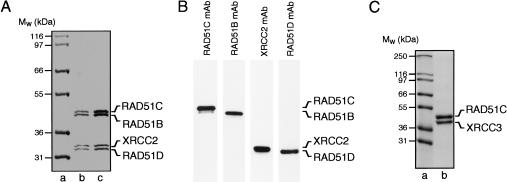
The BCDX2 complex was purified to homogeneity following coexpression of RAD51Bhis6, RAD51Chis10, RAD51D, and XRCC2 in Sf9 cells (Fig. 5A). The presence of all four subunits, which copurified with an apparent 1:1:1:1 stoichiometry at each chromatographic step, was confirmed using mAbs (Fig. 5B). In contrast to the individual subunits, which were generally insoluble when expressed on their own, the coexpression of the proteins resulted in a higher level of solubility. The RAD51Chis10–XRCC3his6 complex was also purified following coexpression in Sf9 cells (Fig. 5C). Both complexes were free of endo- and exonuclease activities, as monitored by the release of acid-soluble counts from 5′-32P-end-labeled single- and double-stranded oligonucleotide substrates (data not shown). Elsewhere, we report the visualization of the RAD51C–XRCC3 complex and describe its ssDNA-binding properties (Masson et al. 2001). In the following sections, we describe the properties of the novel BCDX2 complex.
Figure 5.
Purification of two distinct complexes containing the RAD51 paralogs from baculovirus-infected insect cells. (A) Purification of a complex containing RAD51B, RAD51C, RAD51D, and XRCC2 (BCDX2). The complex was purified from Sf9 cells infected with pBAC51Bhis6, pBAC51Chis10, pBAC51D, and pBACX2, as described in Materials and Methods. Proteins were visualized by SDS-PAGE followed by Coomassie blue staining. (Lane a) Molecular mass markers; (lanes b,c) 2.0 and 3.5 μg of protein. (B) Western blot of purified BCDX2 complex using mAbs raised against RAD51B, RAD51C, RAD51D, and XRCC2. (C) Purified RAD51C–XRCC3 complex. Proteins were purified from Sf9 cells infected with pBAC51Chis10 and pBACX3his6 and visualized by SDS-PAGE followed by Coomassie blue staining. (Lane a) Molecular mass markers; (lane b) 3 μg of RAD51C–XRCC3 complex.
DNA binding and ATPase activity of BCDX2
When purified BCDX2 complex was incubated with single-stranded DNA (100 nucleotides in length), we observed the formation of protein–DNA networks that failed to enter polyacrylamide gels (Fig. 6A, lanes b–f). We concluded that BCDX2 bound preferentially to single-stranded DNA, because little binding to duplex DNA was observed under the same conditions (lanes h–l). Quantification by PhosphorImager analysis indicated that only 1.5% of the input duplex DNA was bound compared with 78% for the ssDNA at 100 nM BCDX2 (Fig. 6B). The BCDX2 complex bound 5′- or 3′-tailed linear DNA substrates at a level that was intermediate of that observed with single- and double-stranded DNA (Fig. 6A, lanes n–r and t–y, respectively; Fig. 6B). The BCDX2 complex did not bind synthetic Holliday junctions (data not shown).
Figure 6.
DNA-binding and DNA-stimulated ATPase activities of BCDX2 complex. (A) DNA-binding reactions containing ssDNA, dsDNA, or tailed duplex DNA and the indicated concentrations of BCDX2 were carried out as described in Materials and Methods. Protein–DNA complexes were analyzed by PAGE. 5′-32P-end labels are indicated with asterisks. (B) Quantification of the data shown in A by PhosphorImaging. (C) The ATPase activities of the BCDX2 complex were analyzed in the presence and absence of DNA, as indicated.
The DNA-binding properties of BCDX2 are therefore similar to those shown by the RAD51C–XRCC3 complex and by RAD52 protein, which also aggregate ssDNA (Benson et al. 1998; Masson et al. 2001). ssDNA binding by BCDX2 was dependent on the presence of Mg2+, but was ATP-independent. Optimal binding was observed at 2.5 mM Mg(OAc)2. Inclusion of 150 mM NaCl or 10 mM EDTA abolished DNA binding completely (data not shown).
Because the RAD51B, RAD51C, RAD51D, and XRCC2 amino acid sequences contain Walker motifs (Walker et al. 1982) and are predicted to bind ATP (Thacker 1999), the BCDX2 complex was assayed for its ability to hydrolyze ATP in the presence and absence of DNA. ATPase activity, stimulated by the presence of ssDNA, was observed at a very low level (Fig. 6C). Indeed, in the presence of ssDNA the turnover rate was estimated to be 0.005–0.01 ATP/min per BCDX2 complex, which is approximately one order of magnitude lower than that observed with RAD51 (Baumann et al. 1996). Although the ATPase activity was extremely low, it is likely to be an intrinsic property of the protein complex because the activity coeluted with BCDX2 at the final stages of protein purification.
Electron microscopic visualization of BCDX2
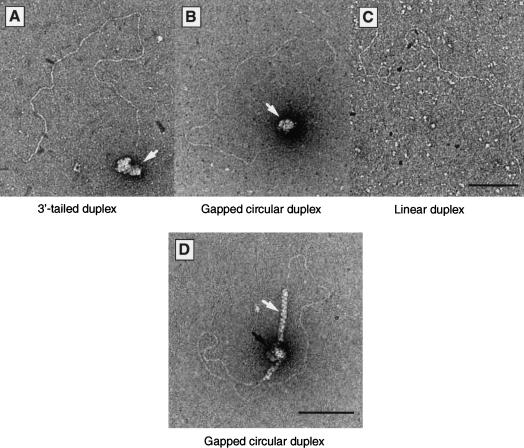
The first step in the repair of a DSB by homologous recombination involves exonuclease resection to expose single-stranded tails that can act as an initiation site for the recruitment of recombination proteins. Because the RAD51 paralogs are required at an early step in recombinational repair, the interaction of BCDX2 complex with resected DSBs was analyzed by electron microscopy. For these experiments, linear plasmid DNA molecules were resected by exonuclease treatment to produce 1.3-kb-long 3′-single-stranded tails. The BCDX2 complex bound specifically to the tailed regions, such that the single-stranded region became bound by BCDX2, whereas the duplex DNA remained essentially protein-free (Fig. 7A, BCDX2 indicated by the white arrow). Similarly, BCDX2 bound to an internal 1.3-kb single-stranded gap present within a circular duplex DNA molecule (Fig. 7B). We did not observe the binding of BCDX2 to linear duplexes that had not undergone resection (Fig. 7C).
Figure 7.
Interaction of BCDX2 complex with single-stranded regions in duplex DNA. (A–C) Electron microscopic visualization of complexes formed between BCDX2 and tailed duplex DNA (A) or gapped circular duplex DNA (B). The bound single-stranded regions are indicated by white arrows. (C) Control indicating the failure of BCDX2 to bind linear duplex DNA. (D) Juxtaposition of BCDX2 complex and RAD51 on gapped circular DNA. The DNA was preincubated with BCDX2 complex (90 nM) for 5 min and then supplemented with RAD51 (0.3 μM). After 5 min at 37°C, the products were visualized by electron microscopy. The white arrow indicates a RAD51 filament, and the black arrow indicates the BCDX2–DNA complexes.
The formation of DNA-damage-induced RAD51 foci is dependent on the presence of the RAD51 paralogs, leading to the suggestion that the proteins may be involved in the recruitment or stabilization of RAD51 filaments at the sites of DNA damage (Bishop et al. 1998; Takata et al. 2000, 2001; Gasior et al. 2001). We therefore analyzed the assembly of RAD51 and BCDX2 complexes at single-stranded gaps in duplex DNA. When subsaturating concentrations of RAD51 were added to preformed complexes of BCDX2 on gapped circular DNA, we observed that 75% of the molecules observed by electron microscopy showed RAD51 filaments in the proximity of BCDX2 (Fig. 7D).
BCDX2 binds to nicks in duplex DNA
Although cells defective in any of the RAD51 paralogs show a mild ionizing radiation sensitivity, the same cell lines are exquisitely sensitive to DNA cross-linking agents, which suggests a role for the paralogs in the repair of DNA cross links (Jones et al. 1987; Tambini et al. 1997; Liu et al. 1998; Takata et al. 2001). The mechanism of cross-link repair in vertebrate systems is presently unknown, but it is thought that incised cross links are either directly or indirectly channeled toward recombinational repair (Bessho et al. 1997; De Silva et al. 2000). We therefore determined whether the BCDX2 complex could bind to nicks in duplex DNA. Using circular or linear duplex DNA molecules containing a single site-specific nick, we observed by electron microscopy the binding of a single BCDX2 complex to each DNA molecule (Fig. 8B,C). Indeed, >80% of the nicked molecules that were well spread showed evidence of a bound BCDX2 complex. Under the same reaction conditions, BCDX2 failed to bind intact duplex DNA (Fig. 8A; data not shown). These studies are consistent with a specific role for BCDX2 in the recognition of nicks during cross-link repair, an interaction that is likely to be important in the initiation of recombination at these sites.
Figure 8.
Interaction of BCDX2 complex with nicks in duplex DNA. Binding reactions contained linear duplex, nicked linear duplex, or nicked circular plasmid DNA, as indicated. Protein–DNA complexes were analyzed by agarose gel electrophoresis (A) or by electron microscopy (B,C). White arrows indicate the binding of BCDX2 to the single nick present in each DNA substrate. The bar represents 100 nm.
Discussion
Biochemical studies of human and yeast proteins involved in homologous recombination are beginning to provide new insights into how mitotic cells repair DNA double-strand breaks (van Gent et al. 2001). Recombinational repair is clearly a highly conserved process, as first indicated by the discovery that human RAD51 is structurally related to bacterial RecA and yeast RAD51 (Ogawa et al. 1993; Story et al. 1993; Benson et al. 1994), and that it has similar biochemical activities in terms of DNA interactions and the initiation of strand exchange (Sung 1994; Sung and Robberson 1995; Baumann et al. 1996; Baumann and West 1997). The in vitro activities of RAD51, however, are considerably reduced in comparison with RecA, indicating a need for other protein cofactors and greater mechanistic complexity. It has been shown that yeast RAD51 is stimulated by RAD52 (Sung 1997a; New et al. 1998; Shinohara and Ogawa 1998), RAD54 (Petukhova et al. 1998, 1999; Mazin et al. 2000; Solinger et al. 2001), RAD55/57 (Sung 1997b), and RP-A (Sung 1994; Sigurdsson et al. 2001). Less is known about possible stimulatory factors for human RAD51 protein, although enhanced activity has been observed in the presence of RAD52 and RP-A (Baumann and West 1997; Benson et al. 1998; McIlwraith et al. 2000).
The vertebrate RAD51 paralogs are related to yeast RAD55/57 by sequence similarity and by functional relationships, leading to the view that the paralogs might also play a role in RAD51-mediated reactions. For example, (1) XRCC2- (irs1) and XRCC3-defective (irsSF) cells show a significant (100-fold and 25-fold, respectively) decrease in the frequency of DSB repair by homologous recombination (Johnson et al. 1999; Pierce et al. 1999). (2) The formation of DNA-damage-induced RAD51 foci is abolished in irs1 (O'Regan et al. 2001) and irs1SF cells (Bishop et al. 1998), and in chicken DT40 cells lacking RAD51B, RAD51C, RAD51D, XRCC2, or XRCC3 (Takata et al. 2001). (3) The radiation sensitivity of the DT40 mutant cell lines can be partially complemented by overexpression of RAD51 (Takata et al. 2001). Like yeast RAD55/57, it is possible that the vertebrate RAD51 paralogs facilitate the formation of RAD51 nucleoprotein filaments on ssDNA, an important early step in the initiation of recombination (Sung 1997b).
To help define the cellular roles of the five vertebrate RAD51 paralogs, we first analyzed their ability to form complexes in HeLa cells. Two distinct complexes were observed: one comprising RAD51B, RAD51C, RAD51D with XRCC2 (BCDX2), and the other RAD51C–XRCC3. The presence of RAD51C in both complexes and the overall architecture of the BCDX2 complex were confirmed using baculovirus expression systems. It is highly unlikely that the interactions we describe are mediated by the presence of DNA because the complexes were stable to high salt. Elsewhere, we and others have reported the properties of the RAD51C–XRCC3 complex: the two subunits are present in a 1:1 stoichiometry, bind preferentially to single-strand DNA, and promote ATP-independent DNA aggregation reactions that can lead to the annealing of complementary DNAs (Kurumizaka et al. 2001; Masson et al. 2001).
Although we do not exclude the possibility that the paralogs may participate in a number of protein subcomplexes, this paper provides the first description of the larger BCDX2 complex. Analyses of the architecture of the complex showed that RAD51B and RAD51C formed a heterodimer that is unable to interact with XRCC2 in the absence of RAD51D, corroborating interactions seen in yeast two-hybrid systems (Schild et al. 2000). The complex showed an Mr of ∼180,000, in which we believe that the four subunits are present in a 1:1:1:1 stoichiometry. The BCDX2 complex binds preferentially to ssDNA and, like RAD51C–XRCC3 and RAD52, promotes DNA aggregation. Recently, using the DT40 system, it was shown that a conditional mutant in RAD52 and XRCC3 is nonviable, whereas either of the single mutants grow reasonably well, indicating that RAD52 and the RAD51 paralogs play overlapping, but nonreciprocal roles in DSBR (Fujimori et al. 2001). Because RAD52 is known to stimulate RAD51 in vitro (Benson et al. 1998; Shinohara and Ogawa 1998; Baumann and West 1999), in part by overcoming inhibitory effects of RP-A (Sung 1997a; New et al. 1998), it is possible that the two paralog complexes play a related role through their ssDNA-binding properties.
Previously, it was shown that the RAD51 paralogs can interact with RAD51 (Liu et al. 1998; Schild et al. 2000). In immunoprecipitation studies carried out in HeLa extracts under stringent conditions, however, we failed to detect direct interactions with RAD51, leading us to infer that the observed interactions may be DNA-mediated. In support of this proposal, using limiting amounts of RAD51, we found that RAD51 colocalized to the gapped region of a circular plasmid that had been prebound by the BCDX2 complex. A role for the RAD51 paralogs in either the stabilization or recruitment of RAD51 nucleoprotein filaments would be consistent with the observed lack of RAD51 foci following DNA damage in all paralog mutants so far examined (Bishop et al. 1998; O'Regan et al. 2001; Takata et al. 2001).
The BCDX2 complex, like RAD51 (Baumann et al. 1996), was shown to promote ATP-hydrolysis in a reaction that was stimulated by the presence of ssDNA. Similar DNA-binding and ATP-hydrolytic properties have been reported for RAD51D alone (Braybrooke et al. 2000). The function of the BCDX2 ATPase is presently unknown because DNA binding appears to be unaffected by the absence of ATP. Moreover, it was recently shown that the substitution of highly conserved residues in the P-loop motif of XRCC2 had little effect on its ability to complement the MMC, X-ray, or genetic instability phenotype of an XRCC2-deficient cell line (O'Regan et al. 2001). Taken together, these studies indicate that the mutation of one ATP-binding subunit may not compromise the function of the multi-subunit complex as a whole.
During the course of these studies, we made the striking observation that the BCDX2 complex binds to nicks in duplex DNA. To our knowledge, this is the first report of nick binding by any recombination protein. The interaction of the paralogs with nicks may be important for the repair of DNA cross links because it has been shown that hamster cell lines defective in XRCC2 and XRCC3 are extremely sensitive (60-fold to 100-fold, respectively) to cross-linking agents such as cisplatin, nitrogen mustard, or mitomycin C (Jones et al. 1987; Fuller and Painter 1988; De Silva et al. 2000). In Escherichia coli and Saccharomyces cerevisiae, cross links are repaired by interlinked nucleotide excision repair (NER) and homologous recombination processes (Jachymczyk et al. 1981; Sladek et al. 1989; Van Houten 1990). In bacteria, the first step in cross-link repair involves incision on either side of the cross link by NER enzymes, followed by exonucleolytic processing to create a gap that serves as a substrate for recombination with homologous duplex DNA. The ensuing strand exchange reaction then transfers in an undamaged strand at the site of the lesion, to allow a second round of NER leading to complete removal of the cross link.
In vertebrate systems, the mechanism of cross-link repair is less clear. Whereas there is again a clear involvement of NER enzymes, in growing cells there appears to be a dose-dependent induction of DSBs (De Silva et al. 2000), which presumably arise from arrested replication forks. The requirement for the RAD51 paralogs in cross-link repair, together with the demonstration that the BCDX2 complex binds nicks, leads us to propose that the RAD51 paralogs bind incised cross links and mediate their progression into recombination pathways. The precise mechanism remains to be determined, but is likely to involve the targeting of RAD51 and/or other recombination enzymes to single-stranded DNA, leading to the initiation of recombination.
Materials and methods
Baculovirus expression vectors
The RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3 genes were PCR-amplified from a human testis cDNA library and cloned into pET11a, pET15b, pET16b, and pET28c (Novagen). In these constructs, the genes were untagged, or linked to N-terminal hexa- or decahistidine tags. The paralog genes from the resultant clones were then subcloned into pFASTBAC1 to produce pBAC51B, pBAC51Bhis6, pBAC51C, pBAC51Chis10, pBAC51D, pBAC51Dhis6, pBACX2, pBACX2his6, and pBACX3his6. The nomenclature used in the naming of the baculoviruses reflects the protein encoded by the virus (e.g., pBAC51Bhis6 expresses RAD51Bhis6 protein). The nucleotide sequences of RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3 were confirmed to be identical to published sequences.
Proteins
Recombinant BCDX2 complex was purified from five 1.25-L spinner flasks of Sf9 cells (1.25 × 106 per milliliter) coinfected with pBAC51Bhis6 (multiplicity of infection, m.o.i. = 10), pBAC51Chis10 (m.o.i. = 10), pBAC51D (m.o.i. = 20), and pBACX2 (m.o.i. = 20) baculoviruses at 27°C for 4 d. Cells were harvested, frozen in dry ice/ethanol, and stored at −20°C. The cell paste was resuspended in 250 mL of phosphate (P) buffer (50 mM NaPO4 at pH 7.0, 0.5 M NaCl, 10% glycerol, 0.02% triton X-100) containing 5 mM imidazole and protease inhibitors. The suspension was lysed using a Dounce homogenizer (20 strokes), sonicated, and then homogenized a second time. Insoluble material was removed by centrifugation (twice at 35,000 rpm for 1 h in a Beckman 45 Ti rotor). The supernatant was loaded on a 20-mL Talon column (Clontech) and washed stepwise with P buffer containing 30 mM (100 mL), 40 mM (100 mL), and 50 mM (50 mL) imidazole. The BCDX2 complex was then eluted with a 200-mL linear gradient of 0.05–1.0 M imidazole in P buffer. The proteins were identified by SDS-PAGE, pooled, dialyzed against H buffer (50 mM NaPO4 at pH 7.6, 150 mM NaCl, 10% glycerol), and loaded on a 20-mL HiPrep 16/10 Heparin FF column (Pharmacia). The column was washed with 100 mL of H buffer containing 0.2 M NaCl, before a linear gradient of 0.2–1.5 M NaCl was applied. Coeluting BCDX2 proteins were pooled and loaded on a 1-mL HiTrap Chelating column (Pharmacia; pre-equilibrated with NiSO4 and P buffer containing 5 mM imidazole) using a Pharmacia Acta FPLC system. The column was washed with P buffer containing 30 mM imidazole, and the proteins were eluted using a linear gradient of 0.03–1 M imidazole in P buffer. Fractions containing BCDX2 complex were pooled, dialyzed for 90 min against 2 L of buffer (20 mM Tris-HCl at pH 7.5, 0.25 M NaCl, 10% glycerol, 1 mM DTT) in Slide-A-Lyzer dialysis cassettes (Pierce) and stored in aliquots at −80°C.
RAD51C-XRCC3 complex (Masson et al. 2001) and RAD51 protein (Baumann et al. 1997) were purified as described. Restriction enzymes were purchased from NEB and used as described by the manufacturer.
Gel filtration
Extracts were prepared from Sf9 cells coinfected with pBAC51B, pBAC51Chis10, pBAC51D, pBACX2, and pBACX3his6. Protein (2 mg) was applied to a 24-mL Superdex 200 HR 10/30 column (Pharmacia) equilibrated with 20 mM Tris-HCl at pH 7.5, 0.5 M NaCl, 10% glycerol, and 1 mM DTT. Fractions (total 72; 500 μL volume) were collected and analyzed by SDS-PAGE followed by Western blotting with mAbs. The gel filtration standards (BioRad) were bovine thyroglobulin (670 kD), bovine gamma globulin (158 kD), chicken ovalbumin (44 kD), horse myoglobin (17 kD), and vitamin B-12 (1.35 kD).
Antibodies
All antibodies were raised against recombinant denatured protein as described (Masson et al. 2001). The monoclonal antibodies 1H3 (RAD51B), 2H11 (RAD51C), 5B3 (RAD51D), 7B7 (XRCC2), and 10F1 (XRCC3) specifically recognized the corresponding protein and not the other paralogs. Similarly, the pAb raised against RAD51D was specific for RAD51D and failed to recognize RAD51B, RAD51C, XRCC2, and XRCC3. The anti-RAD51 mAb (14B4) was purchased from Abcam.
Immunoprecipitation
HeLa S3 cell pellets were resuspended in lysis buffer (50 mM Tris-HCl at pH 7.5, 0.5 M NaCl, 0.5% NP-40) containing protease inhibitors, incubated for 30 min on ice, and then lysed by sonication. Insoluble material was removed by high-speed centrifugation. Protein complexes in the supernatant (equivalent to ∼2 × 107 cells) were pulled down at 4°C for 1.5 h using preimmune serum or pAbs raised against RAD51D cross-linked to Aminolink beads (Pierce). Complexes were washed four times in lysis buffer containing salt as indicated, and visualized by Western blotting using mAbs.
Protein interaction analyses
Spinner flasks (1.25 L) of Sf9 cells (1.25 × 106 cells/mL) were infected with the indicated baculoviruses (m.o.i = 10 for the His-tagged viruses and m.o.i. = 20 for untagged viruses) at 27°C for 4 d. Cells were harvested, frozen in dry ice/ethanol, and stored at −20°C. The cell paste was resuspended in 60 mL of P buffer containing 5 mM imidazole and protease inhibitors. The suspension was lysed using a Dounce homogenizer (20 strokes), sonicated, and then homogenized a second time. Insoluble material was removed by centrifugation (35,000 rpm for 1 h in a Beckman 45 Ti rotor). The supernatant was loaded on a 5-mL Talon column and washed with 70 mL of P buffer containing 30 mM imidazole. Bound protein was eluted with a 40-mL linear gradient of 0.05–1.0 M imidazole in P buffer (40 fractions were collected), and analyzed by Western blotting using mAbs.
DNA
pPB4.3 (4270 bp) form I DNA (Baumann and West 1997) was prepared using QIAGEN Plasmid Mega kits followed by cesium chloride gradient purification. pPB4.3 ssDNA was prepared by standard protocols. pPB4.3 gapped circular DNA or pPB4.3 tailed DNA (both containing a 1.3-kb region of ssDNA) were prepared as described (Davies et al. 2001). Circular pPB4.3 containing a site-specific nick was prepared by annealing heat-denatured NcoI-linearized DNA (150 μg) with ssDNA (200 μg). Nicked circular duplex DNA was purified by gel electrophoresis, electroeluted, and repurified using Qiaquick DNA purification columns (QIAGEN). Nicked linear duplex DNA was produced by treatment of the nicked circular DNA with XmnI. DNA concentrations are expressed in moles of nucleotides, unless indicated otherwise.
DNA-binding assays
Binding assays (10 μL) contained oligonucleotide DNA substrates (10 nM) and the indicated amounts of BCDX2 complex in binding buffer [20 mM triethanolamine-acetate at pH 7.5, 2 mM ATP, 1 mM Mg(OAc)2, 1 mM DTT, 100 μg/mL BSA]. After 5 min at 37°C, the indicated amount of BCDX2 was added (in 2 μL), and incubation was continued for a further 10 min. Protein–DNA complexes were analyzed by 6% PAGE using TBE buffer followed by autoradiography. The 5′-32P-labeled 100-mer and complementary oligonucleotides used in the preparation of the various substrates are described elsewhere (Masson et al. 2001).
In binding reactions with plasmid DNA substrates, the indicated amounts of BCDX2 were incubated with nicked or linear duplex DNA (0.5 μM). Protein–DNA complexes were analyzed by agarose gel electrophoresis followed by autoradiography.
ATPase assays
Reactions (50 μL) contained excess pPB4.3 ssDNA or form I dsDNA (150 μM), 0.24 μM BCDX2 in 50 mM triethanolamine-acetate at pH 7.5, 1 mM Mg(OAc)2, 1 mM DTT, and 100 μg/mL BSA supplemented with 50 nCi [α-32P]ATP (3000 Ci/mmole). Aliquots (5 μL) were removed at the indicated times, stopped by addition of EDTA, and the percentage ATP hydrolyzed was determined by thin layer chromatography.
Electron microscopy
Reactions contained 5 μM DNA and 0.25 μM BCDX2 in 20 mM triethanolamine-acetate at pH 7.5, 2 mM ATP, 1 mM Mg(OAc)2, and 1 mM DTT. Incubation and visualization of protein–DNA complexes were carried out as described (Masson et al. 2001). In some reactions, the DNA was prebound by BCDX2 before addition of RAD51. BCDX2–RAD51–DNA complexes were fixed with 0.2% glutaraldehyde and visualized as described.
Acknowledgments
We thank Anthony Baillot and Simon Vincent for help in the initial stages of this study, Angelos Constantinou and other members of the West laboratory for suggestions, Ruth Peat and the ICRF cell production unit for cell culture, Jane Steel for antibody production, and Jacques Dubochet for his interest. J.-Y.M. was supported by a K.M. Hunter postdoctoral fellowship from the National Cancer Institute of Canada, and M.C.T. by a fellowship from the European Molecular Biology Organization. This work was supported by the Imperial Cancer Research Fund (SCW), the Human Frontiers Science Program (SCW & AS), the Swiss National Foundation (AS), and the North West Cancer Research Fund (FEB).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL s.west@icrf.icnet.uk; FAX 44-20-7269-3811.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.947001.
References
- Albala JS, Thelan MP, Prange C, Fan W, Christensen M, Thompson LH, Lennon GG. Identification of a novel human RAD51 homolog, RAD51B. Genomics. 1997;46:476–479. doi: 10.1006/geno.1997.5062. [DOI] [PubMed] [Google Scholar]
- Baumann P, West SC. The human RAD51 protein: Polarity of strand transfer and stimulation by hRP-A. EMBO J. 1997;16:5198–5206. doi: 10.1093/emboj/16.17.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Heteroduplex formation by human RAD51 protein: Effects of DNA end-structure, hRP-A and hRAD52. J Mol Biol. 1999;291:363–374. doi: 10.1006/jmbi.1999.2954. [DOI] [PubMed] [Google Scholar]
- Baumann P, Benson FE, West SC. Human RAD51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- Baumann P, Benson FE, Hajibagheri N, West SC. Purification of human RAD51 protein by selective spermidine precipitation. Mut Res DNA Repair. 1997;384:65–72. doi: 10.1016/s0921-8777(97)00028-1. [DOI] [PubMed] [Google Scholar]
- Benson FE, Stasiak A, West SC. Purification and characterisation of the human RAD51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson FE, Baumann P, West SC. Synergistic actions of RAD51 and RAD52 in genetic recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- Bessho T, Mu D, Sancar A. Initiation of DNA interstrand cross-link repair in humans: The nucleotide excision repair system makes dual incisions 5′ to the cross-linked base and removes a 22 nucleotide long to 28 nucleotide long damage-free strand. Mol Cell Biol. 1997;17:6822–6830. doi: 10.1128/mcb.17.12.6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Ear U, Bhattacharyya A, Calderone C, Beckett M, Weichselbaum RR, Shinohara A. XRCC3 is required for assembly of RAD51 complexes in vivo. J Biol Chem. 1998;273:21482–21488. doi: 10.1074/jbc.273.34.21482. [DOI] [PubMed] [Google Scholar]
- Braybrooke JP, Spink KG, Thacker J, Hickson ID. The RAD51 family member, RAD51L3, is a DNA-stimulated ATPase that forms a complex with XRCC2. J Biol Chem. 2000;275:29100–29106. doi: 10.1074/jbc.M002075200. [DOI] [PubMed] [Google Scholar]
- Brenneman MA, Weiss AE, Nickoloff JA, Chen DJ. XRCC3 is required for efficient repair of chromosome breaks by homologous recombination. Mutat Res DNA Repair. 2000;459:89–97. doi: 10.1016/s0921-8777(00)00002-1. [DOI] [PubMed] [Google Scholar]
- Cartwright R, Dunn AM, Simpson PJ, Tambini CE, Thacker J. Isolation of novel human and mouse genes of the recA/RAD51 recombination/repair gene family. Nucl Acids Res. 1998a;26:1653–1659. doi: 10.1093/nar/26.7.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright R, Tambini CE, Simpson PJ, Thacker J. The XRCC2 DNA repair gene from human and mouse encodes a novel member of the recA/RAD51 family. Nucl Acids Res. 1998b;26:3084–3089. doi: 10.1093/nar/26.13.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- Cui X, Brenneman M, Meyne J, Oshimura M, Goodwin EH, Chen DJ. The XRCC2 and XRCC3 repair genes are required for chromosome stability in mammalian cells. Mutat Res DNA Repair. 1999;434:75–88. doi: 10.1016/s0921-8777(99)00010-5. [DOI] [PubMed] [Google Scholar]
- Davies AA, Masson J-Y, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- Deans B, Griffin CS, Maconochie M, Thacker J. XRCC2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J. 2000;19:6675–6685. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosanjh MK, Collins DW, Fan WF, Lennon GG, Albala JS, Shen ZY, Schild D. Isolation and characterization of RAD51C, a new human member of the RAD51 family of related genes. Nucl Acids Res. 1998;26:1179–1184. doi: 10.1093/nar/26.5.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J, van Steeg H, de Wit J, Swagemakers SMA, Vermeij M, Hoeijmakers JHJ, Kanaar R. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J. 2000;19:1703–1710. doi: 10.1093/emboj/19.7.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori A, Tachiiri S, Sonoda E, Thompson LH, Dhar PK, Hiraoka M, Takeda S, Zhang YM, Reth M, Takata M. RAD52 partially substitutes for the RAD51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J. 2001;20:5513–5520. doi: 10.1093/emboj/20.19.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller LF, Painter RB. A Chinese hamster ovary cell line hypersensitive to ionizing radiation and deficient in repair replication. Mutat Res. 1988;193:109–121. doi: 10.1016/0167-8817(88)90041-7. [DOI] [PubMed] [Google Scholar]
- Gasior SL, Olivares H, Ear US, Hari DM, Weichelbaum R, Bishop DK. Assembly of RecA-like recombinases: Distinct roles from mediator proteins in mitosis and meiosis. Proc Natl Acad Sci. 2001;98:8411–8418. doi: 10.1073/pnas.121046198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays SL, Firmenich AA, Berg P. Complex formation in yeast double-strand break repair: Participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc Natl Acad Sci. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachymczyk WJ, von Borstel RC, Mowat MR, Hastings PJ. Repair of interstrand cross-links in DNA of Saccharomyces cerevisiae requires two systems for DNA repair: The RAD3 system and the RAD51 system. Mol Gen Genet. 1981;182:196–205. doi: 10.1007/BF00269658. [DOI] [PubMed] [Google Scholar]
- Johnson RD, Liu N, Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- Jones NJ, Cox R, Thacker J. Isolation and cross-sensitivity of X-ray-sensitive mutants of V79-4 hamster cells. Mutat Res. 1987;183:279–286. doi: 10.1016/0167-8817(87)90011-3. [DOI] [PubMed] [Google Scholar]
- Kurumizaka H, Ikawa S, Nakada M, Eda K, Kagawa W, Takata M, Takeda S, Yokoyama S, Shibata T. Homologous-pairing activity of the human DNA-repair proteins XRCC3-RAD51C. Proc Natl Acad Sci. 2001;98:5538–5543. doi: 10.1073/pnas.091603098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Han MG, Romanienko PJ, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, Brookman KW, Siciliano MJ, Walter CA, Fan WF, et al. XRCC2 and XRCC3, new human RAD51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- Masson J-Y, Stasiak AZ, Stasiak A, Benson FE, West SC. Complex formation by the human RAD51C and XRCC3 recombination repair proteins. Proc Natl Acad Sci. 2001;98:8440–8446. doi: 10.1073/pnas.111005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin AV, Bornarth CJ, Solinger JA, Heyer W-D, Kowalczykowski SC. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol Cell. 2000;6:583–592. doi: 10.1016/s1097-2765(00)00057-5. [DOI] [PubMed] [Google Scholar]
- McIlwraith MJ, Van Dyck E, Masson J-Y, Stasiak AZ, Stasiak A, West SC. Reconstitution of the strand invasion step of double-strand break repair using human RAD51, RAD52 and RPA proteins. J Mol Biol. 2000;304:151–164. doi: 10.1006/jmbi.2000.4180. [DOI] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein-A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast Rad51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- O'Regan P, Wilson CR, Townsend S, Thacker J. XRCC2 is a nuclear RAD51-like protein required for damage-dependent RAD51 focus formation without need for ATP hydrolysis. J Biol Chem. 2001;276:22148–22153. doi: 10.1074/jbc.M102396200. [DOI] [PubMed] [Google Scholar]
- Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J Biol Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes & Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman DL, Schimenti JC. Mid-gestation lethality in mice deficient for the recA-related gene, RAD51D/RAD51L3. Genesis. 2000;26:167–173. doi: 10.1002/(sici)1526-968x(200003)26:3<167::aid-gene1>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Pittman DL, Weinberg LR, Schimenti JC. Identification, characterization, and genetic mapping of RAD51D, a new mouse and human RAD51/recA-related gene. Genomics. 1998;49:103–111. doi: 10.1006/geno.1998.5226. [DOI] [PubMed] [Google Scholar]
- Rice MC, Smith ST, Bullrich F, Havre P, Kmiec EB. Isolation of human and mouse genes based on homology to REC2, a recombinational repair gene from the fungus Ustilago maydis. Proc Natl Acad Sci. 1997;94:7417–7422. doi: 10.1073/pnas.94.14.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D, Lio YC, Collins DW, Tsomondo T, Chen DJ. Evidence for simultaneous protein interactions between human RAD51 paralogs. J Biol Chem. 2000;275:16443–16449. doi: 10.1074/jbc.M001473200. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- Shu ZG, Smith S, Wang LJ, Rice MC, Kmiec EB. Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can be partially rescued in a p53(−/−) background. Mol Cell Biol. 1999;19:8686–8693. doi: 10.1128/mcb.19.12.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson S, Trujillo K, Song BW, Stratton S, Sung P. Basis for avid homologous DNA strand exchange by human RAD51 and RPA. J Biol Chem. 2001;276:8798–8806. doi: 10.1074/jbc.M010011200. [DOI] [PubMed] [Google Scholar]
- Sladek FM, Munn MM, Rupp WD, Howard-Flanders P. In vitro repair of psoralen–DNA cross-links by RecA, UvrABC, and the 5′-exonuclease of DNA polymerase I. J Biol Chem. 1989;264:6755–6765. [PubMed] [Google Scholar]
- Solinger JA, Lutz G, Sugiyama T, Kowalczykowski SC, Heyer WD. Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament. J Mol Biol. 2001;307:1207–1221. doi: 10.1006/jmbi.2001.4555. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Takata M, Yamashita YM, Morrison C, Takeda S. Homologous DNA recombination in vertebrate cells. Proc Natl Acad Sci. 2001;98:8388–8394. doi: 10.1073/pnas.111006398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story RM, Bishop DK, Kleckner N, Steitz TA. Structural relationship of bacterial RecA proteins to recombination proteins from bacteriophage T4 and yeast. Science. 1993;259:1892–1896. doi: 10.1126/science.8456313. [DOI] [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast Rad51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- ————— Function of yeast Rad52 protein as a mediator between replication protein-A and the Rad51 recombinase. J Biol Chem. 1997a;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- ————— Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein-A to promote DNA strand exchange by Rad51 recombinase. Genes & Dev. 1997b;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- Sung P, Robberson DL. DNA strand exchange mediated by a Rad51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Fukushima T, Morrison C, Albala JS, Swagemakers SMA, Kanaar R, Thompson LH, Takeda S. The RAD51 paralog RAD51B promotes homologous recombinational repair. Mol Cell Biol. 2000;20:6476–6482. doi: 10.1128/mcb.20.17.6476-6482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five RAD51 paralogs. Mol Cell Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambini CE, George AM, Rommens JM, Tsui L-C, Scherer SW, Thacker J. The XRCC2 DNA repair gene: Identification of a positional candidate. Genomics. 1997;41:84–92. doi: 10.1006/geno.1997.4636. [DOI] [PubMed] [Google Scholar]
- Tebbs RS, Zhao Y, Tucker JD, Scheerer JB, Siciliano MJ, Hwang M, Liu N, Legerski RJ, Thompson LH. Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc Natl Acad Sci. 1995;92:6354–6358. doi: 10.1073/pnas.92.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker J. A surfeit of RAD51-like genes? Trends Genet. 1999;15:166–168. doi: 10.1016/s0168-9525(99)01733-3. [DOI] [PubMed] [Google Scholar]
- Tsutsui Y, Morishita T, Iwasaki H, Toh H, Shinagawa H. A recombination repair gene of Schizosaccharomyces pombe, rhp57, is a functional homolog of the Saccharomyces cerevisiae RAD57 gene and is phylogenetically related to the human XRCC3 gene. Genetics. 2000;154:1451–1461. doi: 10.1093/genetics/154.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- Van Houten B. Nucleotide excision repair in Escherichia coli. Microbiol Rev. 1990;54:18–51. doi: 10.1128/mr.54.1.18-51.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]