Abstract
Oxidation and loss of heme in soluble guanylyl/guanylate cyclase (sGC), the nitric oxide receptor, is thought to be a major contributor to cardiovascular disease and is the target of compounds BAY 58-2667 and HMR1766. Using spectroelectrochemical titration, we found a truncated sGC to be highly stable in the ferrous state (+234 mV) and to bind ferrous heme tightly even in the presence of NO, despite the NO-induced release of the proximal histidine. In contrast, oxidized sGC readily loses ferric heme to myoglobin (0.47 ± 0.02 hr−1). Peroxynitrite, the presumed cellular oxidant, readily oxidizes sGC in 5 mM glutathione.
The biological production of nitric oxide (NO), a free-radical molecule, is vital to the development and function of nearly all animals. NO is produced by NO synthase and regulates numerous physiological activities, including blood pressure, wound healing and memory formation. The NO signal is propagated through the heme-containing enzyme soluble guanylyl cyclase (sGC), an ~150 kDa heterodimeric protein that may reside in the same cell as NO synthase, or in nearby cells. The two sGC subunits are gene duplications with each containing H-NOX, PAS, coiled-coil and cyclase domains (1). NO binds to ferrous heme in the β subunit, which leads to rupture of the proximal histidine bond, stimulation of cyclase activity and the conversion of GTP to cGMP. A variety of tissue-specific physiological responses can result, including smooth muscle relaxation and vasodilation. There are no structures of sGC, but several structures of bacterial H-NOX proteins have been determined and reveal a single-domain fold with a deep hydrophobic cleft for heme binding (2–4).
sGC is prone to heme loss during isolation (5) and apparently also in the cell, particularly after oxidation, which can occur, for example, during inflammation or heart failure. Compounds BAY 58-2667 (cinaciguat) and HMR1766 (ataciguat) were designed to rescue apo-sGC by filling the heme pocket, leading to an sGC complex with high catalytic activity and decreased turnover in the cell (6–11). Investigations into the stability of the sGC heme, its reduction potential and its propensity for loss are needed but have been challenging to pursue, due in part to the difficulty in working with the protein.
We have produced several recombinant forms of sGC from Manduca sexta, the tobacco hornworm, that include H-NOX, PAS and coiled-coil domains of both α and β subunits, but not the cyclase domains, and display greater stability than does the full-length protein (12, 13) (Supplementary Fig. S1). These proteins are bacterially expressed with an intact ferrous heme, display CO and NO binding that is similar to that for the full-length protein, and respond to allosteric stimulators 3-(5′-hydroxymethyl-2′-furyl)-1-benzylindazole (YC-1) and BAY 41-2272 (12, 13). Here, we make use of previously described construct Ms sGC-NT1 (α 1-471, β 1-401) to measure the heme reduction potential, and newly developed Ms sGC-NT13 (α 49-450, β 1-380) to measure heme loss kinetics. Construct Ms sGC-NT13 was trimmed to remove the first 48 residues from the a subunit, which are predicted to be disordered, and 21 residues from the C-termini of both α and β subunits, which are predicted to link the coiled-coil and cyclase domains, leading to a protein with increased levels of expression and stability. Ferrous, unliganded Ms sGC-NT13, as well as its complexes with CO and YC-1, is stable at room temperature on the time-scale of days to weeks with no change in Soret absorbance maxima position or intensity. The Soret absorbance maxima for unliganded Ms sGC-NT13 (433 nm), its complexes with CO (423 nm), CO/YC-1 (422 nm) and NO (400 nm), and after oxidation (394 nm), correspond with values previously published for Ms sGC (12) and other mammalian sGC proteins (14).
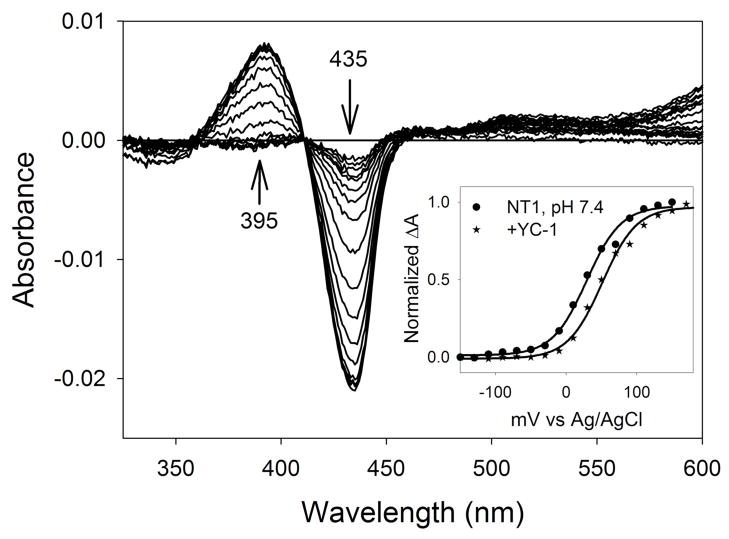
One of the remarkable properties of heme as a protein cofactor is the degree to which heme reduction potential and ligand specificity can be tuned by the protein for the job at hand. We determined the midpoint potential for Ms sGC-NT1 by spectroelectrochemical titration at three pH values (Fig. 1, Table 1). Ms sGC has an unusually high midpoint potential for a b-type heme, +234 mV at pH 7.4, which stabilizes the ferrous state. This value is relatively insensitive to small changes in pH, indicating the absence of ionizable groups near the heme, including heme-ligated water, that have pKa values below 10. Binding of compound YC-1, which allosterically stimulates sGC and blocks escape of both CO and NO from the Ms sGC-NT2 heme pocket (12, 13), leads to a small (22 mV) increase in measured reduction potential (Fig. 1, Table 1), consistent with an increase in heme pocket desolvation upon YC-1 binding.
FIGURE 1.
Spectroelectrochemical titration of Ms sGC-NT1. Applied voltages were from −150 to +170 mV vs. Ag/AgCl in 20 mV increments (add 205 mV for potential vs. NHE). Inset: Nernst plots ± YC-1. YC-1 shifts the Nernst plot right, indicating an increased reduction potential of Ms sGC-NT1. Measurements were carried out in protein buffer containing 50 mM potassium phosphate, pH 7.4, 100 mM KCl and 5% glycerol
Table 1.
Ms sGC-NT1 midpoint potentials (mV)
50 μM.
Not determined.
Although NO binds to both ferri- and ferroheme centers, ferroheme provides at least two functional advantages for sGC. First, binding of NO to ferrous heme is extremely tight, exhibiting dissociation constants in the picomolar to femtomolar range. This allows for very low NO concentrations to initiate the signaling cascade and for avoiding the higher NO concentrations that give rise to nitrosative stress. Recent estimates for typical NO concentrations of importance for signaling in vivo are between 0.1 and 5 nM (15). Second, the trans effect of NO binding is particularly prominent for ferroheme (16), allowing for proximal histidine release and the propagation of an NO-dependent conformational change from the heme pocket to the catalytic center.
The measured reduction potential of Ms sGC-NT1 is approximately 200 mV more positive than that of myoglobin (17), and 67 mV more positive than that of the oxygen-binding Tt H-NOX protein (18), both of which function as ferroheme proteins. The ferriheme nitrophorins from Rhodnius prolixus, which transport NO from the insect saliva to a victim during blood feeding, maintain a midpoint potential of approximately −300 mV (19), over half a volt more negative than that of sGC. The ferriheme allows the nitrophorins to readily bind, transport and release NO. How heme-proteins acquire an appropriate midpoint potential is incompletely understood but likely involves electrostatic stabilization, influence of coordinating ligands, and heme geometry. The Rhodnius nitrophorins achieve their negative reduction potential in part through judicious placement of negatively charged side chains inside the protein, which stabilizes the positively charged ferric heme (20), and through heme ruffling distortion, which mixes porphyrin and metal orbitals to stabilize Fe(III) and disfavor heme reduction (21, 22). Myoglobin has a partially polar heme pocket and a relatively planar heme, even when bound to NO (23). The H-NOX proteins, however, have very hydrophobic heme pockets (2–4), which favor neutral ferrous heme, but are also highly distorted, which should disfavor ferrous heme, but may not, based on results from site-directed mutagenesis of Tt H-NOX (18, 24). In those experiments, mutations leading to decreased heme distortion also lead to decreased reduction potentials at the pH examined. However, the mutations alter the pKa of the water ligand axial to the ferric heme (24), suggesting the reduction potential may have substantial pH dependence. A homology model of the Ms sGC β H-NOX domain (12) suggests that sGC has an even more hydrophobic pocket than Tt H-NOX (Fig. S2), which is consistent with the more positive midpoint potential presented here and its invariance with pH (Table 1).
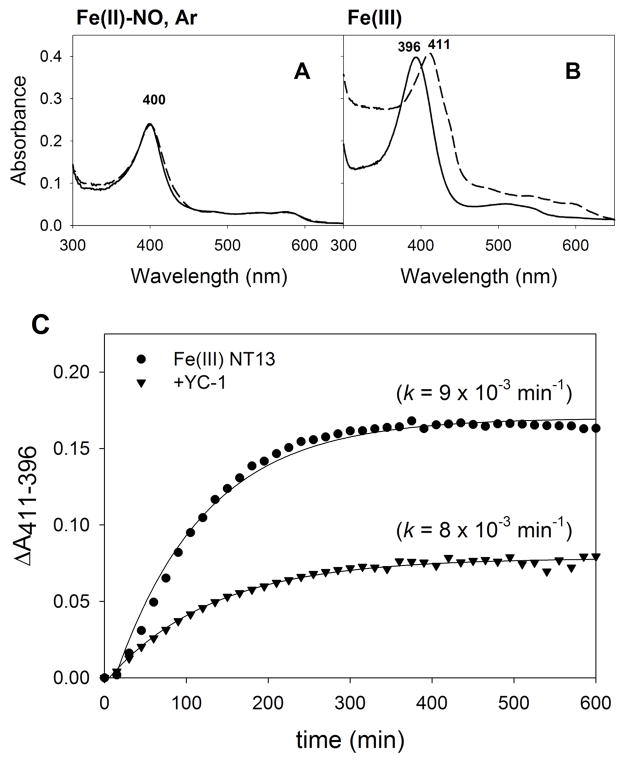
To assess rates of heme loss in Ms sGC, we adapted the approach of Hargrove et al. using a H64Y/V68F myoglobin variant that has a distinct absorbance spectra and a unique green color (25). Addition of hemin to the apo protein led to a ferric (met) myoglobin with Soret and α/β band absorbance maxima of 411 and 600 nm, respectively (Fig. S3A). Excess YC-1 had no effect on hemin uptake by the apo-myoglobin (ApoMb). Addition of ferrous hemin led to a ferrous myoglobin with Soret and α/β band absorbance maxima of 428 and 560 nm respectively.
We investigated the transfer of heme from Ms sGC-NT13 to 10-fold excess ApoMb under a variety of conditions. No loss of heme was detected from ferrous Ms sGC, its CO complex or its CO/YC-1 complex over a 15 hr period (Fig. S3), suggesting that ferrous heme either does not escape from Ms sGC in this time period, or binding is substantially tighter to ferrous Ms sGC than to ApoMb. Interestingly, the ferrous sGC-NO complex, which is five-coordinate after cleavage of the proximal histidine bond, retains high heme affinity and does not lose heme to ApoMb (Fig. 2A). Under anaerobic conditions, the sGC-NO complex is completely stable in both the presence and absence of ApoMb, but will slowly lose NO under aerobic conditions, presumably through NO release and reaction with dioxygen (Fig. S4). Addition of YC-1, which slows NO release, completely stabilizes the sGC-NO complex over 15 hr.
FIGURE 2.
Heme loss measurements. All samples contained 2 μM Ms sGC-NT13 and 20 μM ApoMb H64Y/V68F. A. Ms sGC-NT13 NO-complex (ferrous) formed with 10-fold excess of DEA/NO at 0 (solid line) and 15 hr (dashed line) under saturating Argon. No change in the heme Soret band is observed, indicating no loss of heme to ApoMb. B. Ferric Ms sGC-NT13 (solid line) loses heme to ApoMb, forming metmyoglobin (dashed line). C. Absorbance change (ΔA411-396) plotted versus time for the loss of Ms sGC (Soret maximum 396 nm) and subsequent formation of metmyoglobin (Soret maximum 411 nm), ± YC-1. Rates were determined by a fit to a 3-parameter single exponential using SigmaPlot.
In contrast, oxidation of Ms sGC-NT13 and addition of ApoMb leads to loss of absorbance at 396 nm and gain of absorbance at 411 nm (Fig. 2B), indicating heme transfer from Ms sGC to ApoMb. Transfer is relatively rapid at 20°C and occurs with a first order rate constant of 0.55 ± 0.02 hr−1 (Fig. 2C), which is 20 to 80 times faster than loss of heme from native sperm whale metmyoglobin at 37°C, depending on pH conditions (0.007 hr−1 at pH 7.0, and 0.03 hr−1 at pH 8.0, (25)). A strong increase in absorbance at 280 nm in addition to a large drift in the baseline signal over the course of the experiment, results from precipitation of apo-Ms sGC. In the presence of YC-1, the rate of ferriheme loss and the propensity toward precipitation are unchanged (Fig 2C).
Peroxynitrite (ONOO−) formation from the reaction of NO and superoxide (O2−) is thought to occur in vivo during inflammation and to directly oxidize sGC heme (8, 26). To investigate this possibility, we added ~80 μM peroxynitrite to 5 μM Ms sGC-NT13 while monitoring the sGC spectra. Conversion of the ferrous heme to ferric was rapid and complete (Fig. S5A), and completely reversible with sodium dithionite, a strong reductant. Addition of 5 mM reduced glutathione, the major reductant in the cytosol and present at 1–10 mM, was only able to partially re-reduce oxidized sGC. Addition of ~160 μM peroxynitrite to 5 μM sGC in a buffer containing 5 mM GSH still yielded a small percentage of oxidized sGC, which remained unchanged after 1 hr (Fig. S5B). Taken together, these data suggest that peroxynitrite can lead to oxidized sGC under cellular conditions, supporting proposals of this mechanism for sGC oxidation in vivo.
In summary, Ms sGC is highly stabilized toward the ferrous state (E° = +234 mV) and, when ferrous, is highly resistant to heme loss both in the absence and presence of NO. Heme oxidation leads to a highly unstable protein that readily loses heme (t1/2 = 76 min). Peroxynitrite rapidly oxidizes the ferrous heme and can occur even in the presence of 5 mM glutathione. These data are consistent with the proposed mechanism for compounds BAY 58-2667 and HMR1766 in overcoming loss of sGC activity during oxidative stress in the cell, which is thought to occur by filling the apo-sGC heme pocket and rescuing the protein, leading to an activated and stabilized sGC molecule (6, 8, 9). Our data also highlight the stability of the ferrous heme complex even in the absence of a proximal histidine bond, as occurs in the Fe(II)-NO complex. Loss of Fe(III) heme in sGC appears to be driven by the unfavorable energetics of burying the positively charged heme center (formally +1) in a highly non-polar heme pocket.
Supplementary Material
Acknowledgments
We are grateful to Dr. Katrina Miranda for providing DEA/NO, Dr. John Olson for providing the myoglobin expression plasmid, and Dr. Sue Roberts for homology modeling.
Footnotes
Funding sources: This work was supported by National Institutes of Health grants HL062969 and GM077390 (WRM), HL054826 (FAW) and T32 GM008804 (BGF), and by American Heart Association grants 10PRE2630177 (BGF) and 0515517Z (XH).
ASSOCIATED CONTENT
Supporting Information. Materials and Methods and Tables S1-S5. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Poulos TL. Curr Opin Struct Biol. 2006;16:736–743. doi: 10.1016/j.sbi.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Nioche P, Berka V, Vipond J, Minton N, Tsai AL, Raman CS. Science. 2004;306:1550–1553. doi: 10.1126/science.1103596. [DOI] [PubMed] [Google Scholar]
- 3.Pellicena P, Karow DS, Boon EM, Marletta MA, Kuriyan J. Proc Natl Acad Sci USA. 2004;101:12854–12859. doi: 10.1073/pnas.0405188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma X, Sayed N, Beuve A, van den Akker F. EMBO J. 2007;26:578–588. doi: 10.1038/sj.emboj.7601521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohlstein EH, Wood KS, Ignarro LJ. Arch Biochem Biophys. 1982;218:187–198. doi: 10.1016/0003-9861(82)90335-6. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt PM, Schramm M, Schroder H, Wunder F, Stasch JP. J Biol Chem. 2004;279:3025–3032. doi: 10.1074/jbc.M310141200. [DOI] [PubMed] [Google Scholar]
- 7.Meurer S, Pioch S, Pabst T, Opitz N, Schmidt PM, Beckhaus T, Wagner K, Matt S, Gegenbauer K, Geschka S, Karas M, Stasch JP, Schmidt HH, Muller-Esterl W. Circ Res. 2009;105:33–41. doi: 10.1161/CIRCRESAHA.109.198234. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann LS, Schmidt PM, Keim Y, Schaefer S, Schmidt HH, Stasch JP. Br J Pharmacol. 2009;157:781–795. doi: 10.1111/j.1476-5381.2009.00263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin F, Baskaran P, Ma X, Dunten PW, Schaefer M, Stasch JP, Beuve A, van den Akker F. J Biol Chem. 2010;285:22651–22657. doi: 10.1074/jbc.M110.111559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schindler U, Strobel H, Schonafinger K, Linz W, Lohn M, Martorana PA, Rutten H, Schindler PW, Busch AE, Sohn M, Topfer A, Pistorius A, Jannek C, Mulsch A. Mol Pharmacol. 2006;69:1260–1268. doi: 10.1124/mol.105.018747. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Z, Pyriochou A, Kotanidou A, Dalkas G, van Eickels M, Spyroulias G, Roussos C, Papapetropoulos A. Am J Physiol Heart Circ Physiol. 2008;295:H1763–1771. doi: 10.1152/ajpheart.51.2008. [DOI] [PubMed] [Google Scholar]
- 12.Hu X, Murata LB, Weichsel A, Brailey JL, Roberts SA, Nighorn A, Montfort WR. J Biol Chem. 2008;283:20968–20977. doi: 10.1074/jbc.M801501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu X, Feng C, Hazzard JT, Tollin G, Montfort WR. J Am Chem Soc. 2008;130:15748–15749. doi: 10.1021/ja804103y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karow DS, Pan D, Davis JH, Behrends S, Mathies RA, Marletta MA. Biochemistry. 2005;44:16266–16274. doi: 10.1021/bi051601b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall CN, Garthwaite J. Nitric Oxide. 2009;21:92–103. doi: 10.1016/j.niox.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyllie GR, Scheidt WR. Chem Rev. 2002;102:1067–1090. doi: 10.1021/cr000080p. [DOI] [PubMed] [Google Scholar]
- 17.Ding XD, Weichsel A, Andersen JF, Shokhireva TK, Balfour C, Pierik AJ, Averill BA, Montfort WR, Walker FA. J Am Chem Soc. 1999;121:128–138. [Google Scholar]
- 18.Olea C, Boon EM, Pellicena P, Kuriyan J, Marletta MA. ACS Chem Biol. 2008;3:703–710. doi: 10.1021/cb800185h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersen JF, Ding XD, Balfour C, Shokhireva TK, Champagne DE, Walker FA, Montfort WR. Biochemistry. 2000;39:10118–10131. doi: 10.1021/bi000766b. [DOI] [PubMed] [Google Scholar]
- 20.Berry RE, Shokhirev MN, Ho AY, Yang F, Shokhireva TK, Zhang H, Weichsel A, Montfort WR, Walker FA. J Am Chem Soc. 2009;131:2313–2327. doi: 10.1021/ja808105d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shokhireva T, Berry RE, Uno E, Balfour CA, Zhang H, Walker FA. Proc Natl Acad Sci USA. 2003;100:3778–3783. doi: 10.1073/pnas.0536641100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maes EM, Roberts SA, Weichsel A, Montfort WR. Biochemistry. 2005;44:12690–12699. doi: 10.1021/bi0506573. [DOI] [PubMed] [Google Scholar]
- 23.Schreiter ER, Rodriguez MM, Weichsel A, Montfort WR, Bonaventura J. J Biol Chem. 2007;282:19773–19780. doi: 10.1074/jbc.M701363200. [DOI] [PubMed] [Google Scholar]
- 24.Olea C, Jr, Kuriyan J, Marletta MA. J Am Chem Soc. 132:12794–12795. doi: 10.1021/ja106252b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hargrove MS, Singleton EW, Quillin ML, Ortiz LA, Phillips GN, Jr, Olson JS, Mathews AJ. J Biol Chem. 1994;269:4207–4214. doi: 10.2210/pdb1mgn/pdb. [DOI] [PubMed] [Google Scholar]
- 26.Kagota S, Tada Y, Nejime N, Nakamura K, Kunitomo M, Shinozuka K. J Pharmacol Sci. 2009;109:556–564. doi: 10.1254/jphs.08273fp. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.