Abstract
Heparanomics is the study of all the biologically active oligosaccharide domain structures in the entire heparanome and the nature of interactions among these domains and their protein ligands. Structural elucidation of heparan sulfate and heparin oligosaccharides is a major obstacle in advancing structure-function relationships and the study of heparanomics. There are several factors that exacerbate challenges involved in the structural elucidation of heparin and heparan sulfate. Therefore, there is a great interest in developing novel strategies and analytical tools to overcome the barriers in decoding the enigmatic heparanome. This review article focuses on the applications of isotopes, both radioisotopes and stable isotopes, in the structural elucidation of the complex heparanome at the disaccharide or oligosaccharide level using liquid chromatography, nuclear magnetic resonance spectroscopy and mass spectrometry. This review article also outlines the utility of isotopes in determining the substrate specificity of biosynthetic enzymes that eventually dictate the emergence of biologically active oligosaccharides.
Keywords: Heparin; Heparan sulfate; Proteoglycans; Glycosaminoglycans; HPLC; NMR; MS; Stable isotopes (13C, 33S, 34S); Radio isotopes (3H, 14C, 35S); Sulfotransferases; Antithrombin III; Fibroblast growth factor
Introduction
Heparan sulfate (HS), a sulfated linear polysaccharide chain, is a major structural/functional entity of various proteoglycans (PGs) and is found on the cell surface and in the extracellular matrix. Mounting evidence derived from numerous studies conducted over the past 50 years suggest that HS plays a remarkable role in many cellular, physiological, and pathological processes through interactions with a wide array of proteins including proteases, protease inhibitors, growth factors, morphogens, chemokines, lipoprotein lipases, neurotransmitters and their regulators, and adhesive proteins [1–6]. The system wide roles of HS in many species are attributed to the great structural diversity of HS in terms of size, domain organization, epimer content, sulfation pattern and chain valency.
HS chains and related glycosaminoglycan (GAG) chains are thought to be synthesized in the Golgi by GAGOSOMES, elusive biosynthetic multi-enzyme complexes [7]. HS chain synthesis is initiated on a tetrasaccharide unit that is linked to certain serine residues of core proteins. Chain elongation and subsequent modifications require activated sugar donors such as uridine diphosphate-N-acetylglucosamine (UDP-GlcNAc) and uridine diphosphate-glucuronic acid (UDP-GlcA), and a sulfate donor, 3′-phosphoadenosine 5′-phosphosulfate (PAPS). It is interesting to note that HS chains consist of highly sulfated regions (NS), unmodified regions (NA), and less sulfated variable regions (NA/NS). However, it is unclear how such combinatorial modifications occur seamlessly in vivo [8]. Heparin is structurally similar to HS. However, heparin chains carry a higher percentage of iduronic acid (IdoA) residues and sulfate groups than HS chains do. Furthermore, unlike HS, heparin lacks domain organization. Despite these structural differences between heparin and HS, heparin is widely used as a substitute for HS in biological and chemical studies because of its commercial availability. The disaccharide repeating units of heparin/HS have differing degrees of sulfation, distinct sulfation patterns, and two different uronyl epimers, all of which contribute to sequence microheterogeneity. In addition, these molecules are highly polydisperse in terms of their sizes. Microheterogeneity and polydispersity make it very difficult to sequence the entire heparanome. To advance our knowledge of heparanomics, it is critical to elucidate both the primary sequence and the three-dimensional conformation of HS structures of biological origin. However, these structures are available only in a limited quantity and this scarcity challenges the detection limits of traditional analytical techniques. Therefore, it is essential to develop novel strategies and molecular tools to improve the detection limits of biologically active heparin/HS.
Nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS), two widely used techniques, facilitate the structural analysis of heparin and HS, and also provide complementary information [9,10]. Many primary research articles and review articles provide a detailed description of recent developments and applications of these two mainstay techniques in the structural analysis of Heparin/HS [11–22]. These techniques can be utilized either alone or in combination with on-line separation techniques such as capillary electrophoresis and liquid chromatography (LC) to elucidate biologically significant structures of heparin/HS following a variety of fragmentation methods (Figure 1). This article, therefore, will focus on emphasizing the utility of isotopes in expediting the structural analysis of heparin and HS and discuss the future prospects for the use of stable isotopes in this emerging area of heparanomics.
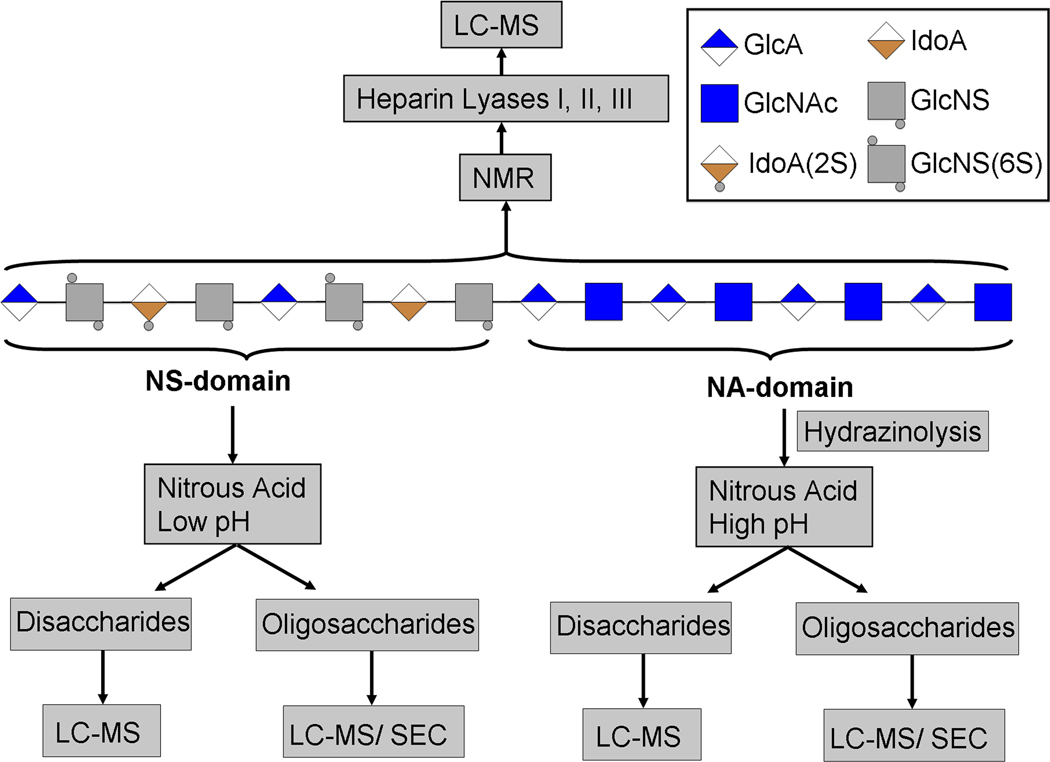
Figure 1.
Overview of HS depolymerization using chemical and enzymatic approaches to prepare HS disaccharides and oligosaccharides for subsequent analysis by LC-MS, SEC-HPLC and IPRP-HPLC. HS chains are composed of a highly sulfated NS region and an unmodified NA region. The intact HS chains are amenable for NMR analysis. However, current MS instruments can not facilitate the structural analysis of the intact HS chains. Therefore, HS chains are depolymerized enzymatically by treating them with heparitinases that fragment both the NA and NS regions into disaccharides for LC/MS analysis. High pH nitrous acid specifically cleaves the HS chains at glucosamine residues. The NA region is fragmented by a two step process in which N-acetyglucosamine residues of NA region are deacetylated by hydrazinolysis and the resulting free amine containing glucosamine residues are subsequently subjected to fragmention by nitrous acid at pH 4.0. Low pH (pH 1.5) nitrous acid treatment specially cleaves NS regions into disaccharides. HS oligosaccharides that are resistant to nitrous acid treatment can be analyzed by size exclusion chromatography to map the heparome.
Radioisotopes in elucidating enzyme specificity and HS / heparin modifications
HS chains consist of repeated disaccharide units of glucosamine (GlcN) and uronic acid (GlcA / IdoA). The large structural diversity of HS chains is created by a series of modifications catalyzed by numerous biosynthetic enzymes in the Golgi [8, 23]. The first modifications are N-deacetylation and subsequent N-sulfonation of GlcNAc residues by N-deacetylase-N-sulfotransferase (NDST) isoforms. GlcA residues are then epimerized to IdoA residues by HS C5-epimerase immediately following N-sulfonation but prior to other sulfonations. A variety of O-sulfotransferase (OST) enzymes subsequently add sulfate groups to C6 (by 6-OST enzyme isoforms) and C3 (by 3-OST isoforms) of GlcN residues, and to C2 (by 2-OST) of IdoA residues. Four NDST isoforms, one C5-epimerase, one 2-OST, seven 3-OST isoforms and three 6-OST isoforms have been found in humans and other organisms. Therefore, it is crucial to understand the specificity of each enzyme isoform that eventually contributes to HS structural diversity. Radioisotope labeling is a robust approach to determine both the activity and specificity of these biosynthetic enzymes. For example, the lyase treatment of radiolabeled HS chains, after enzymatic modification of HS chain with a sulfotransferase in the presence of [35S]-PAPS, can generate several distinct radiolabeled monosulfated disaccharides [ΔU-GlcNAc(6S), ΔU2S-GlcNAc, ΔU-GlcNS, ΔU-GlcN(3S), ΔU-GlcN(6S)], disulfated disaccharides [ΔU-GlcNS6S; ΔU2S-GlcNS; ΔU-GlcNS3S; ΔU2S-GlcN(6S); ΔU2S-GlcN(3S); ΔU-GlcN (3S6S)], trisulfated disaccharides [ΔU-GlcNS3S6S; ΔU2S-GlcNS6S; ΔU2S-GlcNS3S; ΔU2S-GlcN(3S36S)] and a tetrasulfated disaccharide [ΔU2S-GlcNS3S6S]. HPLC disaccharide compositional analysis of the resulting radiolabeled disaccharides can clearly indicate the specificity of a given HS biosynthetic enzyme or its isoforms. The following sections outline several applications of radiolabeling in advancing structural and functional heparanomics.
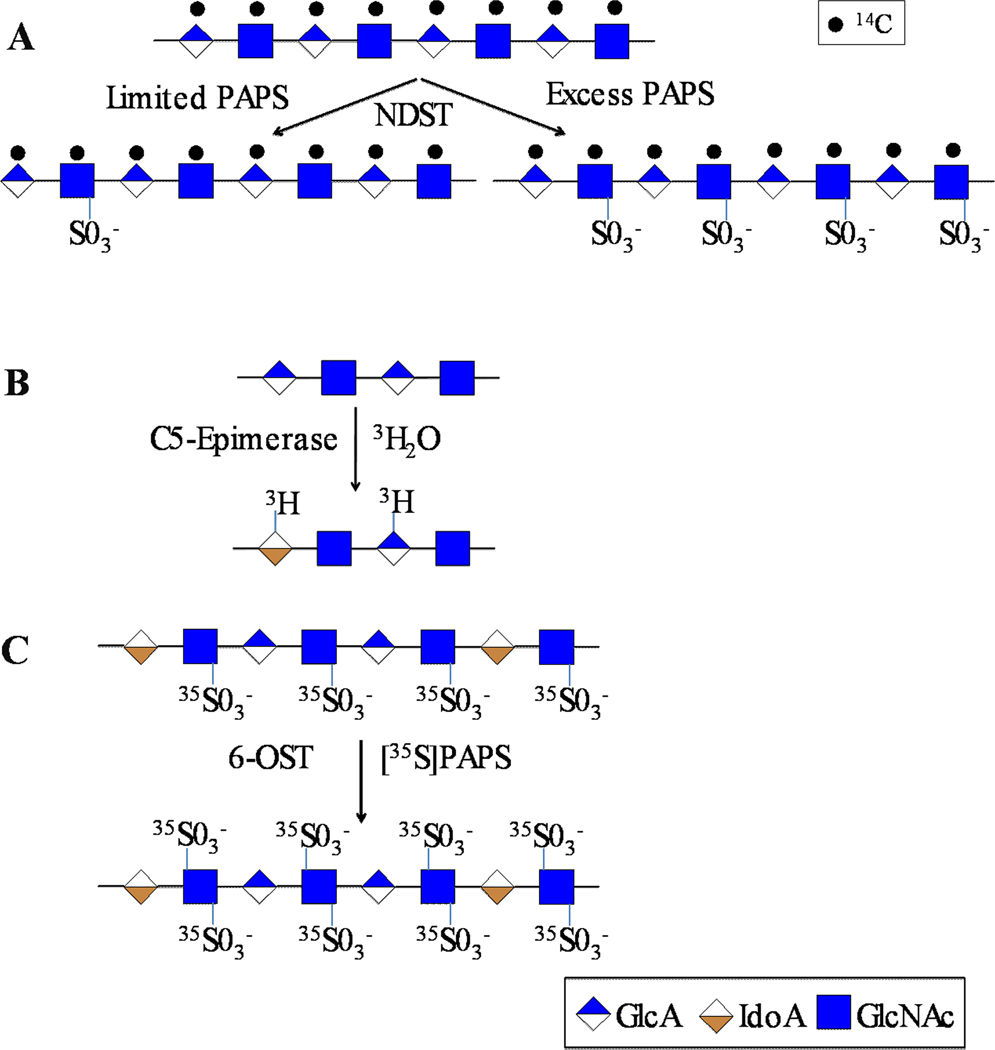
A number of radioisotopes can be used in the preparation/modification of HS fine structures for studying the nature of action of HS biosynthetic enzymes (Table 1). N-acetyl heparosan, a capsular polysaccharide produced by E.coli K5 strain, is the same structure as the unmodified HS /heparin polysaccharide backbone [24]. Therefore, it has been used as a substrate in the enzymatic synthesis of the antithrombin III binding pentasaccharide and in the chemoenzymatic synthesis of heparin-like anticoagulants [25–28]. Furthermore, E.coli K5 strain was grown in the presence of 14C- or 3H-labeled glucose (Glc) to prepare 14C- or 3H-labeled N-acetylheparosan for the study of various HS biosynthetic enzymes. Carlsson et al. extensively used the 14C-labeled N-acetylheparosan as a substrate to determine the nature of action of NDST-1 and NDST-2 under differing amounts of PAPS [29]. It has been shown that PAPS concentration affects the N-deacetylation and N-sulfonation steps and that each NDST isoform has different ratios of N-deacetylation and N-sulfonation relative activity. It has also been shown that these enzymes act on the substrate in a processive or non-processive manner depending on PAPS concentration. In the presence of low amounts of PAPS or in the absence of PAPS, free amine containing GlcN residues were detected. In the presence of an excess of PAPS, extended N-sulfonated domains were synthesized (Figure 2A). This study offers insights into the likely mechanism of differential assembly of heparin and HS. These experiments were performed using a radiolabeled HS precursor backbone that facilitated subsequent structural analyses and allowed researchers to draw these major conclusions.
Table 1.
Common precursors used in the preparation of heparan sulfate / heparin carrying isotopes for various functional and structural biology studies
| Isotope Enriched Precursors for Metabolic Labeling or Modifications |
In Vitro Modifications |
E. coli K5 |
Mammalian Cells |
Ref. |
|---|---|---|---|---|
| [13C]-glucose | − | + | + | [45] |
| [14C]-glucose | − | + | + | [29] [56] |
| [14C]-glucosamine | − | − | + | [31] |
| [14C]-galactose | − | − | + | [31] |
| [3H]-glucose | − | + | + | [29] |
| [3H]-glucosamine | − | − | + | [36] |
| [3H]-galactose | − | − | + | [31] |
| [15N]-ammonium sulfate | − | + | − | [55] |
| [15N]-ammonium chloride | − | + | − | [45] |
| [15N]-glutamine | − | − | + | [52],[57] |
| [35S]-sodium sulfate | − | − | + | [58] |
| [33S]-3'-phosphoadenosine 5'-phosphosulfate | + | − | − | [41] |
| [34S]-3'-phosphoadenosine 5'-phosphosulfate | + | − | − | [41–44] |
| [35S]-3'-phosphoadenosine 5'-phosphosulfate | + | − | − | [33–40] |
Figure 2.
Schematic outline of experiments designed to determine the specificity and activity of heparan sulfate biosynthetic enzymes. A) The scheme illustrates the influence of PAPS concentration on the action of N-deacetylase-N-sulfotransferase (NDST); B) The scheme illustrates the selective incorporation of tritium (3H) at C5-carbon of IdoA generated upon epimerization of GlcA catalyzed by C5-epimerase. Reverse epimerization leads to incorporation of 3H into GlcA residues as well; C) the scheme outlines the incorporation of radioactive sulfates at C6-carbon of GlcNS residues catalyzed by various 6-OST isoforms.
HS C5-epimerase catalyzes the conversion of GlcA residues to IdoA residues. This process regulates the generation of IdoA residues that eventually control the biological actions of HS chains. However, it is difficult to ascertain the stereochemical nature of the uronyl residue and the activity of C5-epimerase using MS because there is no net change in mass. Furthermore, there is significant signal overlap in 1H-NMR spectra which precludes the quantification of epimer content using NMR. Therefore, it has been difficult to study the biochemical action of HS C5-epimerase in vitro. Hagner-Mcwhirter et al. overcame these barriers by using radioisotope labeling to study the epimerization catalyzed by HS C5-epimerase [30–32]. They incubated the appropriate substrate, N-sulfoheparosan, with HS C5-epimerase in 3H2O which led to selective incorporation of 3H at C5 carbon upon epimerization (Figure 2B). Using this method, they could measure the increase in the radioactivity as a read out for the enzyme activity. The product formed was subjected to a nitrous acid degradation procedure that preserves epimer content and the resulting disaccharide pool was analyzed by high performance liquid chromatography (HPLC) coupled to a radiometric detector. Thus, they found that the reaction involves the abstraction of the proton at the C5 carbon of GlcA followed by introduction of a 3H from the medium to generate IdoA carrying 3H at C5 carbon. It is interesting to note that 3H was also incorporated into GlcA containing disaccharide products. These observations suggest that C5-epimerase acts in a reversible manner in a cell free system whereas it acts in an irreversible manner in vivo. This is possibly due to the coupling of epimerization with other modifications that block the reverse reaction in vivo. Only by using radio isotope labeling were these studies able to provide insights into the mechanism of HS C5-epimerase. To study whether the reversibility of the epimerase reaction occurs in a cellular system, Hagner-McWhirter et al. also used a different radio isotope strategy in their studies. They incubated human embryonic kidney 293 cells with [5-3H]-galactose (Gal), a precursor for GlcA, and identified that the GlcA residues of HS were labeled with 3H at the C5 carbon. This method identified epimerized and non-epimerized residues because the 5-3H label would be lost upon C5-epimerization to IdoA. The enzyme activity was closely monitored with the aid of a second isotope by using [14C]-GlcN or [14C]-Gal as a precursor for labeling of cellular HS chains. Incubation of 3H-labeled, mature HS chains with recombinant C5-epimerase failed to release 3H-labels suggesting that C5-epimerization is irreversible in vivo due to the presence of O-sulfate groups in the vicinity.
35S-labeled PAPS, [35S]-PAPS, has been commonly used to investigate specific activities and specificity of various HS sulfotransferases. Rong et al. utilized [35S]-PAPS and [3H]-GlcN to prove that 2-OST catalyzed 2-O-sulfonation of both IdoA and GlcA [33]. The same group used [35S]-PAPS to characterize the substrate specificity of the mouse mastocytoma 2-OST and has shown that 2-OST transfers sulfate more preferentially to IdoA residues than to GlcA residues [33,34]. [35S]-PAPS was also used to characterize the substrate specificity of three isoforms of 6-OST by Habuchi and co-workers (Figure 2C). Based on the generation of specific 35S-labeled disaccharide products by chemical/enzymatic fragmentation of the polymer substrate, they showed that 6-OST1 preferred N-sulfoglucosamine adjacent to IdoA, 6-OST3 targeted N-sulfoglucosamine adjacent to either GlcA or IdoA, and 6-OST2 had a different site preference for modification depending upon the substrate concentrations [35].
Dual isotope labeling by [6-3H]-GlcN and [35S]-PAPS allowed Rosenberg et al. to study the effect of precursor structures on the action of the 3-OST1 enzyme that generates the antithrombin III binding HS anticoagulant structure [36]. They showed that 3-O-sulfonation might occur in the absence of 2-O-sulfonation of IdoA residues. They also showed that 2-O-sulfonation prevented subsequent action of 3-OST1 on GlcN residues located next to the reducing side of IdoA residues. Similarly, Rosenberg et al. used radio isotope labeling technique extensively in vitro to demonstrate the substrate specificity of various isoforms of the 3-OST enzyme family [37,38].
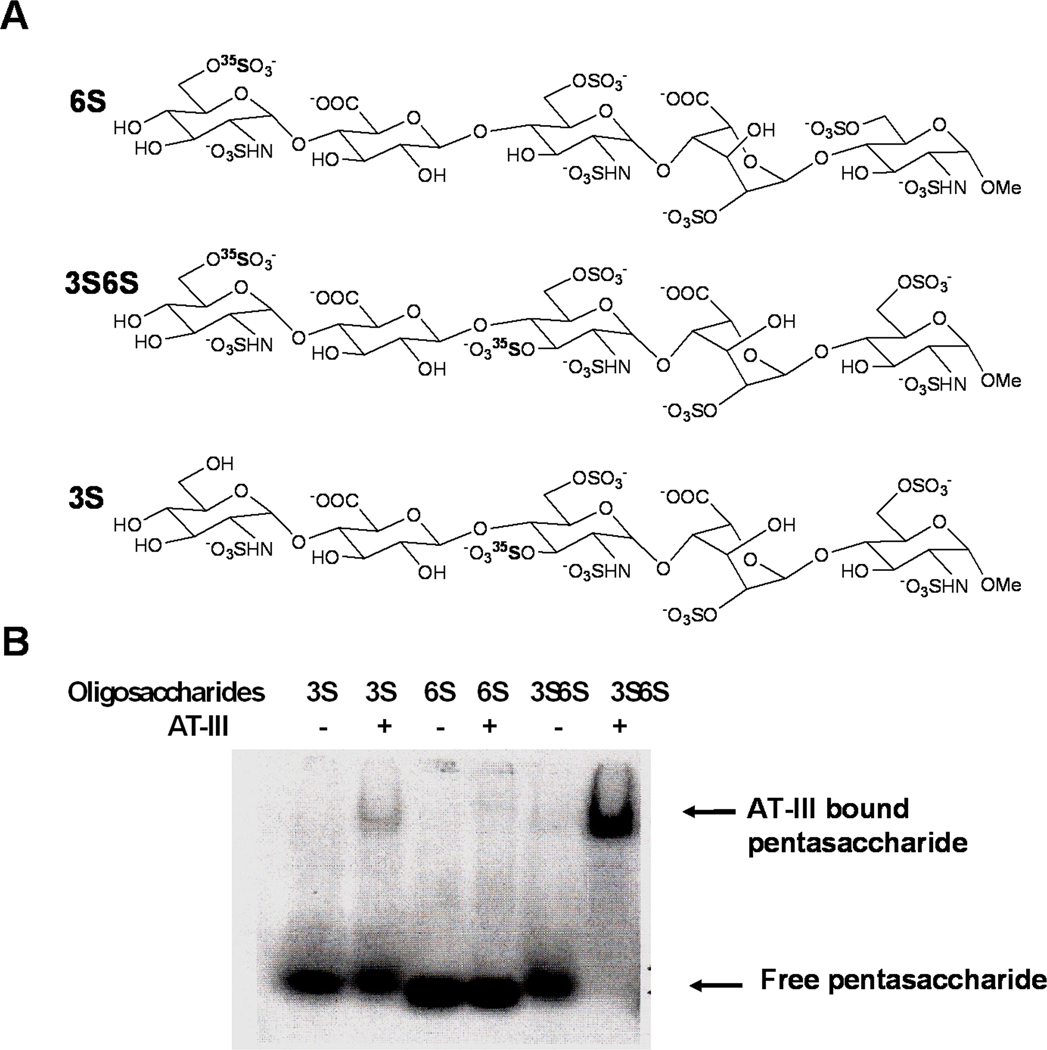
Besides the studies on enzyme specificity, radioisotopes have also been widely used to analyze HS structures and structure-function relationships. To date, it has been difficult to ascertain the fine structural features of HS chains that are required for their biological functions. Therefore, by coupling sulfotransferase reactions with radioisotopes it is possible to label HS chains, analyze enzyme specificity, and to detect HS-protein interactions in a wide variety of binding assays. Wu et al. modified the antithrombin III binding HS pentasaccharide precursor structure with recombinant enzymes, 3-OST and/or 6-OST isoforms, in the presence of [35S]-PAPS to generate radioactive 3-O sulfonated and/or 6-O sulfonated pentasaccharides, 3S, 6S and 3S6S, as shown in Figure 3 [39]. They were then able to use the radiolabeled products in a gel mobility shift assay (GMSA) to study the interactions of different pentasaccharide structures with antithrombin III. After running the electrophoresis under native conditions, the dried gel was autoradiographed by a phosphorImager and subsequently analyzed with the aid of NIH Image and other appropriate software tools to estimate band intensities and calculate the percentage of bound and unbound free heparin pentasaccharides. The pentasaccharides, 6S and 3S, failed to bind to ATIII, i.e. pentasaccharides were not retarded by ATIII, but the pentasaccharide 3S6S could bind to ATIII and was retarded (Figure 3). GMSA experiments proved that 3-O and 6-O sulfates are both equally required to facilitate binding of pentasaccharides to ATIII in a cooperative fashion. They also showed that the GMSA could be applied in any other HS-protein binding studies such as in the study of HS-FGF-FGFR interactions. In another study, heparin oligosaccharides were labeled with [35S]-PAPS by various 3-OST and 6-OST isoforms and the resulting radiolabeled products were utilized to deduce the size, molar ratio, and specific sulfation patterns required for the formation of the HS/FGF1/FGFR1 ternary complex [40].
Figure 3.
Sequence variants of the ATIII binding pentasaccharides. A: 3S, 6S and 3S6S are the pentasaccharides with sulfation at the critical 3-O, 6-O and both positions, respectively. B: Gel mobility shift analysis of labeled pentasaccharides for their ability to bind to ATIII. In each lane, ~ 100, 000 cpm radiolabeled pentasaccharide was incubated with or without ATIII at room temperature for 20 minutes and the complexes were resolved under native conditions.
In summary, radioisotope labeling techniques are clearly powerful in advancing our knowledge of HS biosynthetic enzyme specificity. Radioisotope labeling also facilitates functional studies of specific HS structures. However, this approach does not provide insights into the information pertaining to HS oligosaccharide sequences or specific sulfation patterns at the oligosaccharide level. In addition, radioisotope labeling does not provide insights into three-dimensional structural information. On the other hand, stable isotope enrichment of nuclei with a net spin can provide sequence information and conformational information of HS structures using NMR techniques.
Stable isotopes in the mass spectrometry analysis of heparan sulfate / heparin
Structural analysis of HS/heparin by NMR or HPLC has been inadequate in advancing our knowledge of the fine structural features of the polysaccharide chain. In addition, NMR is limited because it frequently requires large amounts of samples that are very difficult to obtain from samples of biological origin. HPLC analysis is limited because of solvent effects and a need for standards. MS alone overcomes many of the problems associated with NMR and HPLC techniques however it also has several disadvantages. Different levels of sulfation and sulfation patterns affect ionization in MS analysis and make quantitative disaccharide analysis a challenge. Thus, to overcome these inherent problems associated with MS analysis, introduction of stable isotope labeling technique into HS/heparin samples has proven to be a powerful strategy for structural analysis by MS.
The mass spectrum of a given molecular ion typically consists of a sum of signals of various possible naturally occurring isotopic compositions. Thus, the isotopic patterns of molecular ions provide further insights into fine structure. Therefore, it is important to have high resolution mass spectra for which one needs time-of-flight (TOF), orbitrap or fourier-transform ion cyclotron resonance (FT-ICR) MS. The strategy to introduce stable isotopes into HS oligosaccharides usually involves the following steps: 1) synthesis of [34S]-PAPS or [33S]-PAPS; 2) modification of HS oligosaccharides using a recombinant biosynthetic enzyme with the aid of [34 or 33S]-PAPS; and 3) liquid chromatography separation of HS oligosaccharides and subsequent analysis by IPRP-capillary LC-micro ESI-TOF-MS. The stable isotope labeling technique also utilizes the advantages of MS (high resolution, small sample size requirements, and fast analysis) to provide more information about HS structures whereas radioisotope labeling techniques do not provide the same molecular resolution and specific information about HS oligosaccharide sequences. Several studies have provided new insights into heparin structure-function relationships by employing the stable isotope labeling strategy coupled with LC-MS analysis as described in the following sections.
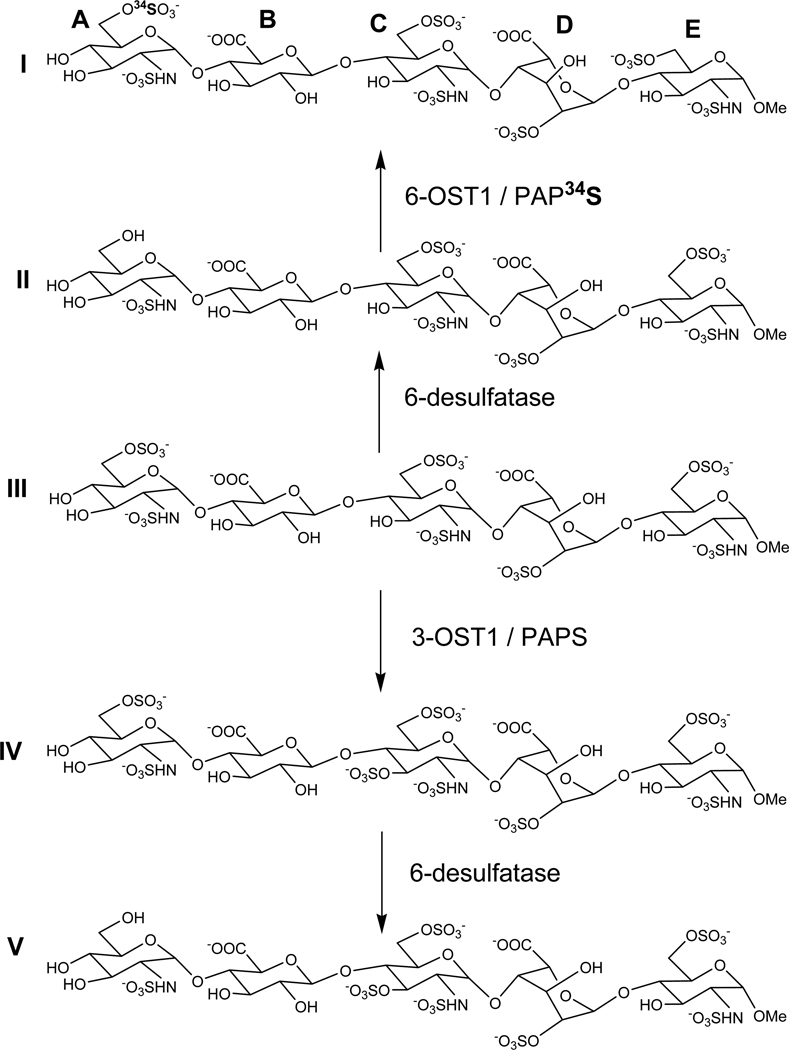
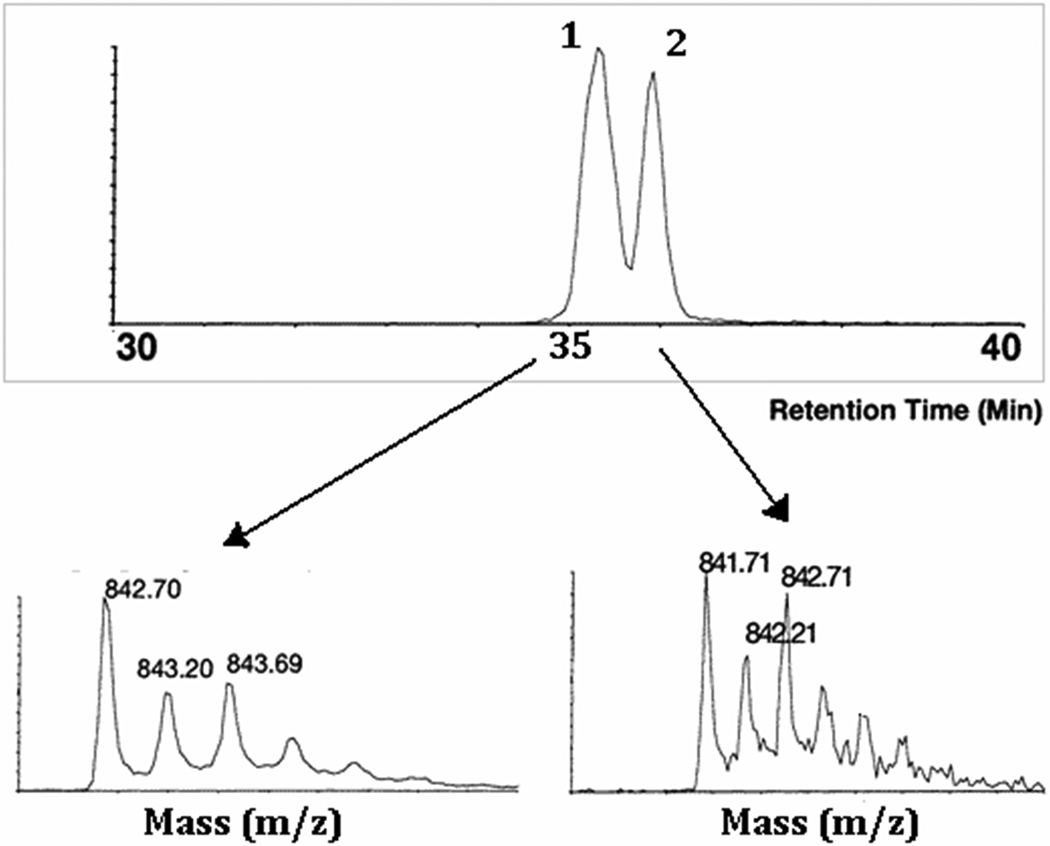
Kuberan et al. utilized this stable isotope labeling strategy to characterize the antithrombin III binding pentasaccharide and its precursors [41]. Heparin elicits its anticoagulant effect through specific binding of the following pentasaccharide sequence, present within the polysaccharide, to antithrombin III (ATIII): GlcNS/Ac(6S)-GlcA-GlcNS(3S ±6S)-IdoA(2S)-GlcNS(6S). Extensive biochemical and biophysical studies carried out over three decades indicate that the 6-O sulfate group of sugar residue A and the 3-O sulfate group of sugar residue C are the most critical ones for the anticoagulant action of heparin. This major discovery could only be made by synthesizing pentasaccharide sequence variants (see scheme 1) and resolving them. However, these variant structures are difficult to characterize because they closely resemble each other. For example, pentasaccharide I and V differ from one another only by the position of sulfate groups as they both carry the same number of sulfate groups. However, these structures were enzymatically synthesized such that pentasaccharide I carry a 34S-isotope enriched sulfate group at the C6-position of glucosamine residue A whereas pentasaccharide V carry a 32S-isotope containing sulfate group at the C3-position of glucosamine residue C. Thus, these pentasaccharides differ in their molecular weights by 2 daltons. The mixture containing both pentasaccharide I and V was resolved through a reverse phase C18 column using dibutyl ammonium acetate (DBA) as an ion-pairing agent. The resolved pentasaccharides were analyzed through online ESI mass spectrometry to identify specific structures. The observed abundant pseudo molecular ion for pentasaccharide I was 842.71 corresponding to [M+2(DBA)-4H]2− and that for pentasaccharide V was 841.71 corresponding to [M+2(DBA)-4H]2 (Figure 4). Adductions with the ion-pairing agent, DBA, were always observed for these pentasaccharides in the abundant double charge state. Thus, these studies demonstrated for the first time the potential utility of isotopes in identifying sequences that are resolved based on sulfation pattern/position of sulfate groups. Resolving and identifying these complex structures with the aid of isotopes will not only improve the confidence about the homogeneity of unknown structures but also the identity of heparin/HS oligosaccharide sequence variants.
Scheme 1.
Enzymatic synthesis of differentially 34S-isotope enriched heparin pentasaccharide isoforms carrying an identical number of sulfate groups
Figure 4.
Total ion chromatogram of pentasaccharide I (peak 1) and pentasaccharide V (peak 2), having the same number of sulfate groups, resolved based on the sulfation pattern using IPRP-capillary HPLC and subsequently characterized by online micro ESI-TOF-MS. Electrospray mass spectra of pentasaccharides I and V are shown in the lower panel. Pentasaccharide I carries a 6-O-34SO3− group as a mass spectral probe to be distinguished from its isomer pentasaccharide V (see scheme 1 for structural information).
In another application of this strategy, Wu and Lech utilized [34S]-PAPS and 6-OST1 labeling to determine the disaccharide profile of the non-reducing end of HS from bovine kidney [42]. This was the first report that described the determination of HS non-reducing end structures using mass spectrometry with the aid of stable isotope labeling. Utilizing this information, the authors showed that the non-reducing end of HS is not cleaved by heparanase and is still capable of forming the HS-FGF-FGFR ternary complex. Wu and Lech also applied this strategy to study the enzymatic actions of 3-OST1 and 6-OST1 on bovine kidney HS [43,44]. In these studies, the authors labeled bovine kidney HS with [34S]-PAPS using 3-OST1 and 6-OST1 and subsequently digested samples with nitrous acid or heparin lyases. The overlapping oligosaccharides containing the stable isotope enrichment could then be identified using changes in m/z profiles. Using the information from the isotope enrichment and LC-MS, they showed a detailed profile of the types and levels of 3-O and 6-O sulfonation sites present in the bovine kidney HS.
Uniformly 13C, 15N- labeled HS backbone can be produced by growing E.coli K5 strain in minimal media containing 13C-Glc and 15N-ammonium chloride or 15N-ammonium sulfate. In fact, our lab has recently produced uniformly 13C, 15N-labeled N-acetyl heparosan for structural biology studies and characterized isotopic enrichment using MS (Figure 5) [55]. More recently, Zhang et al. have utilized 13C, 15N-enriched N-acetyl heparosan as a starting precursor and prepared various disaccharide standards for quantitative disaccharide analysis of HS samples [45]. The isolated precursor was subjected to various enzymatic modifications and was then digested with heparin lyases to obtain disaccharide products. Using these isotope enriched disaccharides as internal MS standards, the authors were able to compare the disaccharide compositions of various HS chains accurately in a fast and reliable manner. The use of isotope-enriched samples as internal standards in MS analysis will greatly improve the speed and accuracy of assessing the quality of pharmaceutical heparin and deducing batch-to-batch variations.
Figure 5.
ESI-MS spectra of N-acetylheparosan disaccharide units of the polymers isolated from E.coli K5 strain grown in minimal medium containing 15N-ammonium sulfate/Uniformly 13C-labeled glucose (bottom panel) and E.coli K5 strain grown in normal LB culture broth (top panel).
Recently, reductive labeling of GAG structures of various species at the reducing end with stable isotopes enabled rapid compositional analysis using tandem MS. Lawrence et al. utilized a labeling strategy, known as glycan reductive isotope labeling (GRIL)-LC-MS, to tag the reducing ends of disaccharides to simultaneously quantify the evolutionary differences in HS fine structures between various species [46]. They were able to differentiate the disaccharide compositions of commercial heparin, keratan sulfate, HS and chondroitin sulfate (CS) from hydra vulgaris, drosophila melanogaster, caenorhabditis elegans, and mammalian cells. They stated that the GRIL-LC-MS method had a sensitivity limit of 1 pmol/disaccharide.
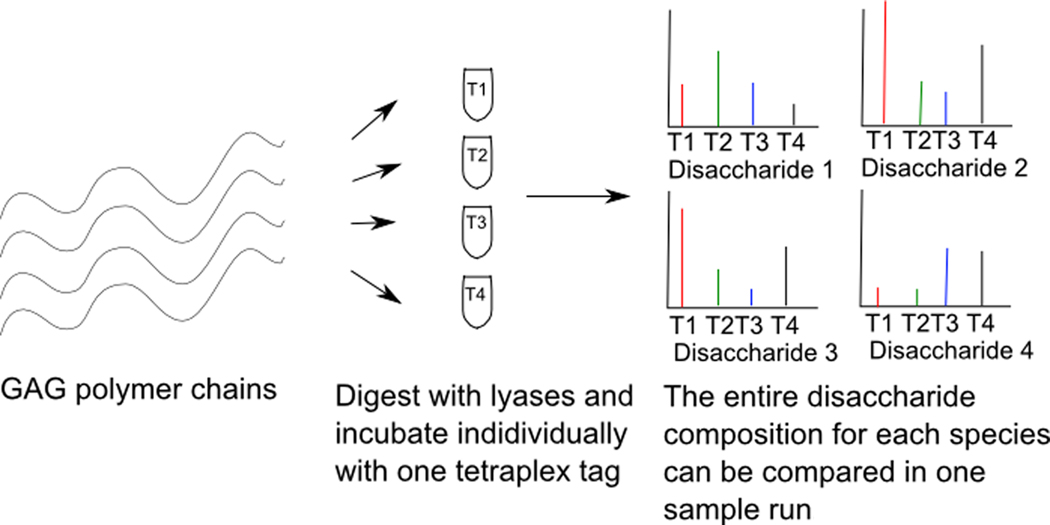
While labeling of each GAG sample with a single isotope and subsequently comparing between samples has been used to effectively show GAG differences between species, the results can sometimes be complicated by the presence of overlapping isotopic peaks. Therefore, samples need to be separately run on LC-MS and compared between two independent runs. To overcome these challenges, Bowman et al. have developed an elegant tetraplex tagging strategy for studying quantitative glycomics [47,48]. By utilizing tags that are four mass units apart, it is possible to reductively label disaccharides from different species with isotopes that do not have overlapping peaks in the mass spectrum (Figure 6). This allows for the simultaneous compositional profiling of disaccharides, which overcomes the error introduced by comparing independent sample runs and also increases sample throughput. Using this technique, the authors were able to compare CS, heparin, and several other glycans of various origins simultaneously.
Figure 6.
Schematic overview of comparative glycosaminoglycan compositional profiling of different species simultaneously using tetraplex isotope coded tags. Total GAG chains isolated from different species are digested into disaccharides using lyases and incubated individually with one tetraplex tag each. After labeling with the tetraplex tags and purifying the GAGs, labeled disaccharides from different species are combined into one sample and analyzed simultaneously on the mass spectrometer.
In summary, the availability of stable isotope enriched disaccharides provides unprecedented opportunity to rapidly and accurately obtain the disaccharide composition in a quantitative manner. In the near future, one can expect the production of rare 3-O sulfonated isotope-enriched disaccharides and oligosaccharides for accelerating the discovery of rare modifications in biological samples and correlating their relative changes with pathophysiological conditions using MS techniques.
Stable isotopes in the nuclear magnetic resonance analysis of heparan sulfate / heparin
NMR and MS are the primary tools for analyzing GAG structures. Each method has different advantages and disadvantages. Even though MS is a sensitive technique that requires a small amount of GAG samples, it can only be used to characterize oligosaccharides and disaccharides in terms of molecular weights, sulfation density, sulfation pattern, and perhaps epimerization. MS is not amenable for characterizing the entire heparanome at the polymer level. Furthermore, NMR is not a highly sensitive method because of the low magnetogyric ratio and low natural abundance of stable isotopes such as 13C (1.1%) and 15N (0.37%). However, NMR can give detailed information regarding conformation, microstructure, and dynamic solution properties of the intact HS chains. Two emerging strategies can be employed to overcome the difficulties associated with the requirement of large amounts of samples for NMR analysis. One approach is to use capillary isotachophoresis hyphenated with on-line microcoil NMR probe technology for improved detection and analysis of microgram quantities of HS chains of biological origin [14,16]. Another approach to overcome the barrier is to produce 15N- and/or 13C- enriched HS precursors from E.coli K5 strain and then enzymatically modify the backbone with recombinant enzymes to obtain HS and heparin polysaccharides for solution structure analysis [49]. However, the enzymatic approach does not have the capability to generate HS structures with discreet domain organizations as found in nature.
The 1H-NMR spectra of HS and heparin provide rapid and semi-qualitative information on monosaccharide constituents and types of substitutions in samples. However, traditional one-dimensional proton NMR spectra are complex and difficult to interpret due to the extensive proton signal overlap. To overcome the problems associated with extensive proton spectral overlap, two-dimensional NMR methods such as heteronuclear single quantum coherence (HSQC) can be employed. By correlating the chemical shifts of heteronuclei in an additional spectral dimension, these methods reduce spectral overlap. Two-dimensional NMR was recently used to identify a contaminant, oversulfated CS that caused several deaths and severe allergic responses, in pharmaceutical heparin preparations [50,51].
[1H, 15N] HSQC technique was recently used for structural characterization of GAG chains of cellular origin. Using this technique, CS, dermatan sulfate (DS) and HS were identified because they gave cross peaks in distinct regions of the NMR spectrum [52]. This was possible because of metabolic labeling of cellular GAG chains by supplementing cell growth media with [15N]-glutamine. The study documented the effect of sulfonation at different sites in CS chains by tracking the positions of 1H-15N cross peaks in HSQC spectra from 15N containing N-acetylgalactosamine in CS-A and CS-C. Another potential application of isotope enrichment is that 2D-HSQC analysis of HS can be used to identify differences in the fine structure and domain organization of HS as a function of cell type. In this recent study, endothelial cells and CHO cells were grown under defined conditions to obtain 15N-enriched HS chains and their HSQC cross peaks were analyzed. It was found that the 1H-15N HSQC cross-peak was less intense near δN = 123.6 ppm/ δH = 8.36 ppm for HS chains isolated from endothelial cells whereas a strong cross peak was observed for HS of CHO cell origin. Thus, this study demonstrated the utility of isotope enrichment in the comparative structural analysis of minute amounts of intact HS chains from two different cellular origins [52]. A potential application of this powerful technique may be to prepare isotope enriched heparanomes of various organ types during the development of an organism and to correlate structural changes in HS to the signaling pathways that are critical for proper development at various time points. The Almond group has purified homogenous oligosaccharides of unsulfated CS (with or without 15N enrichment) from a capsular polysaccharide isolated from E.coli K4 strain and analyzed using high field NMR to compare published CS and hyaluronan 3D structures [53]. The 1H and 13C atoms were assigned using [1H, 13C] HSQC, [1H, 15N] HSQC, [1H, 13C] HMBC and [1H, 1H] COSY experiments. Our group recently reported the preparation of 15N-labeled N-acetylheparosan oligosaccharides of various sizes and assessed their structures by [1H, 15N] HSQC analysis and mass spectrometry (Figure 7) [54]. Our lab also successfully prepared and analyzed uniformly 13C, 15N- labeled N-acetylheparosan using [1H, 13C] HSQC technique (Figure 8). Zhang et al. synthesized uniformly 13C, 15N-labeled heparin-like polysaccharides using chemoenzymatic approaches for the first time and utilized 2D and 3D NMR techniques to examine the solution structures of the uniformly labeled heparin polysaccharides [49]. Uniformly isotope labeled HS oligosaccharides will serve as excellent molecular probes to define HS microstructures and HS-protein interactions with high precision in the coming years. However, one of the disadvantages of uniform 13C-labeling is that direct detection of 13C signals may not be easy. Additionally, it may be difficult to assign the NMR signals due to the presence of one-bond 13C−13C couplings between adjacent carbons that split the signals into multiple peaks. This could also lead to reduced sensitivity. One potential approach to overcome this difficulty may be to produce atom specifically 13C-labeled HS chains [55]. Furthermore, deuterium incorporation can enhance signals for carbons that have faster relaxation rates in the larger heparanome. In fact, our lab has begun to make progress in the production of various isotope labeled HS precursors to address several unresolved issues in the structural biology of the heparanome.
Figure 7.
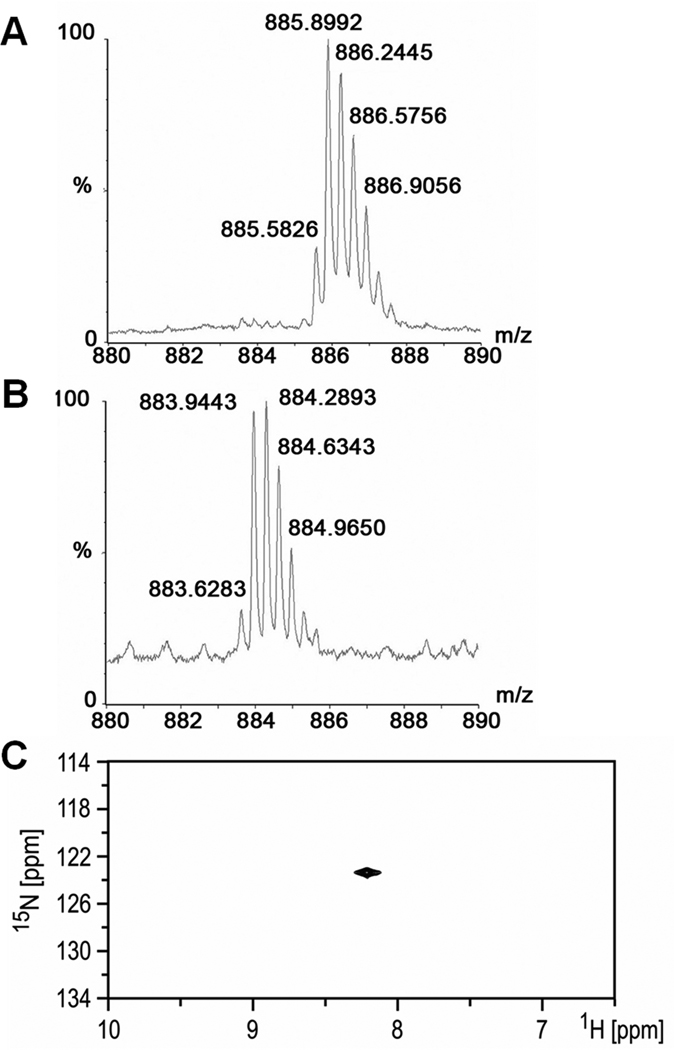
Comparison of the isotopic pattern of molecular ions corresponding to 14N- and 15N- labeled N-acetylheparosan tetradecasaccharide. Analysis of each oligosaccharide was performed using ESI-TOF-MS so as to provide higher isotopic resolution. A: mass spectrum of 15N labeled N-acetylheparosan tetradecasaccharide B:mass spectrum of 14N labeled N-acetylheparosan tetradecasaccharide. C: [15N, 1H] HSQC spectrum of 15N labeled N-acetylheparosan
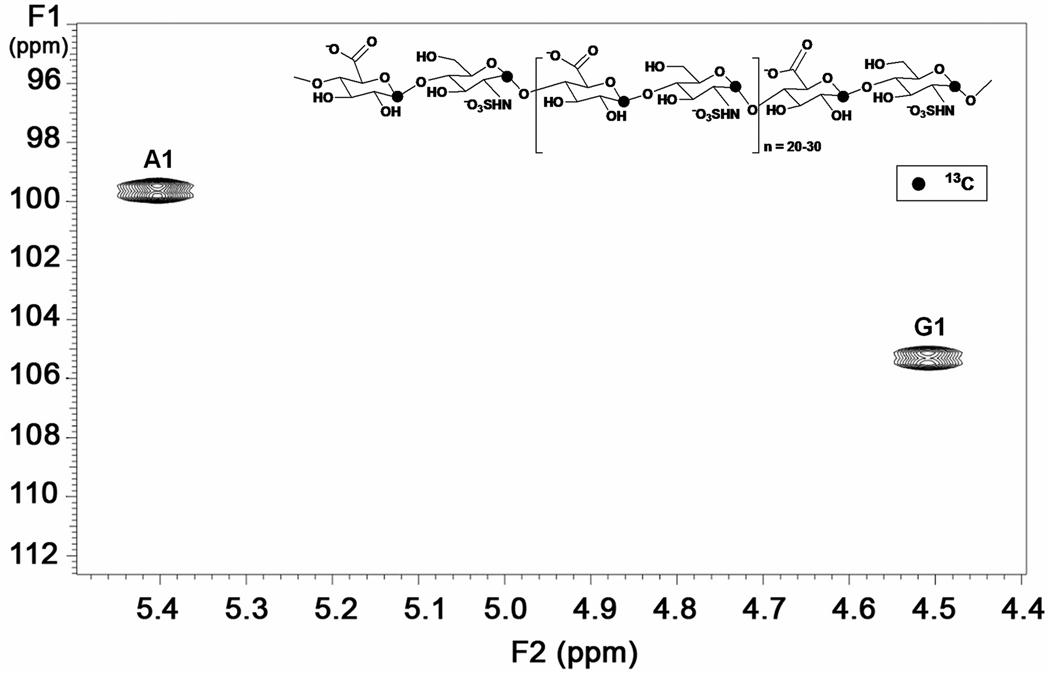
Figure 8.
A selected region of the two-dimensional [13C, 1H] HSQC spectrum of atom-specifically 13C labeled HS precursor polysaccharide at anomeric carbons showing cross peaks for anomeric signals. A1: glucosamine unit, G1: hexuronic acid unit
Conclusions
There are several options available in the preparation of isotope enriched HS and heparin oligosaccharides. Utilizing 15N-labeled HS and uniformly or atom-specifically 13C-labeled HS will undoubtedly yield information on rapid dynamics of higher order structures. Additionally, by utilizing 33S- and 34S-labeled HS oligosaccharides, it may be possible to optimize ionization conditions and fragmentation strategies to deduce sequence information of larger oligosaccharides in MS-based techniques. A library of isotope enriched disaccharides can be applied as internal standards together with GAG samples of various biological origins to enhance the detection of unknown or rare disaccharides. Thus, one can accurately deduce GAG profiles with high confidence and reproducibility with the aid of stable isotopes. We believe that recent advances in the production of recombinantly synthesized isotope enriched HS along with improvements in online nano-MS and microcoil- NMR techniques will lead to further discoveries in the area of structural and functional heparanomics. We look forward to dramatic advances in the coming decade.
Acknowledgements
The research program of B.K is supported by National Institutes of Health grant (GM075168), Human Frontier Science Program grant, American Heart Association National Scientist Development Award and Mizutani Foundation for Glycoscience Award. T.K.N.N. acknowledges a graduate fellowship from the Vietnam Education Foundation.
References
- 1.Raman K, Kuberan B. Curr Chem Biol. 2010;4(1):20–31. doi: 10.2174/187231310790226206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lortat-Jacob H. Curr Opin Struct Biol. 2009;19(5):543–548. doi: 10.1016/j.sbi.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Xian X, Gopal S, Couchman JR. Cell Tissue Res. 2010;339(1):31–46. doi: 10.1007/s00441-009-0829-3. [DOI] [PubMed] [Google Scholar]
- 4.Raghuraman A, Mosier PD, Desai UR. J Med Chem. 2006;49(12):3553–3562. doi: 10.1021/jm060092o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capila I, Linhardt RJ. Angew Chem Int Ed Engl. 2002;41(3):391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 6.Cummings RD. Mol Biosyst. 2009;5(10):1087–1104. doi: 10.1039/b907931a. [DOI] [PubMed] [Google Scholar]
- 7.Victor XV, Nguyen TK, Ethirajan M, Tran VM, Nguyen KV, Kuberan B. J Biol Chem. 2009;284(38):25842–25853. doi: 10.1074/jbc.M109.043208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakato H, Kimata K. Biochim Biophys Acta. 2002;1573(3):312–318. doi: 10.1016/s0304-4165(02)00398-7. [DOI] [PubMed] [Google Scholar]
- 9.Zaia J. Chem Biol. 2008;15(9):881–892. doi: 10.1016/j.chembiol.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaia J. Mass Spectrom Rev. 2009;28(2):254–272. doi: 10.1002/mas.20200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casu B, Lindahl U. Adv Carbohydr Chem Biochem. 2001;57:159–206. doi: 10.1016/s0065-2318(01)57017-1. [DOI] [PubMed] [Google Scholar]
- 12.Cros S, Petitou M, Sizun P, Perez S, Imberty A. Bioorg Med Chem. 1997;5(7):1301–1309. doi: 10.1016/s0968-0896(97)00087-4. [DOI] [PubMed] [Google Scholar]
- 13.Hricovini M, Guerrini M, Bisio A, Torri G, Naggi A, Casu B. Semin Thromb Hemost. 2002;28(4):325–334. doi: 10.1055/s-2002-34301. [DOI] [PubMed] [Google Scholar]
- 14.Korir AK, Almeida VK, Malkin DS, Larive CK. Anal Chem. 2005;77(18):5998–6003. doi: 10.1021/ac050669u. [DOI] [PubMed] [Google Scholar]
- 15.Korir AK, Larive CK. Anal Bioanal Chem. 2009;393(1):155–169. doi: 10.1007/s00216-008-2412-2. [DOI] [PubMed] [Google Scholar]
- 16.Korir AK, Larive CK. Anal Bioanal Chem. 2007;388(8):1707–1716. doi: 10.1007/s00216-007-1400-2. [DOI] [PubMed] [Google Scholar]
- 17.Mulloy B, Forster MJ. Glycobiology. 2000;10(11):1147–1156. doi: 10.1093/glycob/10.11.1147. [DOI] [PubMed] [Google Scholar]
- 18.Przybylski C, Gonnet F, Bonnaffe D, Hersant Y, Lortat-Jacob H, Daniel R. Glycobiology. 2010;20(2):224–234. doi: 10.1093/glycob/cwp169. [DOI] [PubMed] [Google Scholar]
- 19.Saad OM, Leary JA. Anal Chem. 2005;77(18):5902–5911. doi: 10.1021/ac050793d. [DOI] [PubMed] [Google Scholar]
- 20.Tissot B, Gasiunas N, Powell AK, Ahmed Y, Zhi ZL, Haslam SM, Morris HR, Turnbull JE, Gallagher JT, Dell A. Glycobiology. 2007;17(9):972–982. doi: 10.1093/glycob/cwm072. [DOI] [PubMed] [Google Scholar]
- 21.Venkataraman G, Shriver Z, Raman R, Sasisekharan R. Science. 1999;286(5439):537–542. doi: 10.1126/science.286.5439.537. [DOI] [PubMed] [Google Scholar]
- 22.Yates EA, Santini F, Guerrini M, Naggi A, Torri G, Casu B. Carbohydr Res. 1996;294:15–27. doi: 10.1016/s0008-6215(96)90611-4. [DOI] [PubMed] [Google Scholar]
- 23.Sugahara K, Kitagawa H. IUBMB Life. 2002;54(4):163–175. doi: 10.1080/15216540214928. [DOI] [PubMed] [Google Scholar]
- 24.Vann WF, Schmidt MA, Jann B, Jann K. Eur J Biochem. 1981;116(2):359–364. doi: 10.1111/j.1432-1033.1981.tb05343.x. [DOI] [PubMed] [Google Scholar]
- 25.Kuberan B, Lech MZ, Beeler DL, Wu ZL, Rosenberg RD. Nat Biotechnol. 2003;21(11):1343–1346. doi: 10.1038/nbt885. [DOI] [PubMed] [Google Scholar]
- 26.Kuberan B, Beeler DL, Lawrence R, Lech M, Rosenberg RD. J Am Chem Soc. 2003;125(41):12424–12425. doi: 10.1021/ja036737g. [DOI] [PubMed] [Google Scholar]
- 27.Kuberan B, Beeler DL, Lech M, Wu ZL, Rosenberg RD. J Biol Chem. 2003;278(52):52613–52621. doi: 10.1074/jbc.M305029200. [DOI] [PubMed] [Google Scholar]
- 28.Lindahl U, Li JP, Kusche-Gullberg M, Salmivirta M, Alaranta S, Veromaa T, Emeis J, Roberts I, Taylor C, Oreste P, Zoppetti G, Naggi A, Torri G, Casu B. J Med Chem. 2005;48(2):349–352. doi: 10.1021/jm049812m. [DOI] [PubMed] [Google Scholar]
- 29.Carlsson P, Presto J, Spillmann D, Lindahl U, Kjellen L. J Biol Chem. 2008;283(29):20008–20014. doi: 10.1074/jbc.M801652200. [DOI] [PubMed] [Google Scholar]
- 30.Hagner-McWhirter A, Hannesson HH, Campbell P, Westley J, Roden L, Lindahl U, Li JP. Glycobiology. 2000;10(2):159–171. doi: 10.1093/glycob/10.2.159. [DOI] [PubMed] [Google Scholar]
- 31.Hagner-McWhirter A, Li JP, Oscarson S, Lindahl U. J Biol Chem. 2004;279(15):14631–14638. doi: 10.1074/jbc.M313760200. [DOI] [PubMed] [Google Scholar]
- 32.Hagner-Mcwhirter A, Lindahl U, Li J. Biochem J. 2000;347(Pt 1):69–75. [PMC free article] [PubMed] [Google Scholar]
- 33.Rong J, Habuchi H, Kimata K, Lindahl U, Kusche-Gullberg M. Biochemistry. 2001;40(18):5548–5555. doi: 10.1021/bi002926p. [DOI] [PubMed] [Google Scholar]
- 34.Rong J, Habuchi H, Kimata K, Lindahl U, Kusche-Gullberg M. Biochem J. 2000;346(Pt 2):463–468. [PMC free article] [PubMed] [Google Scholar]
- 35.Habuchi H, Tanaka M, Habuchi O, Yoshida K, Suzuki H, Ban K, Kimata K. J Biol Chem. 2000;275(4):2859–2868. doi: 10.1074/jbc.275.4.2859. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, Lawrence R, Schwartz JJ, Bai X, Wei G, Esko JD, Rosenberg RD. J Biol Chem. 2001;276(31):28806–28813. doi: 10.1074/jbc.M100204200. [DOI] [PubMed] [Google Scholar]
- 37.Shworak NW, Liu J, Fritze LM, Schwartz JJ, Zhang L, Logeart D, Rosenberg RD. J Biol Chem. 1997;272(44):28008–28019. doi: 10.1074/jbc.272.44.28008. [DOI] [PubMed] [Google Scholar]
- 38.Shworak NW, Liu J, Petros LM, Zhang L, Kobayashi M, Copeland NG, Jenkins NA, Rosenberg RD. J Biol Chem. 1999;274(8):5170–5184. doi: 10.1074/jbc.274.8.5170. [DOI] [PubMed] [Google Scholar]
- 39.Wu ZL, Zhang L, Beeler DL, Kuberan B, Rosenberg RD. FASEB J. 2002;16(6):539–545. doi: 10.1096/fj.01-0807com. [DOI] [PubMed] [Google Scholar]
- 40.Wu ZL, Zhang L, Yabe T, Kuberan B, Beeler DL, Love A, Rosenberg RD. J Biol Chem. 2003;278(19):17121–17129. doi: 10.1074/jbc.M212590200. [DOI] [PubMed] [Google Scholar]
- 41.Kuberan B, Lech M, Zhang L, Wu ZL, Beeler DL, Rosenberg RD. J Am Chem Soc. 2002;124(29):8707–8718. doi: 10.1021/ja0178867. [DOI] [PubMed] [Google Scholar]
- 42.Wu ZL, Lech M. J Biol Chem. 2005;280(40):33749–33755. doi: 10.1074/jbc.M505677200. [DOI] [PubMed] [Google Scholar]
- 43.Wu ZL, Lech M. Biochem J. 2005;389(Pt 2):383–388. doi: 10.1042/BJ20041827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu ZL, Lech M, Beeler DL, Rosenberg RD. J Biol Chem. 2004;279(3):1861–1866. doi: 10.1074/jbc.M311398200. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Z, Xie J, Liu H, Liu J, Linhardt RJ. Anal Chem. 2009;81(11):4349–4355. doi: 10.1021/ac9001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lawrence R, Olson SK, Steele RE, Wang L, Warrior R, Cummings RD, Esko JD. J Biol Chem. 2008;283(48):33674–33684. doi: 10.1074/jbc.M804288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bowman MJ, Zaia J. Anal Chem. 2010;82(7):3023–3031. doi: 10.1021/ac100108w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowman MJ, Zaia J. Anal Chem. 2007;79(15):5777–5784. doi: 10.1021/ac070581b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z, McCallum SA, Xie J, Nieto L, Corzana F, Jimenez-Barbero J, Chen M, Liu J, Linhardt RJ. J Am Chem Soc. 2008;130(39):12998–13007. doi: 10.1021/ja8026345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blossom DB, Kallen AJ, Patel PR, Elward A, Robinson L, Gao G, Langer R, Perkins KM, Jaeger JL, Kurkjian KM, Jones M, Schillie SF, Shehab N, Ketterer D, Venkataraman G, Kishimoto TK, Shriver Z, McMahon AW, Austen KF, Kozlowski S, Srinivasan A, Turabelidze G, Gould CV, Arduino MJ, Sasisekharan R. N Engl J Med. 2008;359(25):2674–2684. doi: 10.1056/NEJMoa0806450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kishimoto TK, Viswanathan K, Ganguly T, Elankumaran S, Smith S, Pelzer K, Lansing JC, Sriranganathan N, Zhao G, Galcheva-Gargova Z, Al-Hakim A, Bailey GS, Fraser B, Roy S, Rogers-Cotrone T, Buhse L, Whary M, Fox J, Nasr M, Dal Pan GJ, Shriver Z, Langer RS, Venkataraman G, Austen KF, Woodcock J, Sasisekharan R. N Engl J Med. 2008;358(23):2457–2467. doi: 10.1056/NEJMoa0803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pomin VH, Sharp JS, Li X, Wang L, Prestegard JH. Anal Chem. 2010;82(10):4078–4088. doi: 10.1021/ac1001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sattelle BM, Shakeri J, Roberts IS, Almond A. Carbohydr Res. 2010;345(2):291–302. doi: 10.1016/j.carres.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sigulinsky C, Babu P, Victor XV, Kuberan B. Carbohydr Res. 2010;345(2):250–256. doi: 10.1016/j.carres.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen TK, Tran VM, Victor XV, Skalicky JJ, Kuberan B. Carbohydr Res. 2010 doi: 10.1016/j.carres.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roman E, Roberts I, Lidholt K, Kusche-Gullberg M. Biochem J. 2003;374(Pt 3):767–772. doi: 10.1042/BJ20030365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Orlando R, Lim JM, Atwood JA, 3rd, Angel PM, Fang M, Aoki K, Alvarez-Manilla G, Moremen KW, York WS, Tiemeyer M, Pierce M, Dalton S, Wells L. J Proteome Res. 2009;8(8):3816–3823. doi: 10.1021/pr8010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li JP, Gong F, El Darwish K, Jalkanen M, Lindahl U. J Biol Chem. 2001;276(23):20069–20077. doi: 10.1074/jbc.M011783200. [DOI] [PubMed] [Google Scholar]