Abstract
Histone methylation is known to be associated with both transcriptionally active and repressive chromatin states. Recent studies have identified SET domain–containing proteins such as SUV39H1 and Clr4 as mediators of H3 lysine 9 (Lys9) methylation and heterochromatin formation. Interestingly, H3 Lys9 methylation is not observed from bulk histones isolated from asynchronous populations of Saccharomyces cerevisiae or Tetrahymena thermophila. In contrast, H3 lysine 4 (Lys4) methylation is a predominant modification in these smaller eukaryotes. To identify the responsible methyltransferase(s) and to gain insight into the function of H3 Lys4 methylation, we have developed a histone H3 Lys4 methyl-specific antiserum. With this antiserum, we show that deletion of SET1, but not of other putative SET domain–containing genes, in S. cerevisiae, results in the complete abolishment of H3 Lys4 methylation in vivo. Furthermore, loss of H3 Lys4 methylation in a set1Δ strain can be rescued by SET1. Analysis of histone H3 mutations at Lys4 revealed a slow-growth defect similar to a set1Δ strain. Chromatin immunoprecipitation assays show that H3 Lys4 methylation is present at the rDNA locus and that Set1-mediated H3 Lys4 methylation is required for repression of RNA polymerase II transcription within rDNA. Taken together, these data suggest that Set1-mediated H3 Lys4 methylation is required for normal cell growth and transcriptional silencing.
Keywords: SET1, methylation, histone H3, Lys4, rDNA silencing, SET domain
Eukaryotic DNA is assembled with histone proteins to form nucleosomes, which are the basic repeating unit of chromatin. Chromatin assembly and its folding into higher-order structures are dynamic processes that ultimately influence either a permissive or restrictive environment for proteins seeking access to the underlying DNA. Although nucleosomal structure and its association with DNA is well characterized (Luger et al. 1997), it is not fully understood how chromatin functions are regulated at a molecular level. Chromatin remodeling and covalent modification of histones are two means by which variation is introduced into the chromatin polymer, thereby regulating the structure and function of chromatin and ultimately gene expression (Wu et al. 1998; Spencer and Davie 1999).
Several posttranslational modifications are known to exist on the terminal domains of histones, such as acetylation, phosphorylation, ubiquitination, ADP ribosylation, and methylation (van Holde 1989). Histone methylation has been identified on both lysine and arginine residues of histones (van Holde 1989; Gary and Clarke 1998). Lysine methylation has been well documented on histone H3 and H4 in vivo. In H3, lysine residues 4, 9, 27, and 36 are major sites of methylation, whereas in H4, lysine 20 is the only identified site of lysine methylation (van Holde 1989; Strahl et al. 1999). The first histone lysine methyltransferases (HMTs) identified were Su(var) 3–9 family members: SUV39H1 (human), Suv39h1 (mouse), and Clr4 (Schizosaccharomyces pombe; Rea et al. 2000). The methyltransferase activity of SUV39H1 is site specific to H3 lysine 9 (Lys9) and is mediated through its SET domain (Rea et al. 2000). The SET domain is an evolutionarily conserved domain of 130 amino acids named for its appearance in Su(var) 3–9 (suppressor of position effect variegation), E(z) (enhancer of zeste), and Trx (trithorax; Stassen et al. 1995; Jenuwein et al. 1998). Another mammalian SET domain–containing protein, G9a, has also been identified as a lysine methyltransferase for H3 residues 9 and 27 (Tachibana et al. 2001). Currently, the role of G9a in chromatin function is not known.
The well-characterized heterochromatin-associated proteins HP1 and its S. pombe homolog Swi6 (Eissenberg and Elgin 2000) contain chromodomains that have been shown to interact in vitro, with H3 methylated at Lys9 (Bannister et al. 2001; Jacobs et al. 2001; Lachner et al. 2001). In addition, methylation of H3 Lys9 by Clr4 is required for localization of Swi6 to heterochromatic regions in vivo (Nakayama et al. 2001). Furthermore, SUV39H1 and HP1 associate with the tumor suppressor protein retinoblastoma, and together, this complex mediates transcriptional repression (Nielsen et al. 2001). These observations show that H3 Lys9 methylation plays an epigenetic role in establishing silencing in both heterochromatic and euchromatic regions (Jenuwein 2001; Jenuwein and Allis 2001; Rice and Allis 2001).
S. cerevisiae lacks detectable H3 Lys9 methylation on bulk histones isolated from asynchronously grown cells (Strahl et al. 1999), which is consistent with the fact that no apparent SUV39H1 homolog exists in budding yeast. We therefore sought to investigate the role of other SET domain–containing proteins and their corresponding methylation site(s). In S. cerevisiae, seven SET domain–containing genes have been identified (Schultz et al. 2000). These seven genes include the previously identified SET1 (Laible et al. 1997; Nislow et al. 1997) and SET2 (Lutfiyya et al. 1995). Although little is know about the function of these SET domain proteins, disruption of SET1 results in the loss of silencing at telomeres and HML mating-type loci (Laible et al. 1997; Nislow et al. 1997). In addition, Set2 has been reported to be a repressor of GAL4 basal transcription (Lutfiyya et al. 1995). Furthermore, we have determined that Set2 has nucleosomal-specific HMT activity selective for lysine 36 of H3 (Strahl et al. 2001). Until now, the protein products of any of these SET domain–containing genes in S. cerevisiae having site-specific methyltransferase activity had not been determined.
In this study, we focused on the role of H3 lysine 4 (Lys4) methylation and the identity of the responsible HMT in S. cerevisiae. Using an H3 Lys4 methyl-specific antiserum (α-Me[Lys4]H3), we show that deletion of SET1 results in complete abolishment of H3 Lys4 methylation in vivo. Moreover, yeast strains containing histone H3 mutations at Lys4 or a set1Δ both revealed growth defects, indicating that Set1-mediated H3 Lys4 methylation is important for cell growth. Finally, we show by chromatin immunoprecipitation (ChIP) assays that Set1-mediated H3 Lys4 methylation is associated with rDNA and is required for rDNA silencing. Together, these data indicate that Set1-mediated H3 Lys4 methylation is necessary for normal cell growth and transcriptional silencing within rDNA.
Results
Generation and specificity of a histone H3 Lys4 methyl-specific antiserum
Previous work has shown that H3 Lys4 is an evolutionarily conserved site of methylation among a wide range of eukaryotic species (Strahl et al. 1999). Using a ciliate model, H3 Lys4 methylation was found to occur only in a transcriptionally active nucleus, indicating a possible role for H3 Lys4 methylation in facilitating transcription (Strahl et al. 1999). To further investigate the role of H3 Lys4 methylation, we generated an antiserum specific for this methyllysine site. A peptide was synthesized containing histone H3 residues 1–8 with dimethylated Lys4 (Fig. 1A). Following conjugation to carrier protein, this immunogen was used to immunize rabbits (see Materials and Methods).
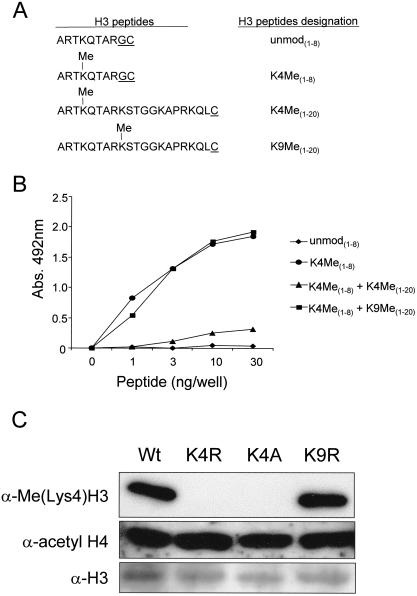
Figure 1.
H3 lysine 4 (Lys4) methyl-specific antiserum recognizes H3 Lys4 methylation on peptides and histones. (A) Amino acid sequences of the different H3 peptides used in the ELISA analyses are shown in B. Underlined amino acids are artificial to the H3 sequence and used for coupling purposes. (B) enzyme-linked immunosorbent assay (ELISA) analyses of the α-H3(Lys4)Me antiserum for its recognition of and specificity toward unmodified (unmod[1–8]) and modified H3 (K4Me[1–8]) peptides. Peptide competitions were performed with 20 μg/mL H3 peptides K4Me(1–20) or K9Me(1–20). (C) Whole cell extracts were isolated from wild-type (Wt) and H3 K4R, K4A, and K9R yeast strains and immunoblotted with α-Me(lys4)H3 or α-acetyl H4 and α-H3 antiserum as a loading controls. Wt, K4R, K4A, and K9R were introduced into the MSY421 background.
Specificity of the obtained antiserum (hereafter designated α-Me[Lys4]H3) was initially analyzed by enzyme-linked immunosorbent assay (ELISA). As shown in Figure 1B, α-Me(Lys4)H3 immunoreacted strongly with the methylated peptide (H3 K4Me[1–8]) and did not detect the corresponding unmodified peptide (H3 unmod[1–8]). To verify that α-Me(Lys4)H3 recognizes the specific H3 Lys4 methyl peptide sequence only, peptide competition assays were performed using H3 Lys4 (K4Me[1–20]) and Lys9 (K9Me (1–20]) methyl peptides. In the presence of 20 μg/mL H3 Lys4 methyl competitor peptide (H3 K4Me[1–20]), little or no detection was observed by the α-Me(Lys4)H3 to the H3 K4Me(1–8) peptide (Fig. 1B). In contrast, α-Me(Lys4)H3 did recognize the H3 K4Me(1–8) peptide in the presence of 20 μg/mL H3 Lys9 methyl competitor peptide (H3 K9Me[1–20]; Fig. 1B), indicating that the α-Me(Lys4)H3 is specific to the H3 Lys4-methylated peptide sequence.
To determine the specificity of the α-Me(Lys4)H3 on yeast histones, histone H3 mutations were introduced into a plasmid construct containing tandem wild-type copies of the histone H3 and H4 genes. H3 Lys4 was converted to either arginine (K4R) or alanine (K4A), and as a control, Lys9 was mutated to arginine (K9R). All mutations were transformed into histone shuffle strains lacking all genomic copies of H3 and H4 (Megee et al. 1990; Hsu et al. 2000). As shown in Figure 1C, no H3 Lys4 methylation was detected with the α-Me(Lys4)H3 on yeast whole cell lysates isolated from both H3 K4R and K4A strains. However, strong reactivity toward H3 Lys4 methylation was detected from whole cell lysates isolated from H3 K9R and isogenic wild-type (MSY421) strains. Together, these data show that α-Me(Lys4)H3 is specific to both appropriately methylated H3 peptides and yeast histone H3.
Conservation and abundance of H3 Lys4 versus Lys9 methylation levels among diverse eukaryotes
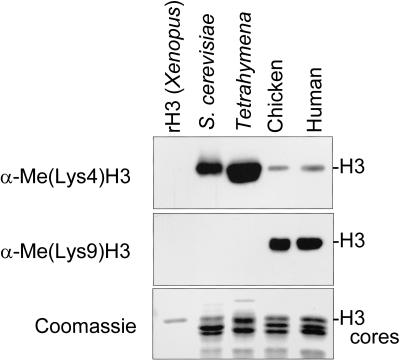
Previous work has shown the existence of H3 Lys4 and Lys9 methylation of bulk histones using metabolic labeling and direct sequencing approaches (van Holde 1989; Strahl et al. 1999). Using the methyl-specific antisera α-Me(Lys4)H3 and α-Me(Lys9)H3 (Jacobs et al. 2001; Nakayama et al. 2001), a direct comparison of the conservation and relative abundance of H3 Lys4 versus Lys9 methylation was made. As shown in Figure 2, immunoblot analyses showed that H3 Lys4 was methylated among all eukaryotes examined. In contrast, H3 Lys9 methylation was detected only in the larger eukaryotes, indicating that H3 Lys9 methylation either is not present or is in extremely low abundance in smaller organisms during asynchronous growth. Intriguingly and in striking contrast, organisms with no apparent H3 Lys9 methylation show high levels of H3 Lys4 methylation (Fig. 2), a result that may reflect a fundamental difference in the ground state of chromatin in simple versus complex organisms (see Discussion).
Figure 2.
Conservation and abundance of H3 lysine 4 (Lys4) and lysine 9 (Lys9) methylation; 1 μg of recombinant histone from Xenopus and 5 μg of total core histones from asynchronously growing Saccharomyces cerevisiae (S. cerevisiae), Tetrahymena thermophila (Tetrahymena), chicken, and human 293T cells (human) were resolved by 15% SDS-PAGE, transferred to PVDF membrane, and probed with α-Me(lys4)H3 or α-Me(Lys9)H3. Note that the Tetrahymena sample represents macronuclear histones. Identical samples were examined in parallel by Coomassie staining to show histone loading.
Set1 mediates H3 Lys4 methylation in vivo
Recently, the SET domains of the Su(var) 3–9 family (SUV39H1, Suv39h1, Suv39h2, and Clr4) and human G9a have been identified as HMTs that are mainly selective in catalyzing H3 Lys9 methylation. Because no apparent Su(var) 3–9 methyltransferase homologs exists in S. cerevisiae, we predicted that another SET domain–containing protein would catalyze H3 Lys4 methylation. Using the SMART (Simple Modular Architecture Research Tool) database (Schultz et al. 2000), at least seven SET domain–containing genes have been identified in S. cerevisiae (see Fig. 3A). Two of these genes are the previously identified SET1 (Laible et al. 1997; Nislow et al. 1997) and SET2 (Lutfiyya et al. 1995). To determine which, if any, of the known SET-containing genes might be responsible for H3 Lys4 methylation, the α-Me(Lys4)H3 antiserum was used to probe yeast whole cell extracts (WCEs) for the loss of H3 Lys4 methylation from strains that harbored individual deletions of each of these genes. Strikingly, H3 Lys4 methylation was completely abolished from cells lacking Set1, indicating that Set1 is responsible for H3 Lys4 methylation in budding yeast (Fig. 3A).
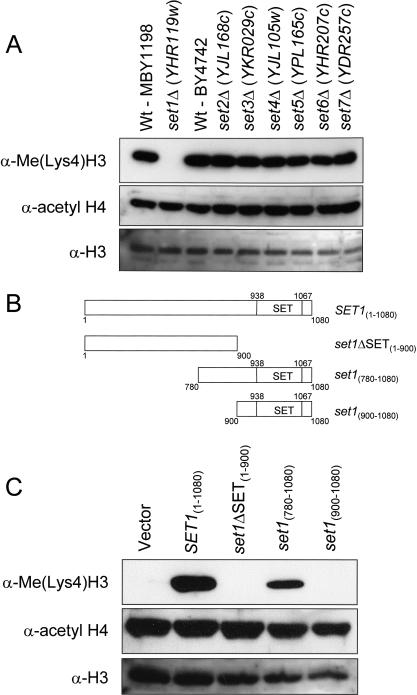
Figure 3.
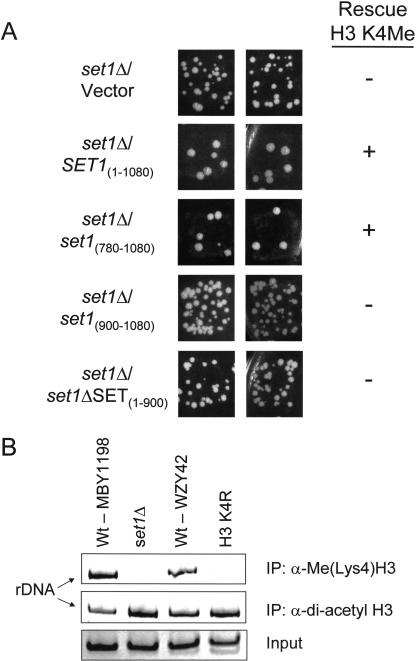
Set1 mediates H3 lysine 4 (Ly4) methylation. (A) Whole cell extracts were isolated from wild-type (Wt; MBY1198), MBY1217 (set1Δ [YHR119w]), Wt (BY4742), and yeast strains carrying individual disruptions in SET domain–containing genes: set2Δ (YJL168c), set3Δ (YKR029c), set4Δ (YJL105w), set5Δ (YPL165c), set6Δ (YHR207c), and set7Δ (YDR257c). MBY1198 is the isogenic Wt strain to set1Δ strain (MBY1217), and BY4742 is the isogenic Wt strain to set2Δ, set3Δ, set4Δ, set5Δ, set6Δ, and set7Δ strains. H3 Lys4 methylation was detected by immunoblotting with α-Me(lys4)H3 antiserum; H4 acetylation, with α-acetyl H4 antiserum; and H3, with α-H3 antiserum. (B) Schematic representation of the Set1 constructs used to rescue H3 Lys4 methylation shown in C. Numbers indicate the amino acids in Set1, and amino acids 938–1067 represent the SET domain of Set1. (C) Set1 constructs shown in B were transformed into a yeast set1Δ strain, MBY1217, and whole extracts of the transformed yeast were generated and probed with α-Me(Lys4)H3 or α-acetyl H4 and α-H3 antiserum as a loading controls.
To confirm that the loss of H3 Lys4 methylation observed from the set1Δ strain was a consequence of deleting the SET1 gene and not because of loss of the epitope in H3 as a result of some unexpected mutation(s), we next asked whether Set1 expression constructs (see Fig. 3B) would restore H3 Lys4 methylation in a set1Δ strain. As shown in Figure 3C, H3 Lys4 methylation was rescued in a set1Δ strain by a Set1 (SET1[1–1080]) expression construct similar to the levels observed in wild-type cells (see Fig. 3A). The same set1Δ strain containing the vector alone did not rescue H3 Lys4 methylation (Fig. 3C). Furthermore, Set1 lacking the SET domain (set1ΔSET[1–900]) did not restore H3 Lys4 methylation, whereas N-terminally truncated Set1, which includes the SET domain (set1[780–1080]), rescued H3 Lys4 methylation (Fig. 3C). Interestingly, a shorter Set1 expression construct mainly encompassing the SET domain (set1[900–1080]) did not restore H3 Lys4 methylation (Fig. 3C), suggesting that regions between amino acids 780 and 900 of the Set1 protein are required for proper function or stability. Thus, these results define a minimal region required for Set1 to mediate H3 Lys4 methylation in vivo.
H3 Lys4 mutations are defective in normal cell growth
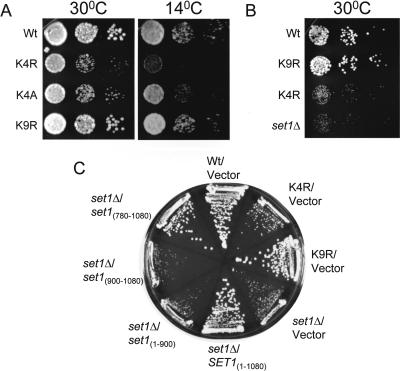
To investigate the role of H3 Lys4 methylation in yeast, mutations were generated in which H3 Lys4 was converted to either arginine (K4R) or alanine (K4A). Spot growth assays show that H3 K4R and K4A yeast strains, identical to the strains used in Figure 1C, have a slow-growth phenotype at permissive temperature (30°C) compared with the isogenic wild-type yeast strain (MSY421; Fig. 4A). This growth defect is specific for H3 Lys4 mutations because no growth defect was detected for a H3 Lys9 mutation (K9R). In addition, when grown at 14°C, the H3 K4R and K4A yeast strains also showed a slow-growth phenotype (Fig. 4A). These results correlate with the loss of H3 Lys4 methylation observed in the H3 Lys4 mutations and not in the H3 K9R or isogenic wild-type strains (see Fig. 1C). Importantly, the H3 K4R and K4A slow-growth phenotype is similar to that observed for a previously published set1Δ strain phenotype (Nislow et al. 1997). In addition, disruption of SET1 in the same yeast background strain as the H3 K4R and K4A mutants revealed a similar slow-growth defect at 30°C (Fig. 4B). Furthermore, the set1Δ slow-growth phenotype was rescued with Set1 constructs that restore H3 Lys4 methylation in vivo (Fig. 4C). Together, these data show that a Set1-mediated H3 Lys4 methylation is important for normal cell growth.
Figure 4.
Histone H3 mutations at lysine 4 (Lys4) and a set1Δ strain (MBY1587) show a slow-growth phenotype. (A) Cell growth between wild-type (Wt), and histone H3 K4R, K4A, and K9R yeast strains was analyzed by spot assays. Tenfold serial dilutions were spotted on YPD and grown for 3 d at 30°C or for 7 d at 14°C. (B) Fivefold serial dilutions of Wt, K9R, K4R, and set1Δ strains were spotted on YPD and grown for 2 d at 30°C. (C) Cell growth of set1Δ strains transformed with Set1 constructs described in Fig. 3B were determined by plating on SC-Ura medium for 3 d at 30°C. Wt, K4R, K4A, K9R, and set1Δ (MBY1587) were all generated in the MSY421 background.
Lys4 methylation is present at rDNA and is required for rDNA silencing
Although seven SET domain–containing genes in S. cerevisiae have been identified, little is known about the function of these SET domain proteins. However, disruption of SET1 results in the loss of silencing at telomeres and HML (Laible et al. 1997; Nislow et al. 1997). Recently, Winston and colleagues (M. Bryk, et al., in prep.), have shown that Set1 is also shown to be required for rDNA silencing by two well-established methods: (1) transposition of Ty1 elements out of the rDNA loci (Bryk et al. 1997) and (2) expression of modified URA3 (mURA3) and LEU2 genes when integrated within rDNA (Smith and Boeke 1997; Smith et al. 1999).
Given these results, we sought to test if Set1 constructs that rescue H3 Lys4 methylation (see Fig. 3B,C) could also complement or restore rDNA silencing within a set1Δ strain. Using the Ty1 transposition assay as described in M. Bryk et al. (in prep.), we show that expression of Set1 (SET1[1–1080]) and N-terminally truncated Set1 (set1[780–1080]) in a set1Δ strain restore rDNA silencing, as indicated by reduced Ty1 transposition compared with the set1Δ strain containing vector alone (Fig. 5A; Table 1). These results indicate that the observed loss of rDNA silencing created by deletion of the SET1 gene is complemented only by Set1 constructs that rescue H3 Lys4 methylation. In contrast, set1Δ strains expressing Set1 lacking the SET domain (set1ΔSET[1–900]) or a shorter Set1 construct (set1[900–1080]) showed similar levels of Ty1 transposition compared with vector control, indicating that these constructs do not complement rDNA silencing (Fig. 5A; Table 1), a result consistent with their inability to rescue H3 Lys4 methylation. In support, the histone H3 K4R mutation causes increased transcription of a Ty1 element in the rDNA (M. Bryk, et al., in prep.). Together, these results indicate that Set1-mediated H3 Lys4 methylation is required for transcriptional silencing within the rDNA locus.
Figure 5.
Set1-mediated H3 lysine 4 (Lys4) methylation at rDNA is required for rDNA silencing. (A) Ty1 transposition patch assays were preformed to determine complementation of rDNA silencing within the set1Δ strain MBY1217 with Set1 constructs SET1(1–1080), set1ΔSET(1–900), set1(780–1080), and set1(900–1080). The ability of Set1 constructs to rescue (+) or not rescue (−) H3 Lys4 methylation in the set1Δ (MBY1217) strain is indicated by the patch assays (see Fig. 3C). (B) Chromatin from wild-type (Wt; MBY1198), set1Δ, and Wt (WZY42), and H3 K4R strains were isolated and immunoprecipitated with α-Me(Lys4)H3 or α-H3 di-acetyl antiserum. The set1Δ strain used was MBY1217, and the H3 K4R was introduced into the WZY42 background. The NTS region of rDNA present in the immunoprecipitated samples were amplified by PCR. Input PCR was shown for loading controls.
Table 1.
Frequency of transposition of Ty1 elements in set1Δ strains with Set1 constructs
| Straina/plasmid
|
Transposition frequencyb of Ty1his3AI-236rc
|
Ratio of frequenciesd
|
|---|---|---|
| Wt/Vectore | 1.7 (±0.7) × 10−8 | 1 |
| set1Δ/Vector | 7.3 (±5.3) × 10−7 | 44 |
| set1Δ/SET1(1–1080)e | 2.4 (±2.0) × 10−8 | 1.4 |
| set1Δ/set1(780–1080)e | 2.0 (±1.3) × 10−8 | 1.2 |
| set1Δ/set1(900–1080) | 3.4 (±2.6) × 10−7 | 20.4 |
| set1Δ set1ΔSET(1–900) | 2.2 (±1.3) × 10−7 | 13.2 |
The relevant genotype of MBY1198 is SET1+ (Wt) and of MBY1217 is set1Δ::TRP1 (set1Δ).
The transposition frequency was measured as described in Materials and Methods.
Ty1his3AI-236r is located in the rDNA and is subject to transcriptional silencing by Set1 (M. Bryk, in prep.).
The frequency of transposition is compared to that of MBY1198/vector.
The one or more of the six cultures showed no detectable transposition events.
To determine if H3 Lys4 methylation is present at rDNA, ChIP was performed using the α-Me(Lys4)H3 antiserum. As shown in Figure 5B, ChIP analyses show that H3 Lys4 methylation is present at the rDNA locus in wild-type yeast but is absent in a set1Δ or H3 K4R control strains. As an internal control for both the set1Δ and H3 K4R strains, we immunoprecipitated identical samples with an α-di-acetyl H3 antiserum (Fig. 5B). The α-di-acetyl H3 antiserum immunoprecipitated acetylated H3 even in the absence of H3 Lys4 methylation, indicating the presence of an intact H3 N terminus. Therefore, the inability to ChIP H3 Lys4 methylation from the set1Δ or H3 K4R strain is a direct consequence of the loss of H3 Lys4 methylation, a result supported by our Western blot analyses (see Figs. 1C, 3A). In addition, immunoprecipitations with the α-di-acetyl histone H3 antiserum revealed no significant difference in H3 acetylation levels between wild-type, set1Δ, or H3 K4R yeast strains, indicating that H3 Lys4 methylation does not influence H3 acetylation at the rDNA locus. The extent to which histone acetylation and methylation influence one another at other sites and/or at other locations outside of rDNA has not yet been determined (see below).
Discussion
In this report, we have developed an antiserum specific to H3 methylated at Lys4 for investigating the role of this modification in S. cerevisiae. Using this antiserum, we show that of the known SET domain–containing proteins, only Set1 is required for histone H3 Lys4 methylation in vivo. Furthermore, Set1 expression constructs that rescue H3 Lys4 methylation also complement an rDNA-silencing defect observed in a set1Δ strain. Together, these results suggest that at least one biological role for H3 Lys4 methylation is to regulate RNA polymerase II transcriptional silencing within rDNA, a specialized chromatin structure known to mediate this biological effect (Bryk et al. 1997; Fritze et al. 1997; Smith and Boeke 1997; Smith et al. 1999; Sun and Hampsey 1999). In addition, mutations in H3 at Lys4 or a set1Δ strain both show similar growth defects, indicating that Set1-mediated H3 Lys4 methylation is also important for cell growth.
Set1 mediates Lys4 methylation
Recent work has shown that the Su(var) 3–9 family members are Lys9-selective HMTs (Rea et al. 2000) and that the HMT activity of this family is mediated by the SET domain (Rea et al. 2000). Although full-length Set1 and a minimal region of Set1 containing amino acids 780–1080 are clearly able to rescue H3 Lys4 methylation in vivo, we have been unable to detect in vitro HMT activity from recombinant Set1 expressing amino acids 780–1080 in bacteria. Although recombinant Set1 was not active in our HMT assays using H3 N-terminal peptides (1–20), yeast or chicken core histones, and chicken mono- or oligonucleosomes as substrates (data not shown), in these same experiments we were able to detect robust HMT activity from recombinant yeast Set2 (Strahl et al. 2001) and human SUV39H1. Currently, it is not clear why Set1 is not active in a recombinant form compared with other recombinantly active histone methyltransferases, SUV39H1 family, G9a, and Set2 (for review, see Zhang and Reinberg 2001). Because the SET domain has also been identified as a protein-protein interaction domain (Rozenblatt-Rosen et al. 1998; Corda et al. 1999; Firestein et al. 2000; Rozovskaia et al. 2000) and is known to be posttranslationally modified (Aagaard et al. 2000; Firestein et al. 2000; Rea et al. 2000), we favor the idea that Set1 requires other associated proteins and/or posttranslational modifications to display enzymatic activity. Recently, the cofactor NAD was shown to be required for the deacetylase activity of Sir2 (Shore 2000). Similarly, it is possible that cofactors yet to be identified may be required for recombinant Set1 methyltransferase activity.
Although the SET domain is the catalytic motif, additional sequences outside of the SET domain, which include flanking cysteine-rich regions termed the pre- and post-SET domain, were found to be necessary for HMT activity of SUV39H1 (Rea et al. 2000). Although Set1 contains only a post-SET domain, it still mediates H3 Lys4 methylation in vivo, suggesting at least one cysteine-rich region is sufficient for Lys4 methylation. Interestingly, a critical region N-terminal to the SET domain of Set1 between amino acids 780–900 is necessary for the function of Set1 in vivo. Therefore, a minimal C-terminal region of Set1 is sufficient for mediating H3 Lys4 methylation, cell growth, and rDNA silencing.
Biological role for Lys4 methylation
Little is known about the functions of SET domain–containing genes in S. cerevisiae. However, a role for Set1 has been previously reported in mediating silencing at telomeres and HML (Laible et al. 1997; Nislow et al. 1997). Studies reported here and in Bryk et al. (in prep.) show that Set1 and H3 Lys4 methylation are coupled in regulating rDNA silencing and maintaining normal cell growth. However, the exact mechanism of Set-mediated H3 Lys4 methylation in regulating cell growth and the specialized chromatin structure of rDNA is not known. In keeping with emerging data, binding of a yet unknown silencing factor(s) to H3 methylated at Lys4 may occur in much the same way as the chromodomain of HP1 or Swi6 recognizes and binds to Lys9-methylated H3 to establish heterochromatin domains (Bannister et al. 2001; Jacobs et al. 2001; Lachner et al. 2001; Nakayama et al. 2001).
Although we show that H3 Lys4 methylation acts as a repressive modification within rDNA chromatin, emerging evidence also suggests that this modification likely has a facilitative role in transcriptional activation. For example, H3 Lys4 methylation has been associated exclusively with transcriptionally active, but not inactive, nuclei of Tetrahymena (Strahl et al. 1999). Furthermore, immunofluorescence studies on human female metaphase chromosomes show that H3 Lys4 methylation is preferentially associated with transcriptionally active regions in autosomal chromosomes but largely excluded from the inactive X chromosome, a chromosome found to be enriched for H3 Lys9 methylation (Boggs et al. 2001). Recently, ChIP studies over large chromosomal domains have shown that H3 Lys4 methylation is associated with chromatin poised for transcription (Litt et al. 2001; Noma et al. 2001). Finally, Set1 has been directly linked to transcriptional regulation of many genes involved in transcription, cell cycle, growth, meiosis, and DNA repair in yeast (Nislow et al. 1997; Corda et al. 1999). Thus, the dualistic nature of a H3 Lys4 methyl mark seems counter-intuitive and might be addressed, at least in part, by the existence of a histone code, wherein additional posttranslational marks on the same or different histone tail(s) could be read as either a repression or activation cassette (Strahl and Allis 2000; Turner 2000). Several reports have suggested that H3 methylation is coupled to acetylation (Hendzel and Davie 1991; Annunziato et al. 1995; Strahl et al. 1999; Litt et al. 2001), but a mechanistic link between H3 Lys4 methylation and H3 lysine acetylation has yet to be shown (Jenuwein and Allis 2001). Alternatively, because Set1-mediated H3 Lys4 methylation may effect global gene expression, we can not rule out an indirect effect whereby Set1 or H3 Lys4 methylation regulates expression of other genes that may act as the direct factors that mediate cell growth or rDNA silencing.
Differential usage of H3 Lys4 versus Lys9 methylation among ukaryotes
Evolutionarily diverse mechanisms of transcriptional regulation have evolved between organisms (Struhl 1999). In support, we document striking differences in the conservation and relative abundance of H3 Lys4 and Lys9 methylation on bulk histones isolated from diverse eukaryotes (see Fig. 2). Interestingly, organisms with less genomic complexities show high levels of H3 Lys4 methylation with little, if any, H3 Lys9 methylation. In contrast, organisms with more genomic complexity show an opposite pattern, with H3 Lys9 methylation predominating. We suggest that these patterns of methylation reflect different chromatin ground states, which correlate well with known differences in the amount of euchromatin and heterochromatin.
Emerging data hint at the intriguing possibility that H3 Lys4 and Lys9 methylation participate in a long-range genomic indexing system that serve to mark transcriptionally poised (Lys4 methylation) from transcriptionally silent (Lys9 methylation) chromatin (Litt et al. 2001; Noma et al. 2001). Perhaps the abundant nature of H3 Lys4 methylation in smaller eukaryotes, mediated by Set1, helps to program most, but not all, of their chromatin in a transcriptionally competent configuration. However, at some loci such as rDNA, it seems clear that H3 Lys4 methylation can also have opposite effects, namely, the facilitation of rDNA silencing of RNA polymerase II genes. Understanding these differences presents a challenge for future investigation and underscores the potential for opposing effects of histone modifications on chromatin structure and function (Jenuwein and Allis 2001).
Materials and methods
Generation of antiserum specific to histone H3 Lys4 methylation
A synthetic H3 peptide was generated containing residues 1–8 (ARTKQTARGC) in which residue 4 was made with dimethyllysine. The underlined amino acid residues, glycine and cysteine, are artificial and were added for coupling purposes (Fig. 1A, K4Me[1–8]). An immunogen was generated by conjugating the H3 K4Me[1–8] peptide to Imject SuperCarrier Immune Modulator (cationized BSA; Pierce) and used to immunize rabbits. Specificity of the resulting rabbit anti-serum was tested by its recognition of H3 unmodified and methylated peptides (see ELISA below; Fig. 1B).
ELISA
Increasing amounts of unmodified and modified peptides (0 to 30 ng) were coated onto ELISA plate wells by incubating in PBS overnight at 4°C. Each well was washed three times in PBS containing 0.5% Tween-20 (PBST) and blocked with 1%BSA in PBST for 1 h. After blocking, all wells were washed three times in PBST, and the α-Me(Lys4)H3 rabbit serum was added at a 1:50,000 dilution in PBST for 2 h, followed by three washes in PBST. A horseradish peroxidase (HRP)-conjugated rabbit secondary antibody (Amersham Pharmacia Biotech) at a 1:5000 dilution in PBST was added to each well and incubated for 2hrs. After three washes in PBST, the bound HRP was detected using o-phenylenediamine dihydrochloride peroxidase substrate (Sigma), and reactions were quantitated by absorbance measurements at 492 nm. For peptide competition, 20 μg/mL of H3 K4Me(1–20) or H3 K9Me(1–20) peptides (see Fig. 1A) were incubated with α-Me(Lys4)H3 serum in the ELISA plate wells and analyzed as described above.
Preparation of WCEs and histones
For isolation of yeast WCEs, yeast cells were grown overnight, diluted the following day, and grown to an O.D.600 between 0.8 and 1.0. Cells were washed with distilled water, pelleted, and frozen overnight at −80°C. The cell pellets were thawed on ice and resuspended in breaking buffer (10 mM Tris at pH 7.4, 300 mM sorbitol, 600 mM NaCl, 5 mM MgCl2, 5 mM EDTA) with addition of fresh protease and phosphatase inhibitors (1 μg/mL aprotinin, leupeptin, and pepstatin A; 1 mM PMSF; 1 μM microcystin-LR; 2 mM p-chloromercuriphenylsulfonic acid). Cells were disrupted by acid-washed glass beads (425 to 600 μM; Sigma) using a mini-beadbeater (Biospec Products) for three 30-sec pulses. The bottoms of the microcentrifuge tubes were punctured, and cell extracts were separated from the beads by brief centrifugation. Separated lysates were clarified by centrifugation at 16,000g for 10 min, and the supernatant (WCE) was used for protein analysis. Approximately 30 to 50 μg of WCEs were resolved by 15% SDS-PAGE, transferred to PVDF membrane, and probed with either the α-Me(Lys4)H3 or the α-acetyl H4 and α-H3 rabbit serum as loading controls.
Tetrahymena thermophila (strain CU 427 or CU 428) was grown in enriched 1% proteose peptone and macronuclear histones isolated from vegetatively growing cells as described in Gorovsky et al. (1975). Chicken histones and nucleosomes were kindly provided by C. Mizzen (University of Virginia Health System, Charlottesville). Yeast histones were isolated from wild-type strain MX4-22A at late log phage as described in Edmondson et al. (1996). Human histones were isolated from asynchronously growing 293T cells as described in Chadee et al. (1999). For analysis of bulk core eukaryotic histones (see Fig. 2A), 5 μg of total core histones and 1 μg of recombinant Xenopus H3 were resolved by 15% SDS-PAGE, transferred to PVDF membrane, and probed with either the α-Me(Lys4)H3 or the α-Me(Lys9)H3 rabbit serum.
Electrophoresis and Western blotting
SDS-PAGE was performed as described by Laemmli (1970). Western blotting analyses were performed using chemiluminescence with the ECL plus kit (Amersham Pharmacia Biotech) as per manufacturer instructions. The primary rabbit serum α-Me(Lys4)H3, α-Me(Lys9)H3, α-acetyl H4, and α-H3 were used at 1:20,000, 1:5000, 1:5000, or 1:200 dilutions, respectively. HRP-conjugated rabbit secondary antibody (Amersham Pharmacia Biotech) was used at a 1:5000 dilution.
Yeast strains
Yeast strains were MBY1198 (MATα, his3Δ200, ade2Δ∷hisG, leu2Δ0, ura2Δ0, met15Δ0, trp1Δ63, Ty1his3AI-236r, Ty1ade2AI-515), MBY1217 (MATα, his3Δ200, ade2Δ∷hisG, leu2Δ0, ura2Δ0, met15Δ0, trp1Δ63, Ty1his3AI-236, Ty1ade2AI-515, set1Δ∷TRP1), WZY42 (MAT a, ura3–52, lys2–801, ade2–101, trp1Δ63, his3Δ200, leu2Δ1, hht1-hhtf1∷LEU2, hht2-hht2∷HIS), YCp50-copyII (ura3+, HHT2-HHF2; Zhang et al. 1998), MSY421 (MAT α, ura3–52, leu2–3,112 trp1, his3, Δ[HHT1-HHF1], Δ[HHT2-HHF2]), pMS329-copyI (ura3+, HHT1-HHF1; Morgan et al. 1991), MBY1587 (MAT α, ura3–52, leu2–3,112 trp1, his3, Δ[HHT1-HHF1], Δ[HHT2-HHF2]), and pMS329-copy I (ura3+, HHT1-HHF1, set1Δ∷KANMX4). Yeast deletion strains YJL168c, YKR029c, YJL105w, YPL165c, YHR207c, YDR257c, and isogenic wild type were obtain from Research Genetics in the BY4742 background.
Construction of H3 mutants and set1Δ strains
To construct yeast strains with histone H3 mutations used to generate Figures 1C, 4, and 5B, standard PCR-based site-directed mutagenesis on the plasmid pJH18 (pRS314/HHT2-HHF2; Hsu et al. 2000) or plasmid pWZ414-F13 (pRS414/HHT2-HHT2; Zhang et al. 1998) was used to convert H3 Lys4 to Arg4 (K4R) or Ala4 (K4A), and Lys9 to Arg9 (K9R). Each mutation was transformed into MX1–4C strain (MSY421) or S288C strain (WZY42) and selected on 5-FOA–containing plates for cells that lost the wild-type copies of HHT1-HHF1 or HHT2-HHF2 (pMS329 or pWZ414-F13). All constructs were sequenced through the coding regions of both H3 and H4. To construct a set1Δ strain for Figure 4, B and C, SET1 was disrupted with a KANMX4 cassette in the MSY421 strain now referred to as MBY1587. The plasmid pJH18 (pRS314/HHT2-HHF2) encoding wild-type H3 and H4 gene products were transformed into MBY1587 and selected on 5-FOA–containing plates for cell that lost wild-type copies of HHT1-HHT1 (pMS329).
Generation of SET1 yeast constructs
The entire SET1 ORF was PCR amplified from genomic DNA from yeast strain JC242 (MATα, hisΔ200, ura3–167, Ty1his3AI-242) using Pfu DNA polymerase (Stratagene) and oligos SET1gr1 (5′-AAAACGCGACTCGAGGTTGGACTTGTTGCATGGAC ATGACTGGT-3′) and SET1gr4 (5′-AAAATGTCCCCGCGGC GTAAGGAAACCCTAGAAATGGGCCGGAATG-3′). The resulting SET1 PCR product was subcloned into pRS416 (Sikorski and Hieter 1989) and completely sequenced. The pRS416-SET1 construct was used to PCR amplify the coding sequence of full-length Set1 containing amino acid residues 1–1080, a set1 deletion lacking the SET domain amino acid residues 1–900, and two N-terminal truncations of Set1 containing amino acid residues 780–1080 or 900–1080. Each construct generated contained an N-terminal Flag epitope (DYKDDDDK); see Figure 3B for schematic representation of constructs. Each PCR fragment was subcloned into a yeast expression plasmid with an ADH1 promoter and a URA3 selectable marker (Mumberg et al. 1995).
rDNA silencing assays
For the transposition patch assay (see Fig. 5A), the set1Δ∷TRP1 strain (MBY1217) was transformed with Vector, SET1(1–1080), set1ΔSET(1–900), set1(780–1080), or set1(900–1080) plasmids (described above). The resulting strains were grown as patches on SC-Ura plates for 3 d at room temperature. These patches were replica-plated to YPD plates and grown at room temperature for an additional day. The patches were then replica plated from the YPD medium to SC-Ura-His plates to measure the level of transposition of the genetically marked Ty1his3AI-236r element in the rDNA. For the transposition frequency assay (see Table 1), the frequency of transposition was measured for two transformants of each strain. For each transformant, 3 × 3 mL SC-Ura medium were inoculated with ∼1000 cells. The cultures were grown for 4 d at 20°C. After 3 d, 1 mL YPD medium was added to each culture, and incubation was continued at 20°C. The number of cells in each culture was determined by plating a 1:1000 dilution of the culture on SC-Ura plates. The number of cells that sustained a transposition event involving Ty1his3AI-236r was determined by plating an aliquot of the culture on SC-Ura-His plates. For each culture, the transposition frequency was calculated by dividing the number of His+Ura+ colonies by the number of Ura+ colonies.
ChIP assays
Yeast immunoprecipitation of formaldehyde crossed-linked chromatin was performed according to Kuo and Allis (1999). Briefly, 50 mL of each yeast strain was grown in YPD medium to a concentration of 1.0 O.D.600 and fixed with 1% formaldehyde for 15 min at room temperature. Cells were harvested, washed twice with distilled water, and lysed in 400 μL FA-lysis buffer (50 mM HEPES-KOH at pH 7.5; 140 mM NaCl; 1 mM EDTA; 1% Triton X-100; 0.1% sodium deoxcholate; 1 mM PMSF; 1 μg/mL leupeptin, aprotinin, and pepstatin A) at 4°C by the glass bead method described above. The extract was sonicated to shear DNA and clarified by centrifuging at 16,000g for 20 min at 4°C. Clarified supernatant was immunoprecipitated with protein A Sepharose and either α-Me(Lys4)H3 or the α-di-acetyl H3 overnight at 4°C. Precipitates were washed sequentially with 1.4 ml FA-lysis buffer, 1.4 ml FA-lysis buffer with 500 mM NaCl, and 1.4 ml LiCl solution (10 mM Tris at pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxcholate, 1 mM EDTA). Antigens were eluted with 1% SDS and 0.1 M NaHCO3,, and the DNA-protein complex was reverse cross-linked for 5 h at 65°C. DNA was purified by phenol-chloroform extraction and recovered by ethanol precipitation. Precipitated DNA was analyzed by PCR using the NTS primers (5′-TCG CATGAAGTACCTCCCAACTAC-3′ and 5′-TCCGCTTCC GCTTCCGCAGTAAAA-3′) as described in Gotta et al. (1997).
Acknowledgments
This research was supported by grants from the National Institutes of Health (NIH) to C.D.A. (GM53512), F.W. (GM32967), and B.D.S. (GM20039). S.D.B is a Leukemia and Lymphoma Society Fellow. M.B. is supported by the American Cancer Society and Leukemia and Lymphoma Society; W.L.C., by the NIH Medical Scientist Training Fellowship. We thank M. Smith for providing the MSY421 yeast strain; R. Rice, M.A. Jelinek, T. Jelinek, and S. Paschke at Upstate Biotechnology for their assistance with the development of the H3 Lys4 methyl-specific antiserum; K. Luger for providing recombinant histone; and all current C.D.A. laboratory members for their helpful discussion, technical advice, and critical reading of this manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL allis@virginia.edu; FAX (804) 924-5069.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.940201.
References
- Aagaard L, Schmid M, Warburton P, Jenuwein T. Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J Cell Sci. 2000;113:817–829. doi: 10.1242/jcs.113.5.817. [DOI] [PubMed] [Google Scholar]
- Annunziato AT, Eason MB, Perry CA. Relationship between methylation and acetylation of arginine-rich histones in cycling and arrested HeLa cells. Biochemistry. 1995;34:2916–2924. doi: 10.1021/bi00009a023. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Boggs, B.A., Cheung, P., Heard, E., Spector, D.L., Chinault, C., and Allis, C.D. 2001. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. (in press). [DOI] [PubMed]
- Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes & Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- Chadee DN, Hendzel MJ, Tylipski CP, Allis CD, Bazett-Jones DP, Wright JA, Davie JR. Increased Ser-10 phosphorylation of histone H3 in mitogen-stimulated and oncogene-transformed mouse fibroblasts. J Biol Chem. 1999;274:24914–24920. doi: 10.1074/jbc.274.35.24914. [DOI] [PubMed] [Google Scholar]
- Corda Y, Schramke V, Longhese MP, Smokvina T, Paciotti V, Brevet V, Gilson E, Geli V. Interaction between Set1p and checkpoint protein Mec3p in DNA repair and telomere functions. Nat Genet. 1999;21:204–208. doi: 10.1038/5991. [DOI] [PubMed] [Google Scholar]
- Edmondson DG, Smith MM, Roth SY. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes & Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Elgin SC. The HP1 protein family: Getting a grip on chromatin. Curr Opin Genet Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- Firestein R, Cui X, Huie P, Cleary ML. Set domain-dependent regulation of transcriptional silencing and growth control by SUV39H1, a mammalian ortholog of Drosophila Su(var)3–9. Mol Cell Biol. 2000;20:4900–4909. doi: 10.1128/mcb.20.13.4900-4909.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritze CE, Verschueren K, Strich R, Easton ER. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- Gorovsky MA, Yao MC, Keevert JB, Pleger GL. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9:311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Gotta M, Strahl-Bolsinger S, Renauld H, Laroche T, Kennedy BK, Grunstein M, Gasser SM. Localization of Sir2p: The nucleolus as a compartment for silent information regulators. EMBO J. 1997;16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel MJ, Davie JR. Dynamically acetylated histones of chicken erythrocytes are selectively methylated. Biochem J. 1991;273:753–758. doi: 10.1042/bj2730753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 2001;11:266–273. doi: 10.1016/s0962-8924(01)02001-3. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Laible G, Dorn R, Reuter G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein/DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T. Mammalian homologs of the Polycomb-group gene enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Lutfiyya L, Hesman T, Johnston M. What activates expression of a transcriptional activator? Analysis of the weak GAL4 promoter. Yeast. 1995;11:S220. (Abstract). [Google Scholar]
- Megee PC, Morgan BA, Mittman BA, Smith MM. Genetic analysis of histone H4: Essential role of lysines subject to reversible acetylation. Science. 1990;247:841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- Morgan BA, Mittman BA, Smith MM. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol Cell Biol. 1991;11:4111–4120. doi: 10.1128/mcb.11.8.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Nislow C, Ray E, Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell. 1997;8:2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma KI, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Rice JC, Allis CD. Histone methylation versus histone acetylation: New insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, Nakamura T, Croce CM, Mazo A, Canaani E. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci. 1998;95:4152–4157. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozovskaia T, Rozenblatt-Rosen O, Sedkov Y, Burakov D, Yano T, Nakamura T, Petruck S, Ben-Simchon L, Croce CM, Mazo A, et al. Self-association of the SET domains of human ALL-1 and of Drosophila TRITHORAX and ASH1 proteins. Oncogene. 2000;19:351–357. doi: 10.1038/sj.onc.1203307. [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D. The Sir2 protein family: A novel deacetylase for gene silencing and more. Proc Natl Acad Sci. 2000;97:14030–14032. doi: 10.1073/pnas.011506198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes & Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- Smith JS, Caputo E, Boeke JD. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol Cell Biol. 1999;19:3184–3197. doi: 10.1128/mcb.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer VA, Davie JR. Role of covalent modifications of histones in regulating gene expression. Gene. 1999;240:1–12. doi: 10.1016/s0378-1119(99)00405-9. [DOI] [PubMed] [Google Scholar]
- Stassen MJ, Bailey D, Nelson S, Chinwalla V, Harte PJ. The Drosophila trithorax proteins contain a novel variant of the nuclear receptor type DNA binding domain and an ancient conserved motif found in other chromosomal proteins. Mech Dev. 1995;52:209–223. doi: 10.1016/0925-4773(95)00402-m. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl, B.D., Grant, P., Briggs, S.D., Sun, Z.W., Bone, J.R., Caldwell, J.A., Mollah, S., Cook, R.G., Shabanowitz, J., Hunt, D.F., et al. 2001. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell Biol. (in press). [DOI] [PMC free article] [PubMed]
- Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Hampsey M. A general requirement for the Sin3-Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics. 1999;152:921–932. doi: 10.1093/genetics/152.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- van Holde KE. The proteins of chromatin I histones. In: Rich A, editor. Chromatin. New York, NY: Springer-Verlag; 1989. pp. 169–180. [Google Scholar]
- Wu C, Tsukiyama T, Gdula D, Georgel P, Martinez-Balbas M, Mizuguchi G, Ossipow V, Sandaltzopoulos R, Wang HM. ATP-dependent remodeling of chromatin. Cold Spring Harb Symp Quant Biol. 1998;63:525–534. doi: 10.1101/sqb.1998.63.525. [DOI] [PubMed] [Google Scholar]
- Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes & Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]