Abstract
The C-terminal domain (CTD) of the RNA polymerase II (Pol II) largest subunit is hyperphosphorylated during transcription. Using an in vivo cross-linking/chromatin immunoprecipitation assay, we found previously that different phosphorylated forms of RNA Pol II predominate at different stages of transcription. At promoters, the Pol II CTD is phosphorylated at Ser 5 by the basal transcription factor TFIIH. However, in coding regions, the CTD is predominantly phosphorylated at Ser 2. Here we show that the elongation-associated phosphorylation of Ser 2 is dependent upon the Ctk1 kinase, a putative yeast homolog of Cdk9/P-TEFb. Furthermore, mutations in the Fcp1 CTD phosphatase lead to increased levels of Ser 2 phosphorylation. Both Ctk1 and Fcp1 cross-link to promoter and coding regions, suggesting that they associate with the elongating polymerase. Both Ctk1 and Fcp1 have been implicated in regulation of transcription elongation. Our results suggest that this regulation may occur by modulating levels of Ser 2 phosphorylation, which in turn, may regulate the association of elongation factors with the polymerase.
Keywords: Transcription, phosphorylation, mRNA processing, mRNA capping enzyme, RNA polymerase II
Eukaryotic RNA polymerase II (Pol II) is a multisubunit complex. The largest subunit (Rpb1) contains a unique C-terminal domain (CTD) which consists of multiple heptapeptide repeats with the consensus of YSPTSPS (Corden 1990). Pol II with an unphosphorylated CTD (Pol IIa) participates in formation of the preinitiation complex, whereas Pol II, in the process of elongation, has a phosphorylated CTD (Pol IIo) (Cadena and Dahmus 1987; Payne et al. 1989; Weeks et al. 1993; O'Brien et al. 1994). Therefore, a transcription cycle involving cyclical phosphorylation and dephosphorylation of the CTD has been proposed. Such a model necessitates the coordinated interaction of CTD kinases and phosphatases (Dahmus 1996; Cho et al. 1999; Komarnitsky et al. 2000; Schroeder et al. 2000).
CTD phosphorylation occurs predominantly at Ser 2 and Ser 5, and these modifications represent an important mechanism for regulating Pol II activity (West and Corden 1995; Bensaude et al. 1999). Ser 2 and Ser 5 are independently essential for viability and behave in genetically and biochemically distinct ways (West and Corden 1995; Yuryev and Corden 1996; Ho and Shuman 1999; Rodriguez et al. 2000). Furthermore, phosphorylation of different serines predominates during different phases of transcription (Komarnitsky et al. 2000). Ser 5 is most strongly phosphorylated at initiation/early elongation phase, whereas Ser 2 is phosphorylated predominantly during the elongation phase. We have proposed that the different phosphorylation patterns serve to mark polymerases at different points in the transcription cycle. The different phosphorylations are predicted to recruit the appropriate RNA processing, elongation, and termination factors. To test this model, it is necessary to identify all of the relevant CTD kinases and phosphatases.
Several kinases are known to phosphorylate the CTD. Ser 5 of CTD is phosphorylated by the Kin28 subunit of general transcription factor TFIIH. The cotranscriptional recruitment of capping enzyme for placement of the 7-methylguanosine cap on pre-mRNA is dependent on Ser 5 phosphorylation (Cho et al. 1997; McCracken et al. 1997; Komarnitsky et al. 2000; Rodriguez et al. 2000; Schroeder et al. 2000). Srb10/Srb11 (Cdk8/Cyclin C) kinase complex, CTD kinase 1 (CTDK-I), and the Bur1/Bur2 complex are also implicated in transcription. The Srb10/Srb11 kinase complex is associated with RNA Pol II holoenzyme (Liao et al. 1995) and functions to negatively regulate initiation (Kuchin and Carlson 1998). Repression may be mediated by phosphorylation of the CTD before preinitiation complex formation (Hengartner et al. 1998) and/or by phosphorylating upstream activating complexes or the Cyclin H subunit of TFIIH (Hirst et al. 1999; Akoulitchev et al. 2000; Chi et al. 2001). CTDK-I is necessary for proper CTD phosphorylation in vivo (Lee and Greenleaf 1991; Sterner et al. 1995; Rodriguez et al. 2000) and proper transcription regulation (Kuchin and Carlson 1998; Patturajan et al. 1999). CTDK-I stimulates efficient elongation by Pol II in vitro, suggesting a role as an elongation factor (Lee and Greenleaf 1997). Mutations in BUR1 and BUR2 can stimulate transcription from a promoter lacking any activator binding sites and also show interactions with transcription elongation factors (Yao et al. 2000; Murray et al. 2001). The physiological phosphorylation targets of the Srb10, Ctk1, and Bur1 kinase complexes on the CTD are not well defined. Srb10/Srb11 kinase complex can phosphorylate both Ser 2 and Ser 5 in vitro (Hengartner et al. 1998; Sun et al. 1998). CTDK-I positively regulates Ser 2 phosphorylation in diauxic growth phase (Patturajan et al. 1999). RNA Pol II coprecipitated with Bur1 becomes phosphorylated at Ser 5 (Murray et al. 2001). Interestingly, Kin28, Srb10, Ctk1, and Bur1 are all members of the cyclin-dependent kinase (CDK) superfamily.
Although several putative kinases are known, only one CTD phosphatase has been identified so far. The Fcp1 phosphatase is conserved in mammalian and yeast cells (Chambers and Dahmus 1994; Chambers and Kane 1996; Archambault et al. 1997, 1998; Cho et al. 1999; Kobor et al. 1999). Fcp1 has been proposed to dephosphorylate Pol IIo at the end of one transcription cycle to regenerate Pol IIa, making polymerase available for another cycle (Cho et al. 1999; Kobor et al. 1999). The catalytic domain of Fcp1 (FCPH) contains motifs found in a new family of small-molecule phosphotransferases and phosphohydrolases (Kobor et al. 1999). Fcp1 also contains a BRCT domain that is important for proper function in vivo (Kobor et al. 2000). Conditional mutants of Fcp1 indicate that the phosphatase is generally required for transcription in Saccharomyces cerevisiae (Kobor et al. 1999, 2000).
We showed previously that different phosphorylated forms of the CTD predominate at different stages of transcription (Komarnitsky et al. 2000). To further explore these modifications, we analyzed the effect of mutations in Srb10, Ctk1, and Fcp1. In vivo cross-linking/immunoprecipitation experiments show that both Ctk1 and Fcp1 cross-link to promoter and coding regions, suggesting that they associate with elongating polymerase. In cells lacking Ctk1, the elongating RNA Pol II does not become highly phosphorylated at CTD Ser 2. This result suggests that Ctk1 is the primary Ser 2 kinase during transcription elongation. In contrast, mutations in Fcp1 lead to an increase in elongation-associated Ser 2 phosphorylation. An Srb10 mutant had little effect on elongating polymerase. These findings are consistent with a model in which CTDK-I and Fcp1 regulate transcription elongation by modulating phosphorylation of the CTD at Ser 2.
Results
Mutations in Fcp1 CTD phosphatase increase levels of CTD phosphorylation at Ser 2
Fcp1 is a CTD phosphatase thought to be important for recycling of Pol II (Cho et al. 1999; Kobor et al. 1999). To determine the effect of Fcp1 depletion on different phosphorylation sites within the CTD, two conditional fcp1 mutants were examined. The fcp1-1 (R250A, P251A) strain carries mutations located near an essential phosphatase motif and is not able to grow at temperatures above 33°C. fcp1-2 (L177A, L181A, H187A) is less severe, showing growth defects at 37°C (Kobor et al. 1999).
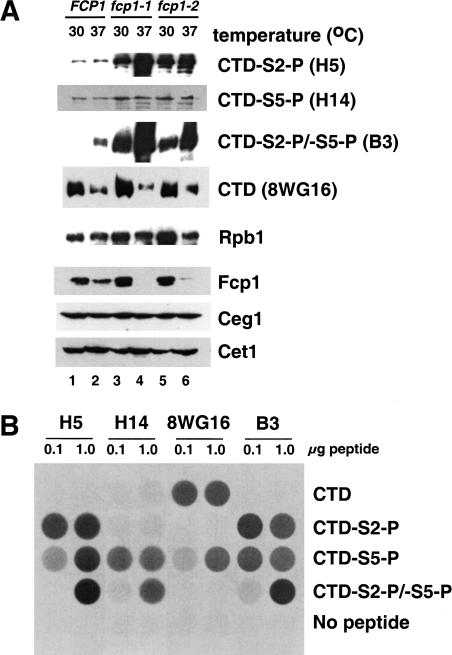
Whole-cell extracts were prepared from yeast strains grown at 30°C and 37°C. Immunoblotting was performed with monoclonal antibodies directed against the CTD (Fig. 1A). The specificities of the antibodies were confirmed by probing immobilized CTD peptides (four heptapeptide repeats) phosphorylated at different serines (Fig. 1B). These antibodies have been characterized previously using CTD peptides with alanine substitutions (Patturajan et al. 1998) and our results largely confirm that study. H5 and H14 (Bregman et al. 1995), recognize CTD repeats phosphorylated at Ser 2 and Ser 5, respectively. At very high antigen concentrations (∼10- to 100-fold higher than would be expected in normal immunoblotting or immunoprecipitations), H5 shows some cross-reactivity with the CTD phosphorylated at Ser 5 (CTD-S5-P). Monoclonal antibody 8WG16 (Thompson et al. 1990) preferentially recognizes the unphosphorylated CTD but can also recognize CTD-S5-P with lower affinity. Importantly, the 8WG16 antigen is completely blocked by phosphorylation at Ser 2. Monoclonal antibody B3 (Mortillaro et al. 1996) reacts well with both CTD-S2-P and CTD-S5-P. All three antibodies that recognize phosphorylated forms of the CTD can recognize doubly phosphorylated CTD-S2-P/S5-P with lower affinity.
Figure 1.
(A) Phosphorylation of RNA Pol II at Ser 2 and Ser 5 are differentially affected by Fcp1 CTD phosphatase. Yeast strains grown at 30°C (OD600 = 0.8) were further incubated at 37°C (even lanes) or 30°C (odd lanes) for 55 min. Whole-cell extracts were prepared from each strain. Extract protein (80 μg) was assayed by immunoblotting with 8WG16 (CTD, recognizing the nonphosphorylated CTD of Rpb1), H14 and H5 (CTD-S5-P and CTD-S2-P, phosphorylated CTD on Ser 5 and Ser 2 position, respectively), B3 (phosphorylated CTD at either Ser 5 or Ser 2), G2 (recognizing Rpb1 outside of CTD) and polyclonal antibodies against Fcp1, Ceg1, and Cet1. Yeast strains used are YMK16α, shuffled with plasmids expressing wild-type FCP1 (pFK1; lanes 1,2), fcp1-1 (pFK4; lanes 3,4) and fcp1-2 (pFK7; lanes 5,6). (B) Specificity of CTD antibodies. Biotinylated peptides (100 ng or 1 μg) containing four CTD repeats with the indicated phosphorylations were coupled to the wells of streptavidin-coated 96-well plates. The indicated antibodies were used to probe the peptides and binding was detected by indirect chemiluminescence.
CTD phosphorylation on Ser 2 was increased in the two fcp1 mutants even at the semipermissive temperature of 30°C (Fig. 1A; cf. lanes 1, 3, and 5). At the nonpermissive temperature of 37°C, the mutant Fcp1 proteins were degraded. This correlated with a further increase in Ser 2 phosphorylation. In the same mutant extracts, phosphorylation at Ser 5 was only mildly increased. Immunoblotting with monoclonal antibody B3 (Mortillaro et al. 1996) showed an increase similar to that of H5. The 8WG16 signal decreases in response to heat in both wild-type and mutant cells as reported previously (Dubois et al. 1999; Kobor et al. 2000). The total amount of polymerase (Rpb1) was monitored using the monoclonal antibody G2, which recognizes an epitope outside of the CTD (R. Burgess, pers. comm.). A moderate increase in total Rpb1 levels was observed in the Fcp1 mutants at 30°C. This is due to excess Rpb1 that is not associated with fully assembled Pol II, and which is degraded at 37°C (M. Kobor and J. Greenblatt, unpubl.). Although the Pol IIa and Pol IIo forms of polymerase were not well separated on these gels, the fcp1 mutant extracts exhibited an additional mobility shift of the CTD band consistent with increased levels of phosphorylation. Taken together, these results suggest that loss of Fcp1 phosphatase activity in vivo lead to a strong and specific increase of phosphorylation at CTD Ser 2.
We found previously that mutations in either the CTD or the CTD kinase Kin28 (which is specific for Ser 5) lead to reduced levels of the capping enzyme guanylyltransferase and the polyadenylation factor Pta1 (Rodriguez et al. 2000). This effect suggests that functional interactions with specific phosphorylated forms of the CTD help regulate levels of certain mRNA processing factors. Comparison of wild-type and Fcp1 mutant extracts failed to reveal any significant changes in protein levels of capping enzyme subunits (Ceg1 and Cet1, Fig. 1) or 3′-processing components (Rna15, Hrp1, Pta1; data not shown).
Fcp1 affects CTD Ser 2 phosphorylation during transcription elongation
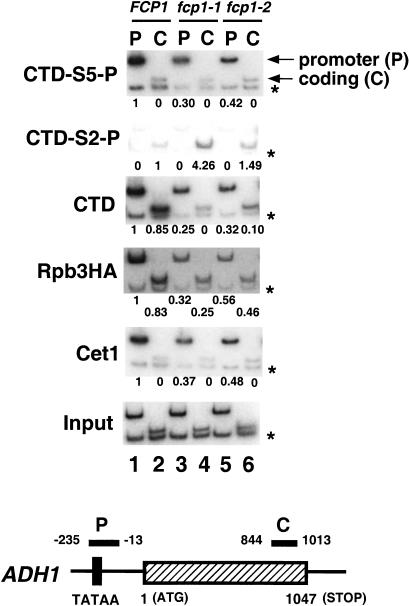
Chromatin cross-linking/immunoprecipitation experiments show that the pattern of CTD phosphorylation changes during transcription, with Ser 5 phosphorylation concentrated at the promoter and Ser 2 phosphorylation predominating in coding regions (Komarnitsky et al. 2000). To determine whether Fcp1 was important for establishing the distribution of Ser 2 and Ser 5 phosphorylation, the same analysis was performed in Fcp1 mutant strains. Yeast cells were grown at the semipermissive temperature of 30°C and sheared chromatin was prepared. The chromatin solution was precipitated with antibodies recognizing specific phosphorylated epitopes of the CTD. Coprecipitated DNA sequences were assayed by PCR performed with primer pairs complementary to different regions of the target gene.
As observed previously, cross-linking of Pol II phosphorylated at Ser 5 is strongest at the promoter (P) of the ADH1 gene and near background in the coding region (C) (Fig. 2, CTD-S5-P). In contrast, CTD phosphorylated at Ser 2 was cross-linked to the coding region of ADH1 gene (Fig. 2, CTD-S2-P). Although the relative distribution of CTD phosphorylation sites was unchanged by Fcp1 mutations, several differences were apparent. First, the total amount of polymerase cross-linked (as monitored by an epitope-tagged Rpb3 subunit and normalization to a nontranscribed internal control) was significantly reduced at both promoters and coding regions. Levels of cross-linked Pol II in fcp1-1 and fcp1-2 mutants were 30%–40% and 30%–50% of wild type, respectively. Most interestingly, the Fcp1 mutants had higher levels of Ser 2 phosphorylation in the coding region. This effect was seen in two ways. First, there was an increase in levels of coding-region DNA precipitated with monoclonal H5 (Fig. 2, CTD-S2-P, cf. lanes 2, 4, and 6) even while total levels of polymerase (RPB3-HA) were decreased. The additional Ser 2-P signal appears modest, but this is because increasing the number of CTD-S2-P epitopes (e.g., from 10 repeats to 27) is unlikely to lead to a linear increase in immunoprecipitation efficiency. The fcp1 mutants also showed a decrease in cross-linking of unphosphorylated, 8WG16-reactive CTD repeats to coding regions (Fig. 2, CTD, lanes 2, 4, and 6). The decrease cannot be accounted for by reduced polymerase levels. There is little Ser 5 phosphorylation in the coding region and the epitope recognized by 8WG16 is blocked by phosphorylation of CTD Ser 2 (Fig. 1B). Therefore, we interpret the drop in 8WG16 reactivity as diagnostic of increased Ser 2 phosphorylation levels.
Figure 2.
Ser 2 and Ser 5 phosphorylations on the ADH1 gene are differentially affected by the Fcp1 CTD phosphatase during transcription. Chromatin immunoprecipitation was performed with YSB763 shuffled with the same Fcp1 plasmids used in Figure 1 (pFK1, pFK4, and pFK7). To monitor the presence of each protein along the ADH1 gene, chromatin was immunoprecipitated with various antibodies and PCR amplified with primer pairs recognizing promoter (P) and coding (C) regions (see schematic at bottom). Each PCR reaction contained a second primer pair that amplifies a region of chromosome V devoid of ORFs, thus providing an internal control for background (*). Each panel shows a different immunoprecipitation with respective antibodies as follows: α-CTD-S5-P (H14), α-CTD-S2-P (H5), α-CTD (8WG16), α-Rpb3HA (12CA5, recognizing an HA epitope on RNA polymerase subunit Rpb3), and α-Cet1 (a polyclonal antibody recognizing the triphosphatase subunit of capping enzyme). Input shows the signal from the chromatin before immunoprecipitation. Signals were quantitated by PhosphorImager and normalized as described previously (Komarnitsky et al. 2000). The signals for the promoter with wild-type chromatin solution were assigned as 1, except for the CTD-S2-P immunoprecipitation, in which the signal from the coding region was taken as 1. A zero indicates that the signal was <0.005. The primer pairs used are (P) ADH1−235 and ADH1−13, (C) ADH1844 and ADH11013, (−) Intergenic V −1 and Intergenic V −2.
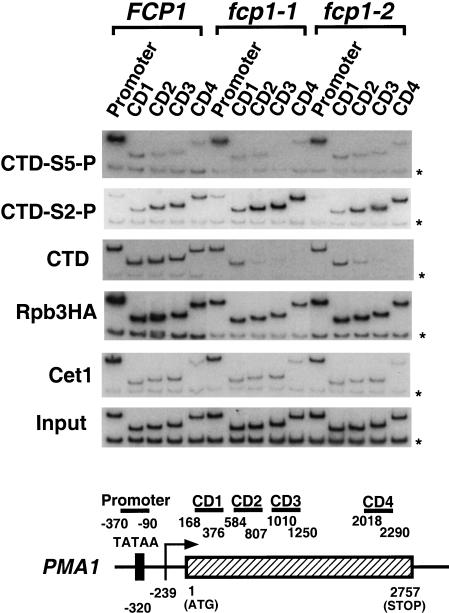
We also analyzed the PMA1 gene for effects of Fcp1 mutations using an extended set of primer pairs that provide greater resolution throughout the gene (Fig. 3). Again, Ser 5 phosphorylation of the CTD is seen at the promoter (Fig. 3, CTD-S5-P), whereas phosphorylation at Ser 2 increases as Pol II passes through the coding region (Fig. 3, CTD-S2-P). As seen with the ADH1 gene, levels of Ser 2 phosphorylation in coding regions increased in the fcp1 mutants. Cross-linking of unphosphorylated CTD immunoprecipitated by 8WG16 is strongly decreased in distal coding regions (Fig. 3, CTD). The total level of Pol II cross-linked to promoter or coding region is maintained throughout transcription (Fig. 3, Rpb3HA).
Figure 3.
Ser 2 and Ser 5 phosphorylations on the PMA1 gene are differentially affected by the Fcp1 CTD phosphatase during transcription. The same chromatin immunoprecipitates shown in A were used for PCR amplification with primer sets that amplify DNA throughout the PMA1 gene (see schematic diagram at bottom). Promoter; PMA1-370 and PMA1-90, Coding region 1 (CD1): PMA1168 and PMA1376; Coding region 2 (CD2): PMA1584 and PMA1807; Coding region 3 (CD3): PMA11010 and PMA11250; Coding region 4 (CD4): PMA12018 and PMA12290.
Our results indicate that Ser 2 becomes phosphorylated during the elongation stage of transcription. However, the Fcp1 CTD phosphatase modulates the number of CTD repeats that are modified. When Fcp1 activity is abrogated, the number of repeats phosphorylated at Ser 2 increases to the point that unphosphorylated repeats are no longer detectable by use of the 8WG16 antibody.
Capping enzyme is properly localized in Fcp1 mutants
In both wild-type and Fcp1 mutant strains, capping enzyme cross-linked to promoter regions but not coding regions (Figs. 2 and 3, Cet1). As observed previously, the capping enzyme pattern mirrored that of Ser 5-phosphorylated CTD (Figs. 2 and 3, CTD-S5-P). This parallel included a drop in promoter cross-linking in the Fcp1 mutants, an effect that is due to a corresponding drop in total transcribing polymerase levels (Rpb3HA). Although interaction of capping enzyme with Pol II is dependent on CTD phosphorylation, all evidence indicates that Ser 5 phosphorylation alone is the physiological signal for recruiting capping enzyme (Ho and Shuman 1999; Komarnitsky et al. 2000; Rodriguez et al. 2000; Schroeder et al. 2000). Therefore, the lack of Fcp1 effect on Ser 5 and capping enzyme are consistent with a specific role for Fcp1 in dephosphorylating Ser 2.
Ctk1, the catalytic subunit of CTDK-I, is the major Ser 2 kinase of elongating polymerase
Several kinases are known to phosphorylate the CTD in vitro and in vivo. We analyzed three kinases with known in vivo connections to Pol II, the TFIIH-associated Kin28, the Pol II holoenzyme component Srb10, and CTDK-I catalytic subunit Ctk1. These three yeast proteins correspond to Cdk7, Cdk8, and possibly Cdk9 in higher eukaryotes. Mutant strains either deleted for the kinase gene (Srb10 and Ctk1, which are not essential) or carrying a partially defective allele (Kin28) were constructed in FCP1 wild-type and fcp1 mutant backgrounds. These strains were also transformed with a plasmid that produces epitope-tagged Rpb1. Chromatin immunoprecipitation was then used to monitor total polymerase (via the HA-epitope on Rpb1) as well as specific phosphorylated forms of the CTD.
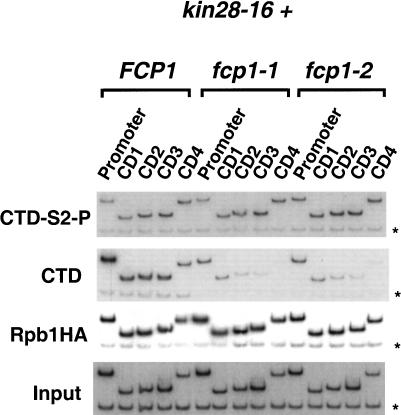
Kin28 is necessary for Ser 5 phosphorylation and recruitment of capping enzyme in vivo (Komarnitsky et al. 2000; Rodriguez et al. 2000; Schroeder et al. 2000). To examine whether Kin28 also has a role in Ser 2 phosphorylation, we analyzed kin28-16, a temperature-sensitive mutant that reduces CTD phosphorylation in vivo (Cismowski et al. 1995). The combination of kin28-16 with fcp1-1 or fcp1-2 resulted in poor growth at 30°C, therefore, cells were grown at room temperature and shifted to 30°C for 2 h before preparing chromatin. As reported previously, cross-linking of CTD-S5-P and the capping enzyme to the promoter region are reduced (see Komarnitsky et al. 2000; Schroeder et al. 2000; data not shown). The kin28-16 mutation does not affect Ser 2 phosphorylation of elongating polymerase (Fig. 4, CTD-S2-P, CD1–CD4). Cross-linking of CTD-S2-P is still increased in fcp1-1 and fcp1-2 strains, whereas nonphosphorylated CTD is decreased (Fig. 4, CTD, CD1–CD4). This indicates that phosphorylation at Ser 2 does not directly require Kin28 kinase activity. Interestingly, an increased level of Ser 2 phosphorylation was seen at the promoter in both wild-type and mutant FCP1 strains. This suggests that Ser 5 phosphorylation may inhibit either Ser 2 phosphorylation or the ability of antibody H5 to recognize the Ser 2 phosphoepitope (see Fig. 1B).
Figure 4.
The TFIIH kinase Kin28 is not required for CTD Ser 2 phosphorylation of elongating polymerase. Chromatin IP/PCR was carried out with kin28-16 mutants combined with FCP1, fcp1-1, or fcp1-2 (YMK223, YMK224, and YMK225, respectively). Strains were also transformed with pY3AtURA to provide an HA epitope-tagged Rpb1 subunit. Immunoprecipitating antibodies are indicated to the left of the autoradiographs and PMA1 primer pairs are as in Figure 3.
Another CTD kinase of interest is the complex of Srb10 and its cyclin partner Srb11. These proteins associate with an RNA Pol II holoenzyme complex, yet their function is clearly different from that of Kin28. Biochemical and genetic data indicate that Srb10/Srb11 negatively regulate transcription at a small set of genes, and that kinase activity is necessary for this repression (Hengartner et al. 1998; Kuchin and Carlson 1998; Sun et al. 1998). The human homologs of yeast Srb10/Srb11 are components of NAT (negative regulator of activated transcription). In in vitro kinase assays, purified yeast Srb10/Srb11 and a mammalian cdk8/cyclin C complex phosphorylate both Ser 2 and Ser 5 of the CTD heptapeptide (Hengartner et al. 1998; Sun et al. 1998).
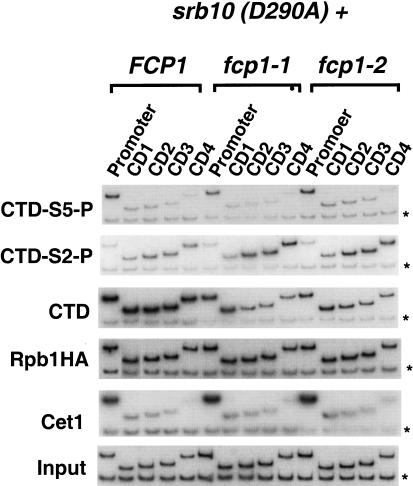
To examine the in vivo role of Srb10, yeast strains carrying a catalytically inactive Srb10 (D290A) protein were created in both wild-type and fcp1 mutant backgrounds. The D290A mutation renders Srb10 protein catalytically inactive but fully capable of being incorporated into the holoenzyme (Liao et al. 1995). Chromatin immunoprecipitations from the srb10 (D290A) strains had normal promoter-localized levels of Ser 5 phosphorylation. Furthermore, Ser 2 still becomes phosphorylated in the elongation complex (Fig. 5). Fcp1 mutations combined with catalytically inactive Srb10 (D290A) still led to an increase in Ser 2 phosphorylation levels and a concomitant reduction of the nonphosphorylated CTD cross-linking in the coding region. Capping enzyme localization was also unaffected. From these results, we conclude that Srb10/Srb11 is not responsible for Ser 2 phosphorylation of elongating polymerase.
Figure 5.
The Srb10 kinase component of RNA Pol II holoenzyme is not required for CTD Ser 2 phosphorylation of elongating polymerase. Chromatin IP/PCR was carried out with Srb10(D290A) combined with FCP1, fcp1-1, or fcp1-2 (YMK162, YMK164, YMK166, respectively). Strains were also transformed with pY3AtURA to provide an HA epitope-tagged Rpb1 subunit. Srb10 is the kinase associated with RNA Pol II holoenzyme. Srb10 (D290A) is catalytically inactive but successfully incorporated into the holoenzyme. Immunoprecipitating antibodies and PMA1 primer pairs are as in previous figures.
Yeast CTDK-I has been implicated in transcription elongation (Lee and Greenleaf 1997). This factor may be functionally related to P-TEFb, a mammalian elongation factor comprised of Cdk9/Cyclin T. P-TEFb is proposed to function by phosphorylating either the CTD or an elongation factor, thereby preventing polymerase arrest (Zhu et al. 1997; Price 2000). Cdk9 has been shown to phosphorylate Ser 2, although its substrate specificity is modified to favor Ser 5 by interaction with the viral Tat protein in vitro (Zhou et al. 2000). Yeast CTDK-I is comprised of three subunits encoded by CTK1, CTK2, and CTK3. CTK1 encodes the catalytic subunit, whereas CTK2 encodes a cyclin subunit. Purified CTDK-I can stimulate productive elongation by human RNA Pol II in vitro (Lee and Greenleaf 1997), and recent genetic results also support a role in elongation (Costa and Arndt 2000; Jona et al. 2001; Murray et al. 2001). At least some yeast genes induced at the diauxic shift require CTDK-I activity for expression (Patturajan et al. 1999).
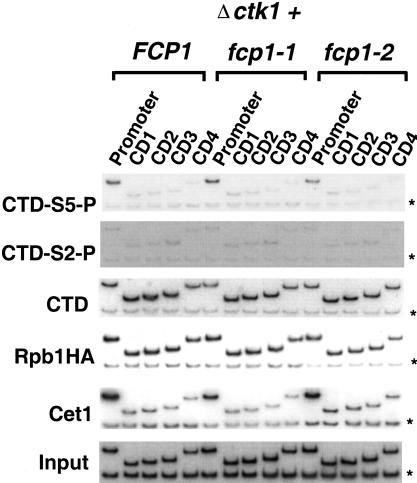
We created Ctk1-deficient strains in FCP1, fcp1-1, and fcp1-2 backgrounds. Chromatin solutions were prepared from cells grown at 30°C and analyzed by chromatin immunoprecipitation as in previous figures. Once again, CTD phosphorylation at Ser 5 was observed at the promoter but not in coding regions (Fig. 6, CTD-S5-P). This pattern was exactly paralleled by the distribution of capping enzyme (Fig. 6, Cet1). Therefore, Ctk1 does not appear to play a role in Ser 5 phosphorylation during transcription. However, in marked contrast to the kin28 or srb10 mutant strains, cross-linking of CTD–Ser 2 phosphorylation was essentially abolished in a Ctk1 deletion background. This finding suggests strongly that Ctk1 is required for Ser 2 phosphorylation of elongating Pol II (Fig. 6, CTD-S2-P). Correlating with loss of CTD-S2-P, the nonphosphorylated CTD epitope was restored to the coding region in the fcp1-1 and fcp1-2 mutant strains (Fig. 6, CTD). The cross-linking of capping enzyme was unaffected by reduced Ser 2 phosphorylation (Fig. 6, Cet1), indicating that this modification of the CTD is not necessary for dissociation of the capping enzyme from the elongation complex. Interestingly, loss of Ser 2 phosphorylation does not reduce overall levels of Pol II at either promoter or coding regions on this gene. It will be of interest to analyze genes strongly regulated by Ctk1 to determine whether they show reduced levels of elongating polymerases.
Figure 6.
The CTDK-I kinase Ctk1 is required for CTD Ser 2 phosphorylation in vivo. Chromatin IP/PCR was carried out with a ctk1 deletion mutant combined with FCP1, fcp1-1, or fcp1-2 (YSB762 shuffled with pFK1, pFK4, and pFK7, respectively). Strains were also transformed with pY3AtURA to provide an HA epitope-tagged Rpb1 subunit. Ctk1 is the catalytic kinase subunit of CTDK-I. Immunoprecipitating antibodies and PMA1 primers are as in previous figures.
Localization of Ctk1 and Fcp1 during transcription
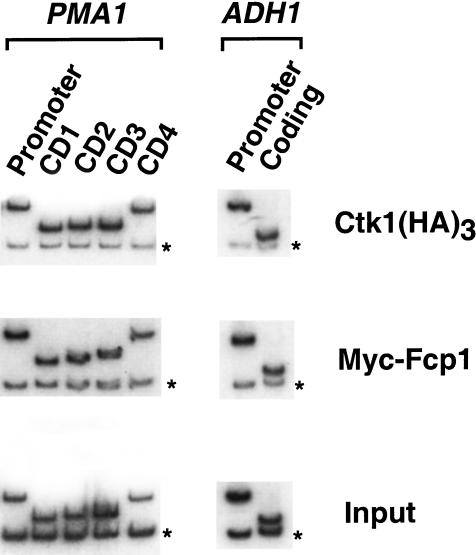
Ser 5 phosphorylation of the CTD correlates with the presence of Kin28/TFIIH at the promoter. The analysis presented in earlier figures implicates the Ctk1 kinase and Fcp1 phosphatase in the regulation of Ser 2 phosphorylation during elongation. To probe the location of these proteins during transcription, chromatin immunoprecipitations were performed using yeast strains expressing either epitope-tagged Ctk1 or Fcp1. Ctk1 was found to cross-link to the promoter and throughout the coding region, but not to a nontranscribed region (Fig. 7). Therefore, the presence of Ctk1 overlaps areas of Ser 2 phosphorylation, although Ser 2 phosphorylation is either not present or very weak near promoter regions. Due to limits in the resolution of the assay (∼300 bp), the Ctk1 cross-linking results cannot distinguish whether Ctk1 is present within the initiation complex or associates with polymerases in early elongation phase. In either case, assuming Ctk1 directly phosphorylates Ser 2, it appears that Ctk1 activity near the promoter may be either inhibited or counteracted by a phosphatase.
Figure 7.
Ctk1and Fcp1 cross-link to both promoter and coding regions. Chromatin IP/PCR was carried out on strains YSB772 containing triple HA-tagged Ctk1 and YMK210 carrying a 13×Myc epitope-tagged Fcp1. 12CA5 monoclonal and 9E10 antibodies were used for immunoprecipitation, respectively. Primers are as in previous figures.
We also monitored the presence of Fcp1 using yeast strains expressing myc- or HA-tagged Fcp1. Fcp1 cross-linked to both promoter and coding regions (Fig. 7). Although the signal was weak by comparison with other factors, it was clearly above levels seen in nontranscribed regions. Fcp1 is known to bind directly to Pol II in a region other than the CTD (Chambers and Kane 1996; M. Kobor and J. Greenblatt, unpubl.). The patterns of Fcp1 and Ctk1 cross-linking are consistent with roles in regulating phosphorylation of the polymerase CTD during elongation.
Discussion
The C-terminal domain of RNA Pol II plays an essential role in gene expression. Although early studies presumed a role in transcription initiation, it has become clear that the CTD is also required for proper elongation and mRNA processing. Many of these functions appear to be mediated by direct interactions between the CTD and the relevant transcription and mRNA processing factors. This might be surprising, given that the CTD consists of a simple repeated heptamer sequence, but it is clear that the CTD is modified by phosphorylation and perhaps other modifications such as glycosylation and ubiquitination (Corden 1990; Bensaude et al. 1999; Mitsui and Sharp 1999; Morris et al. 1999; Wu et al. 2000). We have shown that the phosphorylation pattern of the CTD changes as polymerase moves from initiation to an elongation state (Komarnitsky et al. 2000). This suggests a model in which different transcription and mRNA processing factors can each bind to specifically modified forms of the CTD that characterize various stages of initiation, elongation, and termination.
Phosphorylation of the CTD at Ser 5 occurs at the initiation complex and is dependent upon TFIIH. This modification provides a binding site for the mRNA capping enzyme, which acts on the transcript soon after initiation (Komarnitsky et al. 2000; Schroeder et al. 2000). Soon thereafter, the Ser 5 phosphoepitope disappears and the capping enzyme dissociates from the transcription elongation complex. This change is likely to be mediated by a CTD phosphatase. We suspected that the Fcp1 phosphatase might provide this function, so we tested the effect of Fcp1 mutations on the pattern of Ser 5 phosphorylation. Surprisingly, we find that Ser 5 phosphorylation and capping enzyme patterns are not affected in the fcp1 mutant strains.
Our conclusions are in apparent disagreement with those of Schroeder et al. (2000). A significant difference between their experiments and ours is that we assayed fcp1 mutants at the semipermissive temperature of 30°C, whereas they used the nonpermissive temperature of 37°C. Under our conditions, transcription continues, but clear effects on CTD phosphorylation are seen (Fig. 1). At 37°C, there is a general loss of transcription by Pol II (Kobor et al. 1999, 2000). To explore the difference in temperature, we also performed chromatin immunoprecipitations with Fcp1 mutant strains grown at 37°C (data not shown). In that case, our results were very similar to those of Schroeder et al. (2000). We also observed a dramatic loss of capping enzyme and CTD Ser-5P signals at the promoter. By calculating the ratio of signals between promoter and coding region without factoring in absolute levels, Schroeder et al. (2000) concluded that Fcp1 was required for the change at Ser 5. However, our experiments included an internal control for background cross-linking and it was clear that inactivation of Fcp1 by shift to 37°C caused the cross-linking of capping enzyme and Pol II (phosphorylated and unphosphorylated) to be reduced to background levels. On the basis of this analysis, we believe that complete Fcp1 inactivation results in a block of polymerase recruitment to promoters, not to increased Ser 5 phosphorylation levels associated with coding regions. Therefore, we suspect a phosphatase other than Fcp1 is likely to be responsible for dephosphorylation of Ser 5 in vivo.
In contrast to the lack of effect at Ser 5, fcp1 mutants caused an increased level of CTD Ser 2 phosphorylation in the elongation complex. This agrees well with the fact that Fcp1 itself cross-links to both promoter and coding regions of genes (Fig. 7). Our results suggest that the Fcp1 phosphatase interacts with the elongation complex to remove phosphates from Ser 2. In the absence of Fcp1 activity, Ser 2 phosphorylation levels in transcribing polymerase increase to the point at which very few repeats are nonphosphorylated. Because the epitope recognized by antibody 8WG16 is completely blocked by Ser 2 phosphorylation, the effect of fcp1 mutants is seen as an increase in the H5 signal, but also as a loss of the 8WG16 signal.
To identify the kinase responsible for Ser 2 phosphorylation, we tested deletions of two candidates, Srb10 and Ctk1. Strains lacking Ctk1 showed an essentially complete loss of phosphorylated Ser 2 associated with transcription elongation. Furthermore, Ctk1 itself could be cross-linked to promoter and coding regions. Therefore, we suggest that Ctk1 is the primary Ser 2 kinase during transcription elongation. Although we cannot rule out that Ctk1 is required indirectly for Ser 2 phosphorylation, our in vitro experiments indicate that the immunoprecipitated CTDK-I complex shows a strong preference for Ser 2 in vitro (E.J. Cho and M. Keogh, unpubl.). A role for CTDK-I in elongation is consistent with biochemical experiments (Lee and Greenleaf 1997). There have also been multiple reports of genetic interactions between mutations in CTK1, FCP1, and known and presumed elongation factors (Costa and Arndt 2000; Wu et al. 2000; Jona et al. 2001).
Another CTD kinase designated Bur1/Bur2 has been described recently (Yao et al. 2000; Murray et al. 2001). Bur1, like Ctk1, has strong sequence similarity to mammalian Cdk9. We have not yet examined this kinase, but it is interesting to note that it also has genetic interactions with known elongation factors. Bur1 preferentially phosphorylates CTD Ser 5 in vitro (Murray et al. 2001). It will be interesting to determine what role it plays in CTD phosphorylation in vivo.
Deletion of Srb10 results in only minor effects on the transcribing polymerase. Ser 5 phosphorylation is unaffected. In fcp1, srb10 double mutants, there may be slightly less Ser 2 phosphorylation than in fcp1 mutants alone. The lack of effect is somewhat surprising given that inactivation of Srb10 results in suppression of several Fcp1 mutant phenotypes and vice versa (M. Kobor and J. Greenblatt, in prep.). One model for Srb10 function is that it inhibits transcription by phosphorylating the CTD before the polymerase makes a functional interaction with the promoter (Hengartner et al. 1998). This could explain why the Srb10 deletion shows little effect on polymerase already at promoters or carrying out transcription. We suspect that the Fcp1 CTD phosphatase may function at two distinct points, to counteract Srb10 activity on polymerase not associated with DNA and also on elongating polymerase phosphorylated by Ctk1.
Combining our earlier model (Komarnitsky et al. 2000 and references therein) with the current study, we propose the following scenario. RNA polymerase comes to the promoter in an unphosphorylated state. This step is blocked when the Srb10/Srb11 complex phosphorylates the CTD, but the activity of Fcp1 can reverse the phosphorylation and reactivate polymerase. This is consistent with Srb10 acting as a transcriptional repressor and explains the suppression relationship between Srb10 and Fcp1. Once a preinitiation complex is assembled, the TFIIH kinase subunit phosphorylates the CTD at Ser 5. This modification acts to recruit capping enzyme and perhaps other mRNA processing factors. After the polymerase initiates and moves into elongation phase, the Ser 5 phosphate is removed by an unidentified phosphatase. CTDK-I and Fcp1 bind to polymerase, either at the promoter or in early elongation phase. In this work, we show that these two factors have antagonistic effects on Ser 2 phosphorylation. The chromatin immunoprecipitation technique cannot distinguish whether Ctk1 and Fcp1 are simultaneously present in the same elongation complex or associate either at different times or with distinct elongation complexes. In any of these situations, the competing functions of these two factors may dynamically regulate Ser 2 phosphorylation levels. We have noted a general trend in which Ser2-P levels of the polymerase increase with distance from the promoter (Komarnitsky et al. 2000; this study).
Phosphorylation of the CTD at Ser 2 is likely to recruit specific factors to the polymerase. We speculate that some of these factors regulate elongation and/or termination of transcription. One clear candidate is the prolyl-isomerase Ess1/Pin1, which has been implicated in transcription termination (Hani et al. 1999; Wu et al. 2000). This enzyme has been shown to bind to CTD phosphorylated by CTDK-I and to CTD phosphopeptides (Morris et al. 1999; Myers et al. 2001). Ess1 specifically isomerizes prolines preceded by a phosphorylated serine (Zhou et al. 1999). Remarkably, overexpression of wild-type (but not mutant) Fcp1 suppresses temperature sensitivity caused by missense mutations in Ess1, but Ess1 overexpression does not suppress fcp1 mutant phenotypes (Wu et al. 2000). It has been proposed that Ess1 acts on the CTD to create a CTD conformation that is a substrate for Fcp1 (Hani et al. 1999; Wu et al. 2000). The combined actions of Ess1 and Fcp1 may normally be required for binding of termination and polyadenylation factors to elongating polymerase, but high levels of Fcp1 might compensate for limiting levels of Ess1 activity. In support of this model, Fcp1 overexpression does not suppress a complete deletion of ESS1 (Wu et al. 2000).
It is likely that many aspects of this model are conserved in the higher eukaryotic systems, although there may also be some differences to accommodate the much longer gene sizes found in those organisms. Each of the yeast factors has a highly conserved mammalian counterpart. The mammalian homologs of Srb10 and Srb11 also function to negatively regulate transcription (Sun et al. 1998). Mammalian Fcp1 helps to recycle Pol II in vitro by removing phosphates attached to the CTD during transcription (Chambers and Dahmus 1994; Archambault et al. 1998; Cho et al. 1999). Mammalian Fcp1 can also dephosphorylate elongating polymerase and CTD sensitivity is modulated by interaction with unknown nuclear factors (Lehman and Dahmus 2000; Marshall and Dahmus 2000). One likely homolog of CTDK-I is the mammalian factor P-TEFb (Price 2000). In support of this hypothesis, an RNAi inactivation of the Caenorhabditis elegans Cdk9 gene causes a strong in vivo loss of polymerase recognized by the H5 antibody, but not the H14 antibody (K. Blackwell, pers. comm.). This suggests that Cdk9 may be a major CTD Ser 2 kinase in nematodes. Furthermore, P-TEFb has been shown to phosphorylate CTD Ser 2 in the context of the HIV promoter. The viral TAT protein, which leads to more processive elongation, apparently modifies the specificity of P-TEFb to allow phosphorylation of Ser 5 (Zhou et al. 2000) but also suppresses Fcp1 phosphatase activity (Marshall et al. 1998). How this modulation of CTD phosphorylation changes interactions of the polymerase with negative and positive elongation factors remains to be seen, both in yeast and in higher eukaryotes.
It is surprising that loss of CTD Ser 2 phosphorylation by Ctk1 appears to have a fairly mild effect on overall transcription. Yeast strains deleted for the CTK1 gene grow slowly and are inviable at 15°C (Lee and Greenleaf 1991). In contrast, RNA polymerases in which all of the CTD Ser 2 residues are substituted with either alanine or glutamate cannot normally support viability (West and Corden 1995). Remarkably, the substitution of Ser 2 can be suppressed by several mutations in genes encoding the Srb subunits of the mediator complex (Yuryev and Corden 1996). The mechanism of this suppression remains to be elucidated, but the results show that under certain conditions Ser 2 phosphorylation is not required for viability in yeast. Further insight into the role of Ser 2 phosphorylation will come when proteins that bind specifically to this form of the CTD are identified.
Materials and methods
Plasmids
pSZH: ctk1Δ∷HIS3, AmpR; pMK86: FCP1, URA3, CEN/ARS, AmpR; pFK1: FCP1, TRP1, CEN/ARS, AmpR; pFK4: fcp1-1 (R250A, P251A), TRP1, CEN/ARS, AmpR; pFK7: fcp1-2 (L177A, L181A, H187A), TRP1, CEN/ARS, AmpR; pY3AtURA: RPB1-HA, URA3, CEN/ARS, AmpR. pSZH is described in Sterner et al. (1995). pMK86, pFK1, pFK4, and pFK7 are described in Kobor et al. (1999). pY3AtURA is from West and Corden (1995).
Yeast strains
YSB762
MATα, ura3-, leu2-3, 112, trp1-1, his3-11, 15, fcp1Δ∷LEU2, ade2-1, can1-100, ctk1Δ∷HIS3 {pMK86}.
YSB763
MATα, ura3-, leu2-3, 112, trp1-1, his3Δ200 or his3-11, 15, ade2-1, can1-100 or CAN1, fcp1Δ∷LEU2, rpb3-(HA)3∷HIS3 {pMK86}.
YSB772
MATa, ura3-52, leu2Δ1, trp1Δ63, hisΔ200, lys2Δ202, ctk1-(HA)3∷Kl-TRP1.
YSB771
MATa, ura3-52 or ura3-1, leu2-3, 112, trp1-1, hisΔ200 or his3-11, 15, rpb3-(HA)3∷HIS3, ade2-1.
YMK16α
MATα, ura3-1, leu2-3, 112, trp1-1, his3-11, 15, fcp1Δ∷LEU2, ade2-1, can1-100 {pMK86}.
YMK162
MATα, ura3-1, leu2-3, 112, trp1-1, his3-11, 15, fcp1Δ∷LEU2, ade2-1, can1-100, srb10(D290A), {pFK1}.
YMK164
MATα, ura3-1, leu2-3, 112, trp1-1, his3-11, 15, fcp1Δ∷LEU2, ade2-1, can1-100, srb10(D290A), {pFK4}.
YMK166
MATα, ura3-1, leu2-3, 112, trp1-1, his3-11, 15, fcp1Δ∷LEU2, ade2-1, can1-100, srb10(D290A), {pFK7}.
YMK210
MATa, ura3-1, leu2-3, 112, trp1-1, his3-11, 15, fcp1-13xMYC∷KanR, ade2-1, can1-100.
YMK223
MATα, ura3-1, leu2-3, 112, trp1-1, his3-11, 15, fcp1Δ∷LEU2, ade2-1, can1-100, kin28Δ∷HIS3, p(Fcp1 Ade2 C/A), p(kin28-16 TRP1 C/A).
YMK224
MATα, ura3-1, leu2-3, 112, trp1-1, his3-11, 15, fcp1Δ∷LEU2, ade2-1, can1-100, kin28Δ∷HIS3, p(fcp1-1 Ade2 C/A), p(kin28-16 TRP1 C/A).
YMK225
MATα, ura3-1, leu2-3, 112, trp1-1, his3-11, 15, fcp1Δ∷LEU2, ade2-1, can1-100, kin28Δ∷HIS3, p(fcp1-2 Ade2 C/A), p(kin28-16 TRP1 C/A).
YMK16α is described in Kobor et al. (1999).The construction of YMK162, YMK164, YMK166, YMK223, YMK224, and YMK225 will be described elsewhere (M.S. Kobor and J. Greenblatt, in prep.). YSB762 was generated by transformation of YMK16α with a 2.9-kb SnaBI–AseI fragment from pSZH/ctk1Δ∷HIS. The CTK1 deletion was confirmed by cold and caffeine (7.5 mM)-sensitive phenotypes that are suppressed by a wild-type CTK1 gene. YSB763 was constructed by crossing YMK16α to YSB771 and dissecting tetrads. This strain produces a triple HA-tagged Rpb3 subunit of RNA Pol II in the FCP1 shuffling background. YSB772 carrying a triple HA-tagged Ctk1 gene was generated as follows. Primers carrying Ctk1 3′ sequences were synthesized and PCR was used to amplify a product in which a triple HA tag was fused to the C terminus of the Ctk1 ORF. The fragment also contains the Kluyveromyces lactis TRP1 gene for use as a selectable marker. The PCR product was used to transform YSB726 and the TRP1 marker was used to select for homologous recombination into the chromosome.
Media preparation, yeast transformations, and other yeast manipulations were performed by standard methods as described previously (Guthrie and Fink 1991). In Figures 4, 5, and 6, yeast strains were transformed with pY3AtURA and grown in synthetic complete (SC) medium lacking uracil and tryptophan to maintain the expression of HA-tagged Rpb1. For experiments requiring heat shock, cells were grown at 30°C until A600 reached 0.5–0.8. The culture was diluted 1:1 with fresh medium prewarmed to 55°C and then incubated at 37°C for 55 min. Control cells were diluted 1:1 using fresh medium at room temperature and then incubated at 30°C. The cells were harvested to prepare whole-cell extract as described (Patturajan et al. 1998).
Immunoblotting
For detection of phosphorylated or nonphosphorylated CTD, protein blots were probed with either ascites from monoclonal antibodies H5 and H14 (Covance) at a dilution of 1:1000 or with tissue culture supernatants from monoclonal antibodies 8WG16 or B3 at a dilution of 1:50. Polyclonal antibodies against Ceg1, Cet1, and Fcp1 have been described previously (Kobor et al. 1999; Takase et al. 2000). Monoclonal antibody G2 was a gift from Dick Burgess (University of Wisconsin-Madison). Secondary antibodies conjugated to horseradish peroxidase were from Sigma or Jackson ImmunoResearch and were used as recommended by the manufacturer for enhanced chemiluminescent detection of antigens. For analysis of CTD antibody reactivity (Fig. 1B), biotinylated CTD peptides were synthesized that contained four CTD repeats (YSPTSTS) phosphorylated in all repeats at the indicated serines. Peptides were coupled to streptavidin-coated 96 well plates (Pierce) according to the manufacturer's recommendations. The indicated antibodies were used as 1:1000 dilutions of ascites and processed for chemiluminescence as described above.
Chromatin immunoprecipitation
Chromatin immunoprecipitations and PCR reactions were performed as described in Komarnitsky et al. (2000). All yeast strains were grown at 30°C to an A600 of 0.5–0.8 with the exception of YMK223, YMK224, or YMK225, which were grown at room temperature and shifted to 30°C for 2 h. In this study, PMA1−90 and PMA11250 were used in place of PMA1−70 and PMA11235, respectively; PMA1−90, CTTTTGAATGTGTG TATAAAAGAGAG; PMA11250, TTCTTCTTTCTGGAAGCA GCCAAAC.
Acknowledgments
We thank P. Komarnitsky for help with the chromatin immunoprecipitation assay and yeast strain construction; M. Keogh for help designing oligonucleotides to create the Ctk1-triple HA-tagged strain; C. Dahl of the BCMP Biopolymers Facility for synthesizing CTD peptides; T. Takagi, D. Burgess, C. Moore, A. Greenleaf, B. Blencowe, R. Young, and D. Bregman for plasmids, strains, and antibodies. We thank J. Corden, A. Greenleaf, D. Bregman, and K. Blackwell for helpful discussions. This work was supported by grants GM46498 and GM56663 from NIH to S.B. S.B. is a Scholar of the Leukemia and Lymphoma Society. M.K. is a visiting Scholar under an agreement of international research cooperation between Harvard Medical School and Seoul National University in Korea.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL steveb@hms.harvard.edu; FAX (617) 738-0516.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.935901.
References
- Akoulitchev S, Chuikov S, Reinberg D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature. 2000;407:102–106. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- Archambault J, Chambers RS, Kobor MS, Ho Y, Cartier M, Bolotin D, Andrews B, Kane CM, Greenblatt J. An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1997;94:14300–14305. doi: 10.1073/pnas.94.26.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault J, Pan G, Dahmus GK, Cartier M, Marshall N, Zhang S, Dahmus ME, Greenblatt J. FCP1, the RAP74-interacting subunit of a human protein phosphatase that dephosphorylates the carboxyl-terminal domain of RNA polymerase IIO. J Biol Chem. 1998;273:27593–27601. doi: 10.1074/jbc.273.42.27593. [DOI] [PubMed] [Google Scholar]
- Bensaude O, Bonnet F, Casse C, Dubois MF, Nguyen VT, Palancade B. Regulated phosphorylation of the RNA polymerase II C-terminal domain (CTD) Biochem Cell Biol. 1999;77:249–255. [PubMed] [Google Scholar]
- Bregman DB, Du L, van der Zee S, Warren SL. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadena DL, Dahmus ME. Messenger RNA synthesis in mammalian cells is catalyzed by the phosphorylated form of RNA polymerase II. J Biol Chem. 1987;262:12468–12474. [PubMed] [Google Scholar]
- Chambers RS, Dahmus ME. Purification and characterization of a phosphatase from HeLa cells which dephosphorylates the C-terminal domain of RNA polymerase II. J Biol Chem. 1994;269:26243–26248. [PubMed] [Google Scholar]
- Chambers RS, Kane CM. Purification and characterization of an RNA polymerase II phosphatase from yeast. J Biol Chem. 1996;271:24498–24504. doi: 10.1074/jbc.271.40.24498. [DOI] [PubMed] [Google Scholar]
- Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, Deshaies RJ. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes & Dev. 2001;15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kim TK, Mancebo H, Lane WS, Flores O, Reinberg D. A protein phosphatase functions to recycle RNA polymerase II. Genes & Dev. 1999;13:1540–1552. doi: 10.1101/gad.13.12.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cismowski MJ, Laff GM, Solomon MJ, Reed SI. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol Cell Biol. 1995;15:2983–2992. doi: 10.1128/mcb.15.6.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden JL. Tails of RNA polymerase II. Trends Biochem Sci. 1990;15:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- Costa PJ, Arndt KM. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics. 2000;156:535–547. doi: 10.1093/genetics/156.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- Dubois MF, Marshall NF, Nguyen VT, Dahmus GK, Bonnet F, Dahmus ME, Bensaude O. Heat shock of HeLa cells inactivates a nuclear protein phosphatase specific for dephosphorylation of the C-terminal domain of RNA polymerase II. Nucleic Acids Res. 1999;27:1338–1344. doi: 10.1093/nar/27.5.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–933. [PubMed] [Google Scholar]
- Hani J, Schelbert B, Bernhardt A, Domdey H, Fischer G, Wiebauer K, Rahfeld JU. Mutations in a peptidylprolyl-cis/trans-isomerase gene lead to a defect in 3′-end formation of a pre-mRNA in Saccharomyces cerevisiae. J Biol Chem. 1999;274:108–116. doi: 10.1074/jbc.274.1.108. [DOI] [PubMed] [Google Scholar]
- Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- Hirst M, Kobor MS, Kuriakose N, Greenblatt J, Sadowski I. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol Cell. 1999;3:673–678. doi: 10.1016/s1097-2765(00)80360-3. [DOI] [PubMed] [Google Scholar]
- Ho CK, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- Jona G, Wittschieben BO, Svejstrup JQ, Gileadi O. Involvement of yeast carboxy-terminal domain kinase I (CTDK-I) in transcription elongation in vivo. Gene. 2001;267:31–36. doi: 10.1016/s0378-1119(01)00389-4. [DOI] [PubMed] [Google Scholar]
- Kobor MS, Archambault J, Lester W, Holstege FC, Gileadi O, Jansma DB, Jennings EG, Kouyoumdjian F, Davidson AR, Young RA, et al. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol Cell. 1999;4:55–62. doi: 10.1016/s1097-2765(00)80187-2. [DOI] [PubMed] [Google Scholar]
- Kobor MS, Simon LD, Omichinski J, Zhong G, Archambault J, Greenblatt J. A motif shared by TFIIF and TFIIB mediates their interaction with the RNA polymerase II carboxy-terminal domain phosphatase Fcp1p in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:7438–7449. doi: 10.1128/mcb.20.20.7438-7449.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchin S, Carlson M. Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6-Tup1. Mol Cell Biol. 1998;18:1163–1171. doi: 10.1128/mcb.18.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Greenleaf AL. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1991;1:149–167. [PMC free article] [PubMed] [Google Scholar]
- ————— Modulation of RNA polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J Biol Chem. 1997;272:10990–10993. doi: 10.1074/jbc.272.17.10990. [DOI] [PubMed] [Google Scholar]
- Lehman AL, Dahmus ME. The sensitivity of RNA polymerase II in elongation complexes to C- terminal domain phosphatase. J Biol Chem. 2000;275:14923–14932. doi: 10.1074/jbc.275.20.14923. [DOI] [PubMed] [Google Scholar]
- Liao SM, Zhang J, Jeffery DA, Koleske AJ, Thompson CM, Chao DM, Viljoen M, van Vuuren HJ, Young RA. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Dahmus ME. C-terminal domain phosphatase sensitivity of RNA polymerase II in early elongation complexes on the HIV-1 and adenovirus 2 major late templates. J Biol Chem. 2000;275:32430–32437. doi: 10.1074/jbc.M005898200. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Dahmus GK, Dahmus ME. Regulation of carboxyl-terminal domain phosphatase by HIV-1 tat protein. J Biol Chem. 1998;273:31726–31730. doi: 10.1074/jbc.273.48.31726. [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui A, Sharp PA. Ubiquitination of RNA polymerase II large subunit signaled by phosphorylation of carboxyl-terminal domain. Proc Natl Acad Sci. 1999;96:6054–6059. doi: 10.1073/pnas.96.11.6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DP, Phatnani HP, Greenleaf AL. Phospho-carboxyl-terminal domain binding and the role of a prolyl isomerase in pre-mRNA 3′-end formation. J Biol Chem. 1999;274:31583–31587. doi: 10.1074/jbc.274.44.31583. [DOI] [PubMed] [Google Scholar]
- Mortillaro MJ, Blencowe BJ, Wei X, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray S, Udupa R, Yao S, Hartzog G, Prelich G. Phosphorylation of the RNA polymerase II carboxy-terminal domain by the Bur1 cyclin-dependent kinase. Mol Cell Biol. 2001;21:4089–4096. doi: 10.1128/MCB.21.13.4089-4096.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JK, Morris DP, Greenleaf AL, Oas TG. Phosphorylation of RNA polymerase II CTD fragments results in tight binding to the WW domain from the yeast prolyl isomerase Ess1. Biochemistry. 2001;40:8479–8486. doi: 10.1021/bi0027884. [DOI] [PubMed] [Google Scholar]
- O'Brien T, Hardin S, Greenleaf A, Lis JT. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- Patturajan M, Conrad NK, Bregman DB, Corden JL. Yeast carboxyl-terminal domain kinase I positively and negatively regulates RNA polymerase II carboxyl-terminal domain phosphorylation. J Biol Chem. 1999;274:27823–27828. doi: 10.1074/jbc.274.39.27823. [DOI] [PubMed] [Google Scholar]
- Payne JM, Laybourn PJ, Dahmus ME. The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIa. J Biol Chem. 1989;264:19621–19629. [PubMed] [Google Scholar]
- Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol Cell Biol. 2000;20:104–112. doi: 10.1128/mcb.20.1.104-112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes & Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Lee JM, Hardin SE, Greenleaf AL. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol Cell Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Zhang Y, Cho H, Rickert P, Lees E, Lane W, Reinberg D. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- Takase Y, Takagi T, Komarnitsky PB, Buratowski S. The essential interaction between yeast mRNA capping enzyme subunits is not required for triphosphatase function in vivo. Mol Cell Biol. 2000;20:9307–9316. doi: 10.1128/mcb.20.24.9307-9316.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson NE, Aronson DB, Burgess RR. Purification of eukaryotic RNA polymerase II by immunoaffinity chromatography. Elution of active enzyme with protein stabilizing agents from a polyol-responsive monoclonal antibody. J Biol Chem. 1990;265:7069–7077. [PubMed] [Google Scholar]
- Weeks JR, Hardin SE, Shen J, Lee JM, Greenleaf AL. Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: Correlations with gene activity and transcript processing. Genes & Dev. 1993;7:2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wilcox CB, Devasahayam G, Hackett RL, Arevalo-Rodriguez M, Cardenas ME, Heitman J, Hanes SD. The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery. EMBO J. 2000;19:3727–3738. doi: 10.1093/emboj/19.14.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Neiman A, Prelich G. BUR1 and BUR2 encode a divergent cyclin-dependent kinase-cyclin complex important for transcription in vivo. Mol Cell Biol. 2000;20:7080–7087. doi: 10.1128/mcb.20.19.7080-7087.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuryev A, Corden JL. Suppression analysis reveals a functional difference between the Sers in positions two and five in the consensus sequence of the C-terminal domain of yeast RNA polymerase II. Genetics. 1996;143:661–671. doi: 10.1093/genetics/143.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Halanski MA, Radonovich MF, Kashanchi F, Peng J, Price DH, Brady JN. Tat modifies the activity of CDK9 to phosphorylate Ser 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol Cell Biol. 2000;20:5077–5086. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XZ, Lu PJ, Wulf G, Lu KP. Phosphorylation-dependent prolyl isomerization: A novel signaling regulatory mechanism. Cell Mol Life Sci. 1999;56:788–806. doi: 10.1007/s000180050026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes & Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]