Abstract
The development of novel anti-bacterial treatment strategies will be aided by an increased understanding of the interactions that take place between bacteria and host cells during infection. Global expression profiling using microarray technologies can help to describe and define the mechanisms required by bacterial pathogens to cause disease, and the host responses required to defeat bacterial infection.
Introduction
Exploring the RNA profiles of both host and pathogen through the course of infection promises to illuminate much about the infectious process and aid in the development of successful treatment strategies. This review focuses on the advances in whole genome transcriptional profiling of bacterial pathogens and host cells within the contexts of tissue culture, animal model and human disease. The bacterial transcriptional response to infection offers insight into the physiological state of infecting bacteria and the mechanisms required by bacteria to successfully survive infection [1]; this information could be used to define novel drug development strategies. The intracellular bacterial transcriptome might also be exploited as a bioprobe, to describe the microenvironments encountered by bacteria through the course of infection [2,3••]. Additionally RNA profiling might be employed to identify novel vaccine candidates [4]. The transcriptional response of host cells to bacterial infection enables the intracellular and intercellular interactions to be explored throughout disease progression, facilitating the discovery of bacterial immunomodulatory actions. Furthermore the transcriptional signature of human non-invasive samples also promises to reveal novel diagnostic or predictive applications [5]. Here we review the recent advances in whole genome transcriptional profiling of both host and pathogen in ex-vivo, animal model and human disease contexts. Earlier reviews have been published in this field and might be found useful [6•,7,8].
RNA methodologies
The ability to successfully monitor changes in transcript abundance is dependent on the development of RNA extraction techniques capable of purifying representative RNA populations from a variety of disease settings. The use of RNA stabilizing solutions is paramount in prokaryotic expression analyses, as the bacterial transcriptional response to the extraction process might mask relevant changes in gene expression [9]. An additional problem in studying the bacterial transcriptome during infection is the requirement to separate eukaryotic from bacterial gene expression patterns; this is especially important in paucibacillary infections where the specific activity of labelled bacterial cDNA will be low relative to the background of host cDNA in a total RNA extraction reaction. Four strategies have been employed to overcome this problem, all of which, if validated correctly, enable the transcriptional response of bacteria from mixed RNA populations (host and pathogen) to be examined.
First is the development of differential lysis methods of bacterial RNA extraction, whereupon the host cells are lysed, whilst the bacterial cells remain intact to be recovered for RNA extraction, thereby enriching the bacterial mRNA several thousand-fold. This method was originally developed for Mycobacterium tuberculosis [3••,10,11] and a modified differential lysis approach has similarly been used for Salmonella [12•]. For a review about RNA extraction issues during infection, see [13]. Second is the use of negative selection methods to remove eukaryotic RNA from a population of mixed total RNA [14]. The third involves selective capture hybridisation (SCOTS) strategies which select for specific prokaryotic message [15] and the fourth is the utilization of DNA microarrays to discriminate between bacterial and host transcriptional profiles [16••].
The study of eukaryotic and bacterial mRNA populations has been enabled for individual genes by techniques such as northern blotting, SAGE, nuclease protection, primer extension, in situ hybridisation and particularly quantitative reverse-transcription PCR (RTq–PCR using chemistries such as Taqman or molecular beacons). More population-based mRNA analysis was facilitated before genome sequence availability by methods such as RNA arbitrarily primed (RAP)-PCR and differential display (DD)-RT–PCR; now, however, whole genome expression levels can be simultaneously measured using microarray technology. This review details the impact of whole genome expression profiling on the study of host–pathogen dynamics during infection.
Complexities of host–pathogen models
Clearly the transcriptional response to infection measured is dependent on the system investigated; the interactions between infecting bacteria and the complex mixture of cell types in vivo are likely to be different to those with a single cell-type cultured in vitro. The simplistic interface of host and pathogen in vitro might not reflect the heterogeneous cell types and microenvironments encountered in vivo, but can be used to define bacterial interactions with key cell types that provide a mechanistic insight into bacterial disease progression. Moreover, interpreting the gene expression data from mixed tissues or from bacteria in multiple micro-environments, as would be seen in complex tissues with bacteria in each location expressing different sets of overlapping genes, poses a considerable hurdle to understanding the complex network of interactions that occur during human disease. These datasets might however provide a global perspective of infection, highlighting diagnostic or predictive gene expression signatures.
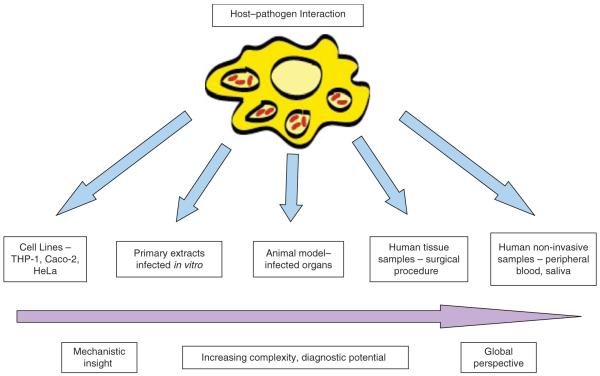
Here, we divide the host–pathogen transcriptional literature into three sections dependent on the infection model used, from in vitro tissue culture studies using cell lines or extracted primary cells, to animal models of infection, and to human patient samples (Figure 1).
Figure 1.
An illustration of the options available to study the RNA profiling of host–pathogen interactions and the alternative perspectives offered.
Tissue culture or primary extracts
Advantages
Many of the global expression analyses have been performed on cell lines or primary extracts in ex vivo model systems. This is largely a result of three factors: the ability to extract sufficient bacterial RNA from in vitro infection models; the availability of host cell types or primary extracts for multiple timepoints/comparisons; and the opportunity to explore the relatively ‘simple’ interaction between a single bacterial species and a fixed cell type or cellular composition.
Inferring bacterial metabolic states and defining virulence mechanisms
The transcriptional patterns of intracellular bacteria have been used to define the responses required for survival and successful infection. Schnappinger et al. [3••], using a differential lysis method for RNA extraction, described the switch in M. tuberculosis metabolism from using glucose and glycerol as a carbon source to using fatty acids in the murine macrophage, together with an induction of genes implicated in the adoption of an anaerobic respiratory state. Comparison with in vitro transcriptomes under defined conditions reveals that the bacterial intracellular transcriptome can act as a bioprobe for the intracellular compartment in which the pathogen resides, which for M. tuberculosis suggested that the endosome is low in iron, oxidative, nitrosative and functionally hypoxic [3••]. The intracellular transcriptomes of Salmonella enterica [12•] and Shigella flexneri [17•] have also been described after phagocytosis of macrophage-like cell lines using differential lysis methodologies, and indicate that genes associated with type III secretion systems appear to be repressed on infection. The bacterial reaction to the intracellular environment also suggests that both magnesium and phosphate are limited during macrophage infection, but interestingly for Salmonella the vacuole was not limiting for iron [12•]. The significance of bacterial type III secretion systems has also been investigated by defining the response of murine macrophage or epithelial cell lines to infection with Yersinia enterocolitica [18] or Pseudomonas aeruginosa [19]. Additionally the role of Helicobacter pylori type IV secretion systems at the interface with gastric epithelial cells has been demonstrated [20]. Belland et al. [14] defined the transcriptional pattern associated with Chlamydia trachomatis growth in epithelial cells by removing the growth-inhibitory effect of interferon-γ (IFN-γ) from the culture medium. The relationship between global mediators of macrophage activation and M. tuberculosis infection has been investigated by Shi et al. [21]. The shift to an alternate metabolic state has also been characterised after macrophage or epithelial cell infection by Listeria monocytogenes [22,23]. Conversely, McCaffrey et al. [24] have identified a cluster of interferon-responsive genes induced in murine macrophages after infection with cytosol-localized compared to vacuole-restricted L. monocytogenes.
Cell adhesion and invasion
The changing pattern of bacterial gene expression might also be used to identify factors required for cell adhesion and entry; Dietrich et al. [25] analysed the transcriptome of Neisseria meningitidis during epithelial and endothelial infection, identifying genes that were differentially regulated in a single cell type only. Similarly Jain et al. [26] demonstrated the induction of a cluster of M. tuberculosis genes of unknown function involved in the invasion of brain endothelial cells, necessary if M. tuberculosis is to cross the blood–brain barrier in central nervous system (CNS) infection. Factors affecting successful cell entry might also be characterised by following host cell responses; Pedron et al. [27•] compared the expression pattern of epithelial cells after infection with invasive or non-invasive S. flexneri strains.
Pathogen clearance or survival
Microarray analysis has also been used to investigate why some bacterial pathogens are not eradicated successfully from blood; Voyich et al. [28] identified genes involved in capsule biosynthesis and oxidative stress as induced in Staphylococcus aureus in response to human polymorphonuclear leukocytes. In addition, several genes of unknown function were observed to be differentially regulated in strains more resistant to killing.
Host responses
Whole genome transcriptomics of the host has enabled pathogen-specific gene expression responses to be recognized in purified or complex cellular environments [29–31]. Comparative microarray analyses enable distinct transcriptional responses to be characterised, dependent on the infecting bacterium [32], highlighting potential immunomodulatory features such as the limited interleukin-12 (IL-12) production in macrophages infected with M. tuberculosis [33]. This comparative approach also reveals differences in the way host immune cells respond to the same infecting bacteria [34]; Granucci et al. [35] demonstrated that IL-2 is produced by dendritic cells but not macrophages after Escherichia coli infection. The transcriptional profiles of host cells in response to infection has been reviewed recently by Jenner and Young [6•].
Animal models
Animal models of infection can be used to profile host and pathogen transcriptomes in complex environments which cannot be recreated in vitro and for which human samples are largely unavailable. Interpretation of the RNA profiles is dependent primarily on the relevance of the animal system chosen.
Complexities of multi-cellular tissue environments
The rabbit ileal loop model of infection has been used to explore the transcriptional responses of Vibrio cholerae [36] and Campylobacter jejuni [37•] to the intestinal environment. Both studies characterize the environment encountered to be nutrient-limiting and oxygen-limiting and identify putative virulence genes induced by bacteria in the rabbit intestine. The expression profiling of host tissues after bacterial infection is complicated by the changing cellular composition of organs after bacterial infection, and the selection of suitable control samples. Huff et al. [38], however, used biopsies from the antrum and corpus of nonhuman primates to describe the gastric transcription pattern through the course of H. pylori infection. The application of laser microdissection microscopy permits distinct cellular populations to be separated from complex tissues; Mueller at al. [39] compared the responses of parietal, mucus-producing and chief cell epithelial lineages in the murine stomach to H. pylori infection and demonstrated that a response to H. pylori is only detected in the mucus-producing cell type.
The environmental niche that bacteria occupy through the course of disease often defines the animal model selected for expression profiling. The gene expression pattern of the syphilis spirochete Treponema pallidum, which is intractable to RNA profiling in vitro, has been described in rabbit testicular tissue [40], and the importance in Y. pestis of a protective response to reactive nitrogen species was demonstrated in the rat bubo, the disruption of which causes attenuated virulence [41]. Indeed, Y. pestis virulence genes involved in type III secretion have been identified to be induced in the murine lung [42].
The response of M. tuberculosis has also been profiled in murine lung tissue, and Talaat et al. [43] also compared the expression pattern of M. tuberculosis in the lungs of immune-competent and immune-deficient mice. This approach has also been adopted to investigate lipoprotein expression by Borrelia burgdorferi in mice [44].
Advantages of animal models
The use of animal models enables three aspects of infection to be investigated that cannot easily be assessed by alternative methods: the interaction of bacteria with complex environments; the comparison of bacterial and/or host expression profiles from different sites of infection, for example Orihuela et al. [45] looked at the differential regulation of Streptococcus pneumoniae genes in whole blood and cerebrospinal fluid (CSF), and identified tissue-specific expression of bacterial genes; and the correlation of bacterial or host RNA profiles with clinical parameters through the course of infection [46•].
Problems with low bacterial abundance
The low number of bacteria and the difficulty in isolating bacterial RNA from host tissue has lead to the development of several systems designed to contain bacteria in an in vivo environment. Yarwood et al. [47] used subcutaneously implanted perforated hollow golf balls to model the S. aureus adaptive response, Karakousis et al. [48] used a hollow fiber assay to describe the interaction between M. tuberculosis and the artificial murine granuloma created, and the study of B. burgdorgeri transcriptomics during mammalian infection has been aided by the use of implanted dialysis membrane chambers [49].
Human disease
Reports of global gene expression profiling in human tissue or non-invasive patient samples suffering from bacterial disease are understandably limited. Rachman et al. [50•] have described the M. tuberculosis transcriptome in lung tissues extracted during surgery for untreatable tuberculosis, and were able to compare gene expression signatures of M. tuberculosis in the granuloma, pericavital tissue and macroscopically normal lung. The gene expression profile of V. cholerae in human stool samples reinforces the model that this organism reaches a hyper-infectious state after colonisation of the human intestine, with genes involved in nutrient acquisition and motility induced [51]; indeed V. cholerae transcriptomes from human stool and vomitus have been compared to model the changing expression pattern from early to late stage infection [52]. The transcriptional profiles of human gastric biopsies before and after elimination of H. pylori infection were compared after laser capture microdissection by Resnick et al. [53••], revealing the differential expression of established virulence genes and genes of unknown functional significance in H. pylori disease. The global expression profiling of host responses in patient blood [54] or saliva [55] has great diagnostic potential, as well as affording the opportunity to define host gene expression patterns in primary settings. Furthermore Ramilo et al. [56•] have recently identified transcriptional signatures able to discriminate between E. coli and S. aureus infection in patient peripheral blood leukocytes.
Conclusions
Much of the power of transcriptome studies to identify changes in global gene expression patterns comes from the ability to compare RNA profiles from different bacterial strains and/or cell types or microenvironments. To this end, transcriptional data from in vitro studies detailing the responses of bacteria/host cells to different conditions/treatments enables the complex in vivo patterns of gene expression to be interpreted. A microarray experimental strategy, including the use of a reference control channel (such as genomic DNA in bacterial systems) and the adoption of ‘minimum information about a microarray experiment’ (MIAME)-compliant microarray databases [57] has also helped in this respect. The comparative nature of microarray analyses however also raises the question of what is the appropriate control RNA to compare in vivo transcriptional data against? Should bacterial in vivo expression data be compared to RNA extracted from in vitro logarithmic grown bacteria, bacteria resuspended in culture medium, or washed off the infected cells? How are differences in the cellular composition of infected compared to uninfected samples accounted for in eukaryotic transcriptional data, and do they need to be? A variety of strategies have been employed, dependent on the aspect of the host–pathogen relationship to be explored. The comparative biasing of microarray data, however, must be considered as more transcriptional datasets are generated and compared.
The development of bacterial amplification techniques [16••] and laser microdissection microscopy promises to ensure that the transcriptional profiling of distinct and previously intractable host–pathogen interactions continues to play an important role in understanding bacterial disease processes.
Acknowledgements
SJW was funded by an EU Sixth Framework Programme Priority (LHP-CT-2004-012187). We would like to acknowledge the Wellcome Trust and its Functional Genomics Resources Initiative for funding the multi-collaborative microbial pathogen microarray facility at St George’s (BμG@S).
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Jansen A, Yu J. Differential gene expression of pathogens inside infected hosts. Curr Opin Microbiol. 2006;9:138–142. doi: 10.1016/j.mib.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Staudinger BJ, Oberdoerster MA, Lewis PJ, Rosen H. mRNA expression profiles for Escherichia coli ingested by normal and phagocyte oxidase-deficient human neutrophils. J Clin Invest. 2002;110:1151–1163. doi: 10.1172/JCI15268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••3.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]; A differential lysis method of RNA extraction was used to define the metabolic and respiratory state of M. tuberculosis after murine macrophage infection and demonstrated a nitric oxide-dependent transcriptome in IFN-γ stimulated macrophages. The pathogen profile was used as a bioprobe for the intracellular environment.
- 4.Grifantini R, Bartolini E, Muzzi A, Draghi M, Frigimelica E, Berger J, Ratti G, Petracca R, Galli G, Agnusdei M, et al. Previously unrecognized vaccine candidates against group B meningococcus identified by DNA microarrays. Nat Biotechnol. 2002;20:914–921. doi: 10.1038/nbt728. [DOI] [PubMed] [Google Scholar]
- 5.Liu M, Popper SJ, Rubins KH, Relman DA. Early days: genomics and human responses to infection. Curr Opin Microbiol. 2006;9:312–319. doi: 10.1016/j.mib.2006.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •6.Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]; A detailed review comparing of the global transcriptional responses of host cells to infection.
- 7.Kato-Maeda M, Gao Q, Small PM. Microarray analysis of pathogens and their interaction with hosts. Cell Microbiol. 2007;3:713–719. doi: 10.1046/j.1462-5822.2001.00152.x. [DOI] [PubMed] [Google Scholar]
- 8.Bryant P, Venter D, Robins-Browne R, Curtis N. Chips with everything: DNA microarrays in infectious diseases. Lancet Infect Dis. 2004;4:100–111. doi: 10.1016/S1473-3099(04)00930-2. [DOI] [PubMed] [Google Scholar]
- 9.Mangan JA, Sole KM, Mitchison DA, Butcher PD. An effective method of RNA extraction from bacteria refractory to disruption, including mycobacteria. Nucleic Acids Res. 1997;25:675–676. doi: 10.1093/nar/25.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li MS, Monahan IM, Waddell SJ, Mangan JA, Martin SL, Everett MJ, Butcher PD. cDNA-RNA subtractive hybridization reveals increased expression of mycocerosic acid synthase in intracellular Mycobacterium bovis BCG. Microbiol. 2001;147:2293–2305. doi: 10.1099/00221287-147-8-2293. [DOI] [PubMed] [Google Scholar]
- 11.Li MS, Waddell SJ, Monahan IM, Mangan JA, Martin SL, Everett MJ, Butcher PD. Increased transcription of a potential sigma factor regulatory gene Rv1364c in Mycobacterium bovis BCG while residing in macrophages indicates use of alternative promoters. FEMS Microbiol Lett. 2004;233:333–339. doi: 10.1016/j.femsle.2004.02.028. [DOI] [PubMed] [Google Scholar]
- •12.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]; The first paper to define the intracellular transcriptome of a bacterial pathogen and showed the intracellular compartment to be different from that predicted for other intracellular bacteria.
- 13.Hinton JC, Hautefort I, Eriksson S, Thompson A, Rhen M. Benefits and pitfalls of using microarrays to monitor bacterial gene expression during infection. Curr Opin Microbiol. 2004;7:277–282. doi: 10.1016/j.mib.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Belland RJ, Nelson DE, Virok D, Crane DD, Hogan D, Sturdevant D, Beatty WL, Caldwell HD. Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc Natl Acad Sci USA. 2003;100:15971–15976. doi: 10.1073/pnas.2535394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faucher SP, Porwollik S, Dozois CM, McClelland M, Daigle F. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc Natl Acad Sci USA. 2006;103:1906–1911. doi: 10.1073/pnas.0509183103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••16.Motley ST, Morrow BJ, Liu X, Dodge IL, Vitiello A, Ward CK, Shaw KJ. Simultaneous analysis of host and pathogen interactions during an in vivo infection reveals local induction of host acute phase response proteins, a novel bacterial stress response, and evidence of a host-imposed metal ion limited environment. Cell Microbiol. 2004;6:849–865. doi: 10.1111/j.1462-5822.2004.00407.x. [DOI] [PubMed] [Google Scholar]; RNA amplification was used to investigate host and pathogen transcriptional profiles from the same RNA extraction in an E. coli murine infection model.
- •17.Lucchini S, Liu H, Jin Q, Hinton JC, Yu J. Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect Immun. 2005;73:88–102. doi: 10.1128/IAI.73.1.88-102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; The transcriptional profile of S. flexneri was compared after infection of two different cell types.
- 18.Sauvonnet N, Pradet-Balade B, Garcia-Sanz JA, Cornelis GR. Regulation of mRNA expression in macrophages after Yersinia enterocolitica infection. Role of different Yop effectors. J Biol Chem. 2002;277:25133–25142. doi: 10.1074/jbc.M203239200. [DOI] [PubMed] [Google Scholar]
- 19.McMorran B, Town L, Costelloe E, Palmer J, Engel J, Hume D, Wainwright B. Effector ExoU from the type III secretion system is an important modulator of gene expression in lung epithelial cells in response to Pseudomonas aeruginosa infection. Infect Immun. 2003;71:6035–6044. doi: 10.1128/IAI.71.10.6035-6044.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guillemin K, Salama NR, Tompkins LS, Falkow S. Cag pathogenicity island-specific responses of gastric epithelial cells to Helicobacter pylori infection. Proc Natl Acad Sci USA. 2002;99:15136–15141. doi: 10.1073/pnas.182558799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi S, Blumenthal A, Hickey CM, Gandotra S, Levy D, Ehrt S. Expression of many immunologically important genes in Mycobacterium tuberculosis-infected macrophages is independent of both TLR2 and TLR4 but dependent on IFN-alphabeta receptor and STAT1. J Immunol. 2005;175:3318–3328. doi: 10.4049/jimmunol.175.5.3318. [DOI] [PubMed] [Google Scholar]
- 22.Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, Machata S, Domann E, Chakraborty T, Hain T. Intracellular gene expression profile of Listeria monocytogenes. Infect Immun. 2006;74:1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joseph B, Przybilla K, Stuhler C, Schauer K, Slaghuis J, Fuchs TM, Goebel W. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 2006;188:556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCaffrey RL, Fawcett P, O’Riordan M, Lee KD, Havell EA, Brown PO, Portnoy DA. A specific gene expression program triggered by Gram-positive bacteria in the cytosol. Proc Natl Acad Sci USA. 2004;101:11386–11391. doi: 10.1073/pnas.0403215101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dietrich G, Kurz S, Hubner C, Aepinus C, Theiss S, Guckenberger M, Panzner U, Weber J, Frosch M. Transcriptome analysis of Neisseria meningitidis during infection. J Bacteriol. 2003;185:155–164. doi: 10.1128/JB.185.1.155-164.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain SK, Paul-Satyaseela M, Lamichhane G, Kim KS, Bishai WR. Mycobacterium tuberculosis invasion and traversal across an in vitro human blood-brain barrier as a pathogenic mechanism for central nervous system tuberculosis. J Infect Dis. 2006;193:1287–1295. doi: 10.1086/502631. [DOI] [PubMed] [Google Scholar]
- •27.Pedron T, Thibault C, Sansonetti PJ. The invasive phenotype of Shigella flexneri directs a distinct gene expression pattern in the human intestinal epithelial cell line Caco-2. J Biol Chem. 2003;278:33878–33886. doi: 10.1074/jbc.M303749200. [DOI] [PubMed] [Google Scholar]; Comparison of the host response to invasive and non-invasive S. flexneri strains was used to define invasion-responsive genes in epithelial cells.
- 28.Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Said-Salim B, Porcella SF, Long RD, Dorward DW, Gardner DJ, Kreiswirth BN, et al. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol. 2005;175:3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 29.Huang Q, Liu D, Majewski P, Schulte LC, Korn JM, Young RA, Lander ES, Hacohen N. The plasticity of dendritic cell responses to pathogens and their components. Science. 2001;294:870–875. doi: 10.1126/science.294.5543.870. [DOI] [PubMed] [Google Scholar]
- 30.Boldrick JC, Alizadeh AA, Diehn M, Dudoit S, Liu CL, Belcher CE, Botstein D, Staudt LM, Brown PO, Relman DA. Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc Natl Acad Sci USA. 2002;99:972–977. doi: 10.1073/pnas.231625398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobayashi SD, Braughton KR, Whitney AR, Voyich JM, Schwan TG, Musser JM, DeLeo FR. Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. Proc Natl Acad Sci USA. 2003;100:10948–10953. doi: 10.1073/pnas.1833375100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feezor RJ, Oberholzer C, Baker HV, Novick D, Rubinstein M, Moldawer LL, Pribble J, Souza S, Dinarello CA, Ertel W, Oberholzer A. Molecular characterization of the acute inflammatory response to infections with gram-negative versus gram-positive bacteria. Infect Immun. 2003;71:5803–5813. doi: 10.1128/IAI.71.10.5803-5813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nau GJ, Richmond JF, Schlesinger A, Jennings EG, Lander ES, Young RA. Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci USA. 2002;99:1503–1508. doi: 10.1073/pnas.022649799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaussabel D, Semnani RT, McDowell MA, Sacks D, Sher A, Nutman TB. Unique gene expression profiles of human macrophages and dendritic cells to phylogenetically distinct parasites. Blood. 2003;102:672–681. doi: 10.1182/blood-2002-10-3232. [DOI] [PubMed] [Google Scholar]
- 35.Granucci F, Vizzardelli C, Pavelka N, Feau S, Persico M, Virzi E, Rescigno M, Moro G, Ricciardi-Castagnoli P. Inducible IL-2 production by dendritic cells revealed by global gene expression analysis. Nat Immunol. 2001;2:882–888. doi: 10.1038/ni0901-882. [DOI] [PubMed] [Google Scholar]
- 36.Xu Q, Dziejman M, Mekalanos JJ. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc Natl Acad Sci USA. 2003;100:1286–1291. doi: 10.1073/pnas.0337479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •37.Stintzi A, Marlow D, Palyada K, Naikare H, Panciera R, Whitworth L, Clarke C. Use of genome-wide expression profiling and mutagenesis to study the intestinal lifestyle of Campylobacter jejuni. Infect Immun. 2005;73:1797–1810. doi: 10.1128/IAI.73.3.1797-1810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; The combination of transcriptional analysis and knockout mutants was used to define gene function in C. jejuni.
- 38.Huff JL, Hansen LM, Solnick JV. Gastric transcription profile of Helicobacter pylori infection in the rhesus macaque. Infect Immun. 2004;72:5216–5226. doi: 10.1128/IAI.72.9.5216-5226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mueller A, Merrell DS, Grimm J, Falkow S. Profiling of microdissected gastric epithelial cells reveals a cell type-specific response to Helicobacter pylori infection. Gastroenterology. 2004;127:1446–1462. doi: 10.1053/j.gastro.2004.08.054. [DOI] [PubMed] [Google Scholar]
- 40.Smajs D, McKevitt M, Howell JK, Norris SJ, Cai WW, Palzkill T, Weinstock GM. Transcriptome of Treponema pallidum: gene expression profile during experimental rabbit infection. J Bacteriol. 2005;187:1866–1874. doi: 10.1128/JB.187.5.1866-1874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sebbane F, Lemaitre N, Sturdevant DE, Rebeil R, Virtaneva K, Porcella SF, Hinnebusch BJ. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc Natl Acad Sci USA. 2006;103:11766–11771. doi: 10.1073/pnas.0601182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lathem WW, Crosby SD, Miller VL, Goldman WE. Progression of primary pneumonic plague: a mouse model of infection, pathology, and bacterial transcriptional activity. Proc Natl Acad Sci USA. 2005;102:17786–17791. doi: 10.1073/pnas.0506840102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talaat AM, Lyons R, Howard ST, Johnston SA. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc Natl Acad Sci USA. 2004;101:4602–4607. doi: 10.1073/pnas.0306023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang FT, Nelson FK, Fikrig E. Molecular adaptation of Borrelia burgdorferi in the murine host. J Exp Med. 2002;196:275–280. doi: 10.1084/jem.20020770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orihuela CJ, Radin JN, Sublett JE, Gao G, Kaushal D, Tuomanen EI. Microarray analysis of pneumococcal gene expression during invasive disease. Infect Immun. 2004;72:5582–5596. doi: 10.1128/IAI.72.10.5582-5596.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •46.Virtaneva K, Porcella SF, Graham MR, Ireland RM, Johnson CA, Ricklefs SM, Babar I, Parkins LD, Romero RA, Corn GJ, et al. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc Natl Acad Sci USA. 2005;102:9014–9019. doi: 10.1073/pnas.0503671102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bacterial transcriptional patterns were correlated with clinical parameters through the course of infection.
- 47.Yarwood JM, McCormick JK, Paustian ML, Kapur V, Schlievert PM. Repression of the Staphylococcus aureus accessory gene regulator in serum and in vivo. J Bacteriol. 2002;184:1095–1101. doi: 10.1128/jb.184.4.1095-1101.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karakousis PC, Yoshimatsu T, Lamichhane G, Woolwine SC, Nuermberger EL, Grosset J, Bishai WR. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J Exp Med. 2004;200:647–657. doi: 10.1084/jem.20040646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Revel AT, Talaat AM, Norgard MV. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc Natl Acad Sci USA. 2002;99:1562–1567. doi: 10.1073/pnas.032667699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •50.Rachman H, Strong M, Ulrichs T, Grode L, Schuchhardt J, Mollenkopf H, Kosmiadi GA, Eisenberg D, Kaufmann SH. Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect Immun. 2006;74:1233–1242. doi: 10.1128/IAI.74.2.1233-1242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; The gene expression profiles of M. tuberculosis in different human lung tissues extracted after surgery were compared.
- 51.Merrell DS, Butler SM, Qadri F, Dolganov NA, Alam A, Cohen MB, Calderwood SB, Schoolnik GK, Camilli A. Host-induced epidemic spread of the cholera bacterium. Nature. 2002;417:642–645. doi: 10.1038/nature00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larocque RC, Harris JB, Dziejman M, Li X, Khan AI, Faruque AS, Faruque SM, Nair GB, Ryan ET, Qadri F, et al. Transcriptional profiling of Vibrio cholerae recovered directly from patient specimens during early and late stages of human infection. Infect Immun. 2005;73:4488–4493. doi: 10.1128/IAI.73.8.4488-4493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••53.Resnick MB, Sabo E, Meitner PA, Kim SS, Cho Y, Kim HK, Tavares R, Moss SF. Global analysis of the human gastric epithelial transcriptome altered by Helicobacter pylori eradication in vivo. Gut. 2006;55:1717–1724. doi: 10.1136/gut.2006.095646. [DOI] [PMC free article] [PubMed] [Google Scholar]; Laser capture dissection was used to profile distinct cell types from human biopsies before and after H. pylori infection in the same patient.
- 54.Griffiths MJ, Shafi MJ, Popper SJ, Hemingway CA, Kortok MM, Wathen A, Rockett KA, Mott R, Levin M, Newton CR, et al. Genomewide analysis of the host response to malaria in Kenyan children. J Infect Dis. 2005;191:1599–1611. doi: 10.1086/429297. [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Zhou X, St JM, Wong DT. RNA profiling of cell-free saliva using microarray technology. J Dent Res. 2004;83:199–203. doi: 10.1177/154405910408300303. [DOI] [PubMed] [Google Scholar]
- •56.Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, Wittkowski KM, Piqueras B, Banchereau J, Palucka AK, Chaussabel D. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gene expression signatures were identified in human patient leukocytes that were able to discriminate between bacterial infections.
- 57.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]