Abstract
It is postulated that the symmetric organization of plant lateral organs is based on two crossed axes, the abaxial–adaxial and the lateral axes. The PRESSED FLOWER (PRS) gene, the expression and function of which are dependent on the lateral axis, is reported in this study. In the prs mutant, growth of the lateral sepals is repressed, and although the size and shape of the abaxial and adaxial sepals are normal, the cell files at the lateral margins are missing. Double-mutant analyses showed that the PRS gene functions independently of the determinations of both floral organ identity and floral meristem size. The PRS gene, encoding a putative transcriptional factor with a homeodomain, was shown to be required for cell proliferation. PRS gene expression is spatially and temporally unique and is expressed in a restricted number of L1 cells at the lateral regions of flower primordia, floral organ primordia, and young leaf primordia. Our study strongly suggests that the PRS gene is involved in the molecular mechanism of lateral axis-dependent development of lateral organs in Arabidopsis.
Keywords: PRESSED FLOWER, Arabidopsis, homeobox, lateral axis, flower development, cell proliferation
Postembryonic lateral organs in aerial parts of plants are generated from shoot apical meristems. Leaf primordia are generated at the peripheral zone of vegetative meristem, and after entering the reproductive phase, the vegetative meristem shifts to an inflorescence meristem, which produces lateral floral meristems from its flank. Floral meristems generate floral organs, sepals, petals, stamens, and carpels in a manner similar to leaf generation. Leaves, floral organs, and the arrangement of floral organs have symmetric structures, which suggests an axis-dependent development mechanism.
The dome-like leaf primordium grows to a flat mature leaf, the two sides of which can be distinguished in Arabidopsis. Epidermal cells on one side of leaves toward the meristem (adaxial side) are shaped like pieces of a jigsaw puzzle and form trichomes. Cells on the other side (abaxial side) are smaller and have fewer trichomes, but form more stomata than do the cells on the adaxial side. Under the epidermis, the palisade and spongy cells align at the adaxial and abaxial sides, respectively. At the lateral ends of a leaf, where epidermal cells of either side meet, long marginal cells are formed (Bowman 1994). The leaf structure reflects, therefore, the abaxial–adaxial axis and the lateral axis against the position of meristem from which their primordia are formed. The structure of floral organs, sepals, petals, stamens, and carpels resembles that of leaves. The center of the floral meristem is the reference point of the postulated axes in the floral organs. The abaxial–adaxial and the lateral axes also exist in the floral meristem. The positions of the floral organs are determined according to the axes arranged in reference to the inflorescence meristem. The shape of lateral organs and the arrangement of floral organs indicate that the pattern of cell growth and differentiation is dependent on the postulated axes. Recent studies revealed that the expression pattern of a set of genes is under the control of the axes.
YABBY (YAB) genes of Arabidopsis, which encode putative transcriptional factors, carry a zinc finger domain and a YABBY (helix-loop-helix) domain. Three members of the YAB gene family—FILAMENTOUS FLOWER (FIL), YABBY2 (YAB2), and YABBY3 (YAB3)—are expressed in tissues at the abaxial side of leaves and floral organs (Sawa et al. 1999; Siegfried et al. 1999). Leaf epidermal cells of the fil-5 yab3-1 double mutant are mixtures of the abaxial and adaxial cell types and miss the distinction of the abaxial and adaxial sides (Siegfried et al. 1999). In the transgenic plants, in which either FIL or YAB3 is ectopically expressed, the cell shape on the adaxial surface of leaves resembles that of the abaxial surface (Sawa et al. 1999; Siegfried et al. 1999). On the other hand, members of a HD-Zip III subfamily—including REVOLUTA (REV), PHABULOSA (PHB), and PHAVOLUTA (PHV)—are suggested to regulate the adaxial cell fates. REV and PHB are expressed in the adaxial side tissue of leaves; REV is also expressed in the adaxial side of young floral organs (McConnell et al. 2001; Otsuga et al. 2001; J. Emery and J.L. Bowman, unpubl.). Leaves of the dominant phb and phv mutants are transformed to radially symmetric filaments, the epidermal cells of which have adaxial characters (McConnell and Barton 1998; McConnell et al. 2001). The results strongly suggest that the abaxial–adaxial axis in the lateral organ primordia determines the clear boundary of the cells expressing genes required for differentiation of either the abaxial or adaxial sides of leaves and floral organs.
The side-specific genes are expressed in flower primordia. In early stages of developing flowers, when no floral organ primordia appear, both FIL and YAB3 are expressed in the abaxial side of flower primordia (Sawa et al. 1999; Siegfried et al. 1999), and in the adaxial side, REV is expressed (Otsuga et al. 2001; J. Emery and J.L. Bowman, unpubl.). The biased expression pattern of FIL, YAB3, and REV is known to be responsible for the normal development of floral meristem. In the fil-1 mutant and the fil-5 yab3-1 double mutant, floral meristems are converted to filamentous structures (Sawa et al. 1999; Siegfried et al. 1999). Similar filamentous structures are formed in the inflorescence of rev-1 (Talbert et al. 1995), and these findings indicate that the side-specific expression of FIL, YAB3, and REV is required to establish a flower primordium with correctly positioned floral organs.
In contrast to the abaxial–adaxial axis-dependent development, little is known about the molecular mechanism of the lateral axis-dependent development of the lateral organs and floral meristems. To investigate the mechanism, we isolated and analyzed a flower mutant of Arabidopsis, pressed flower (prs). Here, we report the phenotype of prs and molecular identification of PRS. This will be the first report of a gene involved in the lateral axis-dependent development of lateral organs and floral meristems of Arabidopsis.
Results
Phenotypes of prs mutant
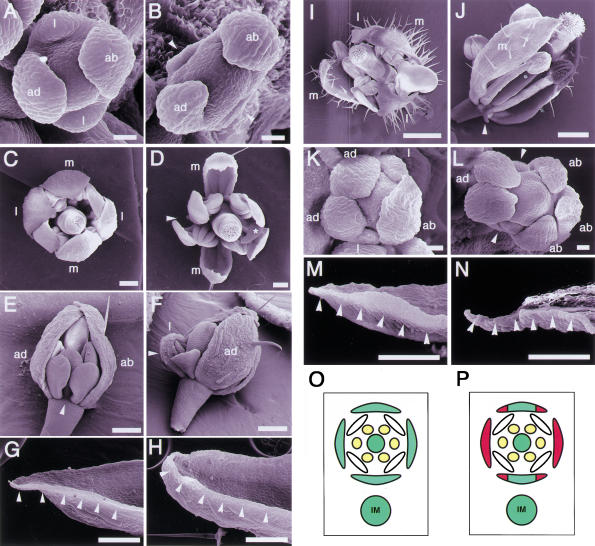
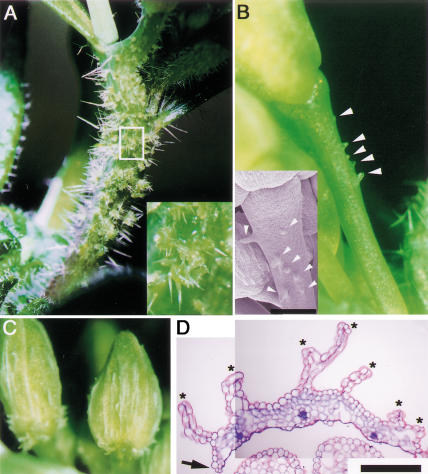
We isolated a recessive and single gene mutant, prs, named because the flower looks flattened, from an M2 population of ethylmethane sulfonate mutagenized seeds of Arabidopsis Landsberg erecta (L. er). In the flower primordia at stages 1 and 2 (Smyth et al. 1990), before the sepal primordia appear, no morphological differences between the wild type and prs were observed. Defects in the lateral sepal development in prs were first detected at stages 3 and 4, when the sepal primordia arise and bulge. Most of the lateral sepal primordia of prs were smaller than those of the wild type and were sometimes absent (Fig. 1A,B). In the mature flower of prs, 70% of the lateral sepals developed into small sepals (Fig. 1C,D,F) or into filamentous organs (data not shown) or were missing when the primordia had not formed (Fig. 1D,E). This phenotype indicates that prs has defects in the formation of the sepals at the lateral positions in relation to the inflorescence meristem. Although the shape and size of the abaxial and adaxial sepals were indistinguishable from the corresponding wild-type sepals (Fig. 1D), more detailed observations revealed defects in the marginal regions. Knife-edge cells, which are made from the L1 layer, reside in the marginal regions of wild-type sepals (Fig. 1G), whereas these cells were missing from the marginal regions of the abaxial and adaxial sepals of prs (Fig. 1H). Other than the sepals in prs, we have not been able to find any difference in the marginal regions of floral organs. The marginal regions of sepals correspond to the lateral positions in sepals in relation to the center of the flower. In other words, prs has defects in the development of the cells at the lateral marginal regions relative to the floral meristem. These phenotypes indicate that PRS functions are highly relevant to the flower and sepal development at the lateral regions in relation to inflorescence and floral meristem, respectively (Fig. 1O,P).
Figure 1.
Phenotypes of mutant flowers. (A) A wild-type flower at stage 4. (B) A prs flower at stage 4. Lateral sepal primordia are not observed (arrowheads). (C) A mature flower of wild type. (D) A mature flower of prs. One lateral sepal is missing (arrowhead), and the other is smaller than normal (asterisk). (E) A young flower of prs. A lateral sepal is completely absent (arrowhead). (F) A young flower of prs. A lateral sepal (arrowhead) is smaller than the adaxial sepal. (G) Knife-edge margin of a wild-type sepal (arrowheads). (H) Margin of a prs sepal. The knife-edge cells are absent (arrowheads). (I) An ap2-1 flower. (J) A prs ap2-1 flower. A transformed leaf-like organ at lateral position remained as a filamentous organ (arrowhead). (K) A clv1-4 flower. (L) A prs clv1-4 flower. Sepal primordia are absent at the lateral regions (arrowheads). (M) Margin of an ap3-5 sepal in the second whorl. The knife-edge cells are shown by arrowheads. (N) Margin of a prs ap3-5 sepal in the second whorl. The knife-edge cells are absent (arrowheads). (O) Diagram of a wild-type flower. (P) Diagram showing positions of defects observed in a prs flower (shown in red). (ab) abaxial; (ad) adaxial; (l) lateral; (m) medial; (IM) inflorescence meristem. A,B,K,L, bar, 20 μm; C–H,M,N, bar, 200 μm; I,J, bar, 500 μm.
Double mutant analyses
To examine whether PRS is also involved in processes of flower formation other than sepal development, we crossed prs with several flower mutants. First, we investigated whether PRS function is related to the determination of sepal identity. APETALA2 (AP2) is a key gene determining the identity of sepals (Bowman et al. 1991). In ap2-1, one of the weak alleles, floral organs in the first whorl lose sepal identity and are homeotically transformed into leaf-like organs with trichomes (Fig. 1I; Bowman et al. 1989). In the prs ap2-1 double mutant, the organs in the lateral position of the first whorl were absent or transformed into filamentous organs. On the other hand, leaf-like organs were formed at the abaxial and adaxial position (Fig. 1J). This phenotype indicates that PRS functions independently of the floral organ identity. Second, we crossed prs with a meristem size mutant, clv1-4 (Fig. 1K; Clark et al. 1993), for investigating the relation of the PRS function with the meristem size. In the prs clv1-4 double mutant, the size of the floral meristem increased enough to initiate five or six sepal primordia, but there were vacant spaces, with no primordia, at the lateral positions in the first whorl (Fig. 1L). However, such openings were not observed in the clv1-4 single-mutant flower. This finding suggests that PRS functions independently of the floral meristem size. Third, we examined the PRS function in the inner-whorl organs, which did not have obvious defects in the prs single mutant. The second whorl organs of apetala3-5 (ap3-5) are homeotically transformed to sepals (Jack et al. 1992). We confirmed the transformed sepals had knife-edge cells, which are indistinguishable from those observed in wild-type sepals (Fig. 1M). The second whorl organs of the prs ap3-5 double mutant looked like sepals, but the margin-specific cells were not observed at the margin of the organs (Fig. 1N). This result indicates that PRS has a role in the formation of the margin cells of the second whorl organs as well as of those of the first whorl organs.
Cloning of the PRS gene
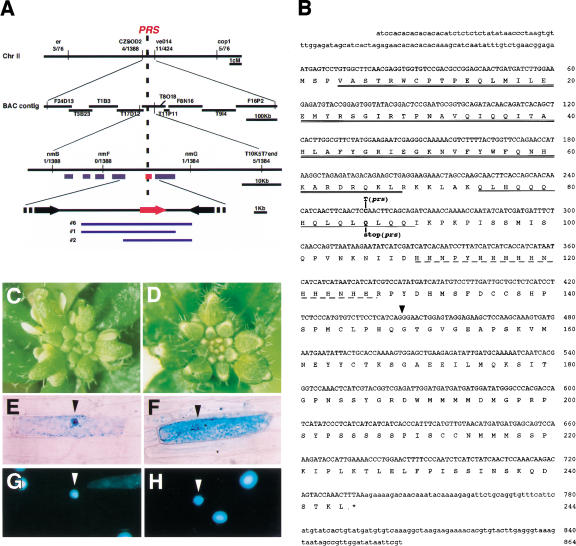
We isolated PRS by positional cloning methods. PRS was mapped on chromosome 2 within a region of ∼30 kb between nmB and nmG markers, made on the basis of the genomic sequence data from The Institute for Genomic Research (TIGR; Fig. 2A). The region is covered with annotated BAC clones, T8O18 and T17D12. We found six predicted open reading frames (ORFs) in this region and compared the genomic sequences between wild type (L. er) and prs. A single-base change was found in one ORF, T8O18.10 (Fig. 2A,B). To confirm that this ORF corresponded to PRS, we used three genomic fragments from a genomic library, including T8O18.10 ORF, to transform prs by a vacuum infiltration method. The T1 plants carrying these genomic clones showed complementation of the floral defects of prs (Fig. 2A,C,D). PRS cDNA, obtained by 5′- and 3′-RACE, is 966 bp, including 5′-UTR of 102 bp and 3′-UTR of 129 bp (Fig. 2B). PRS encodes a protein containing 244 amino acids, with a homeodomain at the N terminus showing an identity of 68% to a homeodomain of WUSCHEL (WUS; Fig. 2B; Mayer et al. 1998). PRS had two low complexity regions; in the middle of PRS, we found a glutamine-rich region and a histidine-rich one (Fig. 2B). There were no genes sharing any similarity to the other regions of PRS in the database. In prs, a nucleotide change from C to T was found at nucleotide 256, which caused a change of glutamine 86 to a stop codon (Fig. 2B).
Figure 2.
Scheme of the positional cloning and the structure of PRS. (A) Genomic structure of the PRS (T8O18.10) region on chromosome 2. PRS was mapped between markers nmB and nmG. Numbers below the markers indicate the recombination frequency between the marker and the PRS locus. The red bar and arrow show the predicted ORF of T8O18.10. Blue lines show the genomic clones that complemented the prs phenotype. (B) cDNA sequence of PRS and the predicted amino acid sequence including a homeodomain (double underline), a glutamine-rich region (single underline), and a histidine-rich region (broken line). Mutation site in prs is shown. The position of the intron (437 bp) is indicated by a triangle (GenBank accession no. AB058920). (C) Inflorescence of prs. (D) Inflorescence of prs transformed with a genomic clone including T8O18.10. (E–H) Subcellular localization of the PRS-GUS fusion protein by particle bombardment of onion epidermal cells. (E,G) Transiently transformed with 35S:PRS-GUS. (F,H) Transiently transformed with 35S:GUS as a control. GUS (E,F) and DAPI (G,H) staining are shown. Arrowheads indicate the nucleus.
PRS was predicted to be a transcriptional factor according to the deduced amino acid sequence from the cDNA sequence. To investigate the cellular localization of PRS, we made a chimeric gene of the full-length PRS cDNA, translationally fused to β-glucuronidase (GUS) coding region driven by the 35S promoter of the cauliflower mosaic virus, and introduced it into onion epidermal cells with the use of a particle bombardment system. The fusion protein was localized in the nucleus (Fig. 2E–H). Although an obvious nuclear localization signal was not found, the motif of homeodomain and the nuclear localization of the fusion protein in onion cells strongly suggest that PRS functions as a transcriptional regulator.
Spatiotemporal pattern of the PRS expression
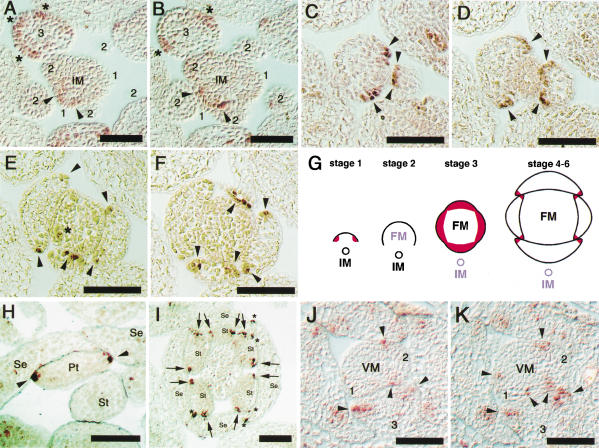
The expression of PRS in flowers was under the detectable level in Northern blot analysis (data not shown). RT–PCR analysis showed that PRS was expressed in aerial parts of seedlings, inflorescences, and flowers (data not shown). To examine the temporal and spatial pattern of the expression of PRS in the inflorescences and flowers, we performed in situ hybridization. To prevent cross-hybridization to other homeobox genes, a part of the cDNA of PRS lacking the homeodomain was used as a probe. The first appearance of PRS expression was in the L1 cells of the lateral regions of a flower primordium at early stage 1, in which the lateral sepals are expected to develop at a later stage (Fig. 3A,B, arrowheads and G). PRS expression rapidly diminished at the late stage 1 and disappeared at stage 2 (Fig. 3A,B,G). At stage 3, PRS expression reappeared in all four-sepal young primordia (Fig. 3A,B, asterisks and G). No expression was detected at the central zone of the inflorescence meristem and the floral meristem (Fig. 3A,B). In stages 4 through 6, when four sepals develop to enclose the flower bud, PRS mRNA was localized at the lateral edges of the four sepals (Fig. 3G). A series of sections of a flower at stage 6 showed that the regions expressing PRS formed an arch of the L1 cells at the margin of sepals (Fig. 3C–F). In addition, PRS began to be expressed in the young primordia of petals (Fig. 3E, asterisks) and stamens (data not shown). As the petals and stamens develop, the expression of PRS was limited at the margins of petals and stamens in a way similar to that of sepals (Fig. 3H,I). Interestingly, no expression was detected in the carpels (Fig. 3I). Figure 5, A–D, are schematic drawings of the expression patterns of PRS in a flower primordium and floral organs. Moreover, in the vegetative phase, PRS was expressed at the lateral regions of young leaf primordia, as well as in flowers and floral organs (Figs. 3J,K and 5C,D).
Figure 3.
Expression patterns of PRS in inflorescence, floral organs, and leaf primordia of wild type. (A,B) PRS expression patterns in an inflorescence in serial sections. Numbers show the stage of flowers. Arrowheads and asterisks show expressions at early stage 1 and at stage 3, respectively. (C–F) PRS expression patterns in sepals in serial sections at stage 5. Arrowheads show the expressing cells at the lateral edges of sepal primordia. In E, the asterisk shows the expression in a petal primordium. (G) Schematic drawings that show the PRS expression patterns. At early stage 1, the PRS expression is restricted at the lateral regions of a flower primordium. At stage 2, the PRS expression is disappeared. At stage 3, the PRS expression is reappeared in all four-sepal young primordia. After stage 4, the PRS expression is restricted at the lateral edges of young sepals. (H) Expression of PRS at the lateral edges of a petal (arrowheads). (I) Expression of PRS at the lateral edges of stamens (arrows) and of sepals (asterisks). No expression is detected in the carpel. (J,K) PRS-expression patterns in a vegetative meristem in serial sections. Numbers show the leaf primordium in order. Arrowheads show the expressions at the lateral edges of primordia. (IM) Inflorescence meristem; (FM) floral meristem; (VM) vegetative meristem; (Se) sepal; (Pt) petal; (St) stamen. Bars, 50 μm.
Figure 5.
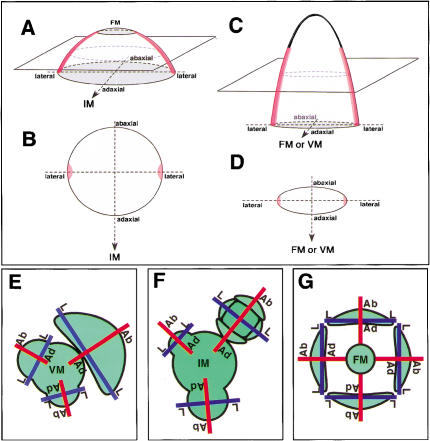
Diagrams of PRS expression and axes in lateral organs and in the flower. Diagrams showing the expression patterns of PRS in a flower primordium at stage 1 (A, B), and in a floral organ or a leaf (C, D). Red color indicates the position of cells expressing PRS. (E) Top view of a vegetative shoot showing abaxial–adaxial and lateral axes in leaf primordia. (F) Top view of an inflorescence showing abaxial–adaxial and lateral axes in early stage flowers. (G) Abaxial–adaxial and lateral axes in floral organs. (Ab-Ad) Abaxial–adaxial axis; (L-L) lateral axis; (VM) vegetative meristem; (IM) inflorescence meristem; (FM) floral meristem.
The unique expression pattern correlates to the phenotypes of the prs mutant. In the process of flower development, PRS expression was detected first at the two presumptive areas to generate the lateral sepals (Fig. 3A,B, arrowheads). The restricted inhibition of the lateral sepal development in prs would be responsible for the loss of PRS function in this stage. The second observation of PRS expression was at the lateral margins of sepals (Fig. 3A,B, asterisks and 3C–F, arrowheads). The missing cell files at the margin of the abaxial and adaxial sepals are most likely to be related to the PRS expression in the cells at the corresponding regions. PRS was also expressed at the margins of petals and stamens (Fig. 3H,I), suggesting that PRS has a function in forming the margin of the inner-whorl organs. This is confirmed by the loss of the marginal cells in the second whorl organs in prs ap3-5 (Fig. 1N). In prs, however, we were not able to find any structural abnormality in petals and stamens, possibly because the marginal cells are indistinguishable from abaxial and/or adaxial cells. In addition, PRS was expressed at the lateral edges of leaf primordia, although distinct defects in the regions were not detected in prs. One explanation is that functionally redundant genes are involved in the development of the cells at the margins of leaves.
Effects of ectopic and overexpression of the PRS gene
To investigate the function of PRS, we also analyzed a phenotype of a gain-of-function mutant of PRS. In 35S:PRS transgenic plants, multicellular bulges with trichomes were observed on the stem (Fig. 4A) and on the peduncle (Fig. 4B). On the sepals, white wrinkle structures were observed (Fig. 4C). Transverse sections showed that the structures were outgrowths of epidermal cells (Fig. 4D). The margin of wild-type sepals was made of similar outgrowths of epidermal cells (Fig. 4D). We interpret these phenotypes as being the result of ectopic and overproliferation of the epidermal cells. This aberrant proliferation of epidermal cells in various organs observed in the transgenic plants strongly suggests that PRS functions to promote the genetic pathways involved in the proliferation of L1 cells.
Figure 4.
Phenotypes of 35S:PRS transgenic plants. (A) Multicellular bulges on the stem. (Inset) Magnification of the white box. (B) Multicellular bulges on the peduncle (arrowheads). (Inset) Magnified picture. (C) White wrinkle structures on sepals. (D) Transverse section of a sepal. The arrow and asterisks indicate the edge of the sepal and the wrinkle structures, respectively. B inset, bar, 250 μm; D, bar, 100 μm.
Discussion
Symmetrical structures of lateral organs and flowers suggest a molecular basis to lateral axis-dependent development. Our results strongly indicate that PRS is involved in the molecular mechanism.
The expression pattern of PRS is regulated by positional information
In this study, we showed that PRS is expressed in a temporally and spatially restricted manner. It strongly indicated that the spatial pattern of PRS expression is under the control of some basic system that determines the topological structure of lateral organs and flower primordia. The expression of PRS was detected at the lateral regions of lateral organs and of flower primordia at very early stages (Fig. 5A–D), when the abaxial and adaxial side-determinant genes, YABs and REV, were expressed in a side-specific manner (Sawa et al. 1999; Siegfried et al. 1999; Otsuga et al. 2001; J. Emery and J.L. Bowman, unpubl.). This observation suggests that both of the lateral side-specific expression patterns of PRS and the abaxial–adaxial side-specific expression patterns of YABs and REV are determined at about the same time.
A question arises as to how the expression patterns of these genes are controlled. In some aspects of animal development, a gradient of small diffusible molecules leads a clear boundary of gene expression in a cell mass, which appears uniform, by inducing the expression of genes that mutually repress their expression (Briscoe et al. 2000). When applying this mechanism to the plant meristem, the radial distribution of signal molecule(s) from the meristem will lead to the formation of the abaxial–adaxial boundary, separating cells expressing either the abaxial side-specific genes or the adaxial side-specific genes, and the boundary will define the cells expressing PRS. Here, we would like to propose a simple model. Because PRS is expressed in the L1 cells located at the lateral ends, where the abaxial–adaxial boundary merges with the L1 cell layer (Fig. 5A–D), the PRS-expressing cells could be defined both by the L1-specific signals and by some signal(s) specifying the abaxial–adaxial boundary.
In Arabidopsis, several L1-specific genes are known. Recent analysis of PROTODERMAL FACTOR1 (PDF1) and other genes revealed that an 8-bp motif, 5′-TAAATG(C/T)A-3′, referred to as the L1 box, in the 5′ upstream regions is essential to induce L1-specific expression (Abe et al. 2001). Interestingly, PRS has the L1 box in the 5′ upstream position (from −1265 to −1258), suggesting that the L1-specific expression of PRS depends on the sequence. It is necessary to examine whether the L1 box of PRS is sufficient for L1-specific expression of PRS. It would also be interesting to investigate the expression pattern of PRS in a series of mutants with abnormal abaxial–adaxial polarity.
Temporal disappearance of PRS expression reflects the independence of the floral meristem from the inflorescence meristem
The developmental process of a flower can be divided into two phases. In the first phase, a flower primordium is generated from the peripheral zone of an inflorescence meristem in a spiral manner similar to leaf formation from a vegetative meristem. In the second phase, a floral meristem, generated in the flower primordium, sets a whorled arrangement of floral organ primordia; the center of the organogenesis is the floral meristem, not the inflorescence meristem. Therefore, a flower primordium could be considered to have acquired independence from the inflorescence meristem.
We noticed that the timing of the shift of the center of organogenesis coincides with the temporal disappearance of the PRS expression in a flower primordium (Fig. 3G). PRS was expressed first in a flower primordium of early stage 1 at the lateral sides relative to the inflorescence meristem. Obviously, the position of the PRS-expressing cells is controlled by the positional relationship with the inflorescence meristem. PRS expression disappears at late stage 1 but reappears at stage 3 at four regions where sepal primordia are formed. The expression continues at the lateral marginal regions of the developing sepal primordia. Apart from the carpels, a similar expression pattern is observed in other floral organs. The lateral position in the floral organs can be determined against the center of the floral meristem. It would be reasonable to assume that the pivoting center determining the regions of PRS expression has shifted from the inflorescence meristem to the floral meristem after the temporal disappearance. Therefore, it is strongly suggested that the floral meristem acquires independence from the inflorescence meristem between early stage 1 and late stage 2 and that the positional signals from the inflorescence meristem is replaced by the signals from the developing floral meristem after late stage 2.
PRS contributes to the formation of flower architecture and lateral sepals
In a flower, floral organs are formed on a receptacle located at the top of a peduncle. In the mutants of YABs (FIL and YAB3) and REV, flowers are transformed to filamentous structures, which lack the receptacle and the floral organs (Talbert et al. 1995; Sawa et al. 1999; Siegfried et al. 1999). The abaxial–adaxial polarity is not observed in the filamentous structures, which indicates that the loss of determinants responsible for either the abaxial or the adaxial side of flower primordia results in the loss of the receptacle and floral organs. This suggests that the determination of the abaxial–adaxial axis at very early stages is required for the normal development of flowers. A flower primordium develops to a flower with a receptacle in the prs single mutant. Our preliminary data show that flower primordia are converted to filaments without receptacles in double mutants of prs fil and prs rev. This suggests that PRS supports the function of FIL and REV at early stages of flower development.
We found that the first expression of PRS was restricted to a few L1 cells at the lateral regions in the flower primordia. In the prs mutant, growth of lateral sepals is inhibited, and in the extreme case, the sepal is completely lost. These results suggest that the PRS expressing cells work as founder cells of lateral sepals or they promote the neighboring cells to become founder cells. A previous report of sector boundary analysis showed the numbers of founder cells giving rise to one sepal could be eight (Bossinger and Smyth 1996), a number similar to the PRS-expressing cells. However, analysis of sepal development using layer-specific markers showed that the major part of sepals is made of L1 and L2 layers and that the vascular is from L3 (Jenik and Irish 2000). In addition, microscopic analysis showed that the formation of sepal primordia initiates from periclinal divisions in the subepidermal cells of L2 layer of flower primordia (Hill and Lord 1989). These results show that sepal development requires cell proliferation of the L2 layer, as well as the L1 layer. Therefore, it is important to see if PRS expression in L1 cells would promote the periclinal cell divisions of L2 cells in a non-cell autonomous manner to promote the formation of lateral sepal primordia. As for the abaxial and adaxial sepals, we can postulate that some unknown gene(s) has a similar function to PRS in triggering the development of sepal primordia.
In this study, we have shown that PRS contributes to flower development in a lateral axis-dependent manner. There are several mutants whose phenotypes are likely to be based on the lateral axis-dependent developmental mechanism. In some alleles of ap2 floral homeotic mutants of Arabidopsis, the abaxial and adaxial organs in the first whorl are transformed to carpel-like structures, but the lateral organs remained as normal sepals (Komaki et al. 1988; Kunst et al. 1989). Although AP2 is expressed uniformly in floral tissue (Jofuku et al. 1994), the mutant phenotype suggests that the function of AP2 is controlled by an axis-dependent mechanism. In Antirrhinum, a flower of the incomposita mutant has petaloid sepals only in the lateral positions of the first whorl and has normal sepals in the abaxial and the adaxial positions (Wilkinson et al. 2000). It would be interesting to examine whether the lateral axis-dependent developmental mechanism is conserved in Arabidopsis, Antirrhinum, and other plant species.
PRS expression and function present the activity of the marginal meristem
The number of cells expressing PRS at the lateral edges of sepal primordia is almost the same at early and later stages of flower development. As shown in continuous transverse sections, the PRS-expressing cells form one or two cell files in the L1 layer at the margin of the sepals (Fig. 5C,D). Similar arched alignment of the PRS-expressing cell files is observed in other floral organs and in leaves. It is interesting that the number of PRS-expressing cells does not increase during organ development. One explanation is that the lateral edge cells expressing PRS divide anticlinally to extend the cell files but do not divide periclinally. In this case, daughter cells of lateral edge cells will express PRS equally. Another explanation is that when the PRS-expressing cells divide periclinally the expression of the PRS gene can occur in one of the daughter cells located at the margin. It is interesting that a similar mechanism is considered to select cells expressing WUS in the shoot apical meristem (Mayer et al. 1998). The phenotypes of both the loss-of-function mutant and the 35S:PRS transgenic plant (gain-of-function mutant) suggest that PRS activates the proliferation of marginal cells. According to the cell division pattern in young leaves of Arabidopsis, cell division is temporally activated at the leaf margins of leaf primordia (Donnelly et al. 1999). Because the timing and location of the PRS expression correlates with the temporal activity of the marginal meristem, PRS would be an activator of the marginal meristem. The NARROW SHEATH1 (NS1) and NARROW SHEATH2 (NS2) genes in maize were also required for the development of leaf margins. Mutations in the two genes cause the deletion of lateral domains of leaves (Scanlon 2000). After cloning of the genes, comparing the functions of NS1 and NS2 with those of PRS could lead us to understand whether there is common and/or specific mechanism(s) of the development of leaf margins in the two species.
In this report, we have shown that PRS has a unique function and expression pattern based on the lateral axis. Our findings provide additional evidence that the structural grand design of flower primordia and of lateral organs is based on the abaxial–adaxial and lateral axes. Expression of a set of genes required for region-specific cell growth and differentiation is controlled by some axes-dependent mechanism, possibly common in flower primordia and lateral organs (Fig. 5E–G). Further analyses should focus on the molecular nature of the positional signal transferred from the central meristem to the lateral primordia, on the mechanisms of the axis formation and of the regulatory system of the axis-dependent gene expression, and on the genetic pathway leading to lateral organ development.
Material and methods
Plant growth conditions
Seeds were sown on the surface of vermiculite in small pots and incubated for three days at 4°C. Plants were grown in a laboratory at 22°C and under continuous illumination of 50 to 100 μE/m2 per sec.
Scanning electron microscopy
For scanning electron microscopy, flowers and inflorescences of the mutants and the transgenic plants were fixed in a carnoa liquid mixture (isoamyl acetate/ethanol, 1:3) overnight. The samples were rinsed twice with ethanol, incubated in an ethanol/isoamyl acetate (1:3) mixture for 15 min, then immersed in isoamyl acetate for 15 min, and dried in liquid carbon dioxide. After trimming, the samples were mounted on scanning electron microscopy stubs, coated with gold, and observed with a scanning electron microscope (JEOL DATUM, JSM-5800LV) at an accelerating voltage of 20 kV. The images were revised with Adobe PhotoShop on a Macintosh computer.
Gene cloning
DNA markers used for positional cloning were based on RFLP (restriction fragment length polymorphism), CAPS (cleaved amplified polymorphic sequence), and SSLP (simple sequence length polymorphism) between the Arabidopsis ecotypes Landsberg erecta and Columbia. Information on the cop1 marker (CAPS) and the CZSOD2 marker (SSLP) were obtained from The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/). We changed the ve014 marker from the RFLP marker to the CAPS marker. Primer sequences are 5′-ACAT TACGTGGAGTAACCTG-3′ and 5′-ACATTCGATACTATC TGGCG-3′, and the enzyme used for digestion is HaeIII. Based on the sequence data from the TIGR Arabidopsis thaliana Database (http://www.tigr.org/tdb/ath1/htmls/ath1.html) and on the RFLPs we found, we made new PCR markers. The T10K5T7 end marker is a CAPS marker; the primer sequences are 5′-TGACAATGACATGTTTCGCG-3′ and 5′-CTACTACAATCT TCAGGAGC-3′; and the enzyme used for digestion is SspI. The nmB marker is a CAPS marker; the primer sequences are 5′-TTCTCTTTCTCTCTCCCGCC-3′ and 5′-AAGACTTGCTAG TTCCTCGG-3′; and the enzyme used for digestion is DraI. The nmF markers is a dCAPS marker (Neff et al. 1998); the primer sequences are 5′-GGTTTATCACCAAACCAGTTTATTG-3′ and 5′-TTGTTTGTTCGGGTCTCTCC-3′; and the enzyme used for digestion is BstXI. The nmG marker is a CAPS marker; the primer sequences are 5′-GGACAGGTAAGAGACAGTAG 3′ and 5′-AGAAGAGGAGCGTGTATGCT-3′; and the enzyme used for digestion is EcoRV.
A genomic library of Arabidopsis (ecotype Columbia) in λFIX II (provided from H. Kaya, Kyoto University, Japan) was screened with a probe of a PCR-amplified DNA fragment, including the ORF of the PRS gene. cDNA cloning was performed by both 5′-RACE and 3′-RACE using the SMART RACE cDNA Amplification Kit (Clontech) and total RNA from inflorescences of the Columbia wild type.
Sequencing
PCR-amplified DNA fragments and subcloned inserts were sequenced with ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kits and an ABI Prism 310 and 377 from Perkin Elmer.
Complementation test
Three genomic clones—1 (∼7 kb), 2 (∼5.4 kb), and 6 (∼9 kb)—were subcloned into a binary vector pPZP211 for the complementation test (Hajdukiewicz et al. 1994). We transformed prs by a vacuum infiltration procedure with the Agrobacterium strain C58C1. Transgenic plants were selected on an agar medium containing a 30 μg/mL kanamycin and 100 μg/mL carbenicilin.
Particle bombardment
The full length of PRS cDNA was ligated into a plasmid pBI221 (Clontech) to construct PRS-GUS fusion protein. For the transient expression of the 35S:PRS–GUS chimeric gene in onion epidermal cells, we used a Biolistic PDS-1000/He Particle Delivery System (Bio-Rad). Samples were stained with X-Gluc solution (5.7 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronide, 1.5 mM K3Fe(CN)6, 1.5 mM K4Fe(CN)6, 0.9% TritonX-100) and DAPI (1 μg/mL).
In situ hybridization
We modified the protocol used in Kathy Barton's laboratory (http://www.wisc.edu/genetics/CATG/barton/protocols.html) to conduct nonradioactive in situ hybridization. Inflorescences were fixed in FAA solution (50% ethanol, 5% acetic acid, 3.7% formaldehyde) for 3–4 h at room temperature, and seedlings were fixed in 4% paraformaldehyde overnight at 4°C. Sections were 7 μm thick, and antisense and sense probes used for in situ hybridization were prepared by subcloning a part of PRS cDNA (corresponding to the sequence at 205–856) into pBluescript SK (Stratagene). Fluorescein RNA Labeling Mix (Roche) was used for labeling. After hybridization at 50 °C, sections were washed three times for 10 min at 50°C in 4× SSPE and 5 mM DTT. After the first wash, the sections were treated with RNase A (20 μg/mL) for 30 min at 37°C and washed in RNase buffer (0.5 M NaCl, 10 mM Tris-HCl at pH 7.5, 1 mM EDTA) for 15 min at 37 °C three times. The final wash was performed twice, for 20 min at 50°C in 0.5× SSPE and 5 mM DTT. Anti-Fluorescein-AP (Roche) and NBT/BCIP stock solution (Roche) were used for fluorescein immunological detection.
35S:PRS transgenic plants
For the experiments of over expression, full-length cDNA of PRS was first ligated into pBI221 digested by XbaI and SacI, lacking the GUS gene, and the EcoRI/HindIII fragment, including 35S:PRS, was ligated into a binary vector, pPZP211. We transformed the Columbia wild type by a vacuum infiltration procedure with the Agrobacterium strain C58C1. We selected the transgenic plants and analyzed their phenotypes. The condition of selection was the same as for the complementation test.
Acknowledgments
We thank Koji Goto, Takashi Araki, Tadashi Uemura, and members of Okada's laboratory for meaningful discussions; Hidetaka Kaya for providing the genomic library; Ohio State University Arabidopsis Biological Resource Center; Arabidopsis Information Resource (TAIR) (http://www.arabidopsis.org) for providing BAC clones, mapping data, sequence data, or molecular markers for gene cloning; and Ryuji Tsugeki for critically reviewing the manuscript. This work was funded by grants from the program Grants-in-Aid for Scientific Research on Priority Areas (No. 10182101) of the Japanese Ministry of Education, Culture, Sports, Science and Technology of Japan (MIXT) and grants from the Human Frontier Science Program and the Mitsubishi Foundation to K.O., and by a fund (9570) from the Japan Society for the Promotion of Science to N.M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
1E-MAIL kiyo@ok-lab.bot.kyoto-u.ac.jp; FAX 81-75-753-4257.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.931001.
References
- Abe M, Takahashi T, Komeda Y. Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 2001;26:487–494. doi: 10.1046/j.1365-313x.2001.01047.x. [DOI] [PubMed] [Google Scholar]
- Bossinger G, Smyth DR. Initiation patterns of flower and floral organ development in Arabidopsis thaliana. Development. 1996;122:1093–1102. doi: 10.1242/dev.122.4.1093. [DOI] [PubMed] [Google Scholar]
- Bowman JL. Morphology of the expanded first leaves. In: Bowman JL, editor. Arabidopsis: An atlas of morphology and development. New York, NY: Springer-Verlag; 1994. pp. 38–39. [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993;119:397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol. 1999;215:407–419. doi: 10.1006/dbio.1999.9443. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Hill JP, Lord EM. Floral development in Arabidopsis thaliana: A comparison of the wild type and the homeotic pistillata mutant. Can J Bot. 1989;67:2922–2936. [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- Jenik PD, Irish VF. Regulation of cell proliferation patterns by homeotic genes during Arabidopsis floral development. Development. 2000;127:1267–1276. doi: 10.1242/dev.127.6.1267. [DOI] [PubMed] [Google Scholar]
- Jofuku KD, den Ber BGW, Van Montagu M, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki MK, Okada K, Nishino E, Shimura Y. Isolation and characterization of novel mutants of Arabidopsis thaliana defective in flower development. Development. 1988;104:195–203. [Google Scholar]
- Kunst L, Klenz JE, Martinez-Zapater J, Haughn GW. AP2 gene determines the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell. 1989;1:1195–1208. doi: 10.1105/tpc.1.12.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KFX, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Barton MK. Leaf polarity and meristem formation in Arabidopsis. Development. 1998;125:2935–2942. doi: 10.1242/dev.125.15.2935. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- Neff MM, Neff JD, Chory J, Pepper AE. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. REVOLUTA regulates meristem initiation at lateral positions. Plant J. 2001;25:223–236. doi: 10.1046/j.1365-313x.2001.00959.x. [DOI] [PubMed] [Google Scholar]
- Sawa S, Watanabe K, Goto K, Liu Y-G, Shibata D, Kanaya E, Morita EH, Okada K. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes & Dev. 1999;13:1079–1088. doi: 10.1101/gad.13.9.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon MJ. NARROW SHEATH1 functions from two meristematic foci during founder-cell recruitment in maize leaf development. Development. 2000;127:4573–4585. doi: 10.1242/dev.127.21.4573. [DOI] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development. 1999;126:4117–4128. doi: 10.1242/dev.126.18.4117. [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Adler HT, Parks DW, Comai L. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development. 1995;121:2723–2735. doi: 10.1242/dev.121.9.2723. [DOI] [PubMed] [Google Scholar]
- Wilkinson M, de Andrade Silva E, Zachgo S, Saedler H, Schwarz-Sommer Z. CHORIPETALA and DESPENTEADO: General regulators during plant development and potential floral targets of FIMBRIATA-mediated degradation. Development. 2000;127:3725–3734. doi: 10.1242/dev.127.17.3725. [DOI] [PubMed] [Google Scholar]