Abstract
The arachidonic acid and prostaglandin pathway has been implicated in prostate carcinogenesis, but comprehensive studies of the individual members in this key pathway are lacking. Here, we first conducted a systematic bioinformatic study of the expression of 36 arachidonic acid pathway genes across 9783 human tissue samples. The results showed that the PLA2G7, HPGD, EPHX2, and CYP4F8 genes are highly expressed in prostate cancer. Functional studies using RNA interference in prostate cancer cells indicated that all four genes are also essential for cell growth and survival. Clinical validation confirmed high PLA2G7 expression, especially in ERG oncogene-positive prostate cancers, and its silencing sensitized ERG-positive prostate cancer cells to oxidative stress. HPGD was highly expressed in androgen receptor (AR)-overexpressing advanced tumors, as well as in metastatic prostate cancers. EPHX2 mRNA correlated with AR in primary prostate cancers, and its inhibition in vitro reduced AR signaling and potentiated the effect of antiandrogen flutamide in cultured prostate cancer cells. In summary, we identified four novel putative therapeutic targets with biomarker potential for different subtypes of prostate cancer. In addition, our results indicate that inhibition of these enzymes may be particularly powerful when combined with other treatments, such as androgen deprivation or induction of oxidative stress.
Androgen deprivation has remained one of the main therapeutic options for prostate cancer; however, hormonal therapy is not curative, often resulting in the development of castration-resistant prostate cancer. Such recurrent and often metastatic tumors remain virtually impossible to treat with current medications.1 Recent studies indicate that prostate tumors may adapt to the reduced levels of testosterone by acquiring hypersensitivity to low steroid levels [eg, by mutations or amplifications of the androgen receptor (AR)], as well as by increased intracrine synthesis of androgens.2–5 Novel drugs targeting de novo intratumoral steroid synthesis are under development to increase the efficacy of hormonal treatments.6 Nonetheless, in addition to these therapies, rationally designed novel therapeutic approaches are needed.
The arachidonic acid (AA) pathway, a key inflammatory pathway involved in cellular signaling, is implicated in prostate carcinogenesis.7 Arachidonic acid is stored in cell membranes, but on stimulation it is mobilized by phospholipase A2 (PLA2) and is converted to various biologically active eicosanoids by cyclooxygenases (COXs), lipoxygenases (LOXs), or P450 cytochromes (CYP). The rate of AA turnover in prostate cancer cells is 10-fold enhanced, compared with the surrounding normal prostate epithelial cells,8 and AA, as well as many eicosanoids, induces prostate cancer proliferation in vitro.7,9,10 The specific molecular mechanisms involved in this process and the role of individual genes along the AA pathway remain poorly understood. Recently, AA synthesis has been shown to induce androgen production in steroid-starved prostate cancer cells, suggesting a contribution also to the activation of AR in castration-resistant prostate cancer progression.11 Previously, prostate carcinogenesis and cancer growth have been linked mostly to aberrant AA metabolism through COX and LOX pathways.12–14 Widely used COX-2 inhibitors suppress the growth of prostate cancer cells in vitro and tumorigenesis in vivo.15–17 However, because of cardiovascular adverse effects, the use of COX-2 inhibitors as cancer drugs raises safety concerns.18 In addition, inhibitors of the ALOX5 (arachidonate 5-lipoxygenase, or 5-LO) and secretory phospholipase A2 (sPLA2) proteins have been shown to reduce prostate cancer cell proliferation in vitro,7,10,19,20 although these inhibitors have not yet progressed to clinical trials in prostate cancer.
The AA pathway is a promising area for translational research, because many targets along this pathway have been already intensively investigated in other indications, such as cardiovascular diseases and pain, providing an opportunity for repositioning of drugs already in clinical development to new indications. Furthermore, understanding the roles of different downstream pathways and individual enzymes in AA metabolism may provide more effective therapeutic opportunities with fewer adverse effects.14 In the present study, we applied bioinformatics to systematically explore the expression patterns of 36 key AA pathway members in vivo, and then performed targeted clinical validation and functional siRNA knockdown studies of the seven most prostate cancer-specific AA pathway genes. The results point to four previously undescribed potential therapeutic targets in prostate cancer: PLA2G7, HPGD, EPHX2, and CYP4F8.
Materials and Methods
In Silico Data Mining and Gene Selection Criteria for Functional Studies
A list of 36 central AA pathway members was collected from multiple studies on AA pathway in prostate cancer.7,21–23 The GeneSapiens database24 was then applied to bioinformatically explore the gene expression levels across 9783 human tissue samples. Briefly, GeneSapiens (http://www.genesapiens.org/) is a collection of 9873 Affymetrix microarray experiments. All data are reannotated and normalized with a custom algorithm. The data are collected from various publicly available sources, including Gene Expression Omnibus and ArrayExpress. The data cover 175 different tissue types.24 Mean expression of each AA pathway gene was determined in prostate cancer (n = 329), healthy prostate (n = 147), and all normal tissue samples (n = 1626). Genes with (1) significantly higher expression in prostate cancer compared with normal prostate (fold change FC > 2, P < 0.001) and (2) genes showing high expression (FC > 1.5 and P << 0.001 or FC > 2 and P < 0.001) in prostate cancer compared with mean expression across all healthy tissues were selected for further studies.
Clinical Material
Primary prostate cancer samples derived from total prostatectomy patients (n = 33, see Supplemental Table S1 at http://ajp.amjpathol.org), nonmalignant samples with normal histology (n = 17) and hyperplastic histology (benign prostatic hyperplasia) (n = 5) and tissue microarrays containing metastatic prostate cancer samples (n = 103) from 62 patients were obtained from the Department of Pathology at Turku University Hospital. The nonmalignant samples used for immunohistochemistry were from patients aged 49–86 years (mean, 70.6 years). The 19 advanced prostate tumor samples used in quantitative reverse transcription PCR (qRT-PCR) have been described previously.25 All tissue samples were used according to contemporary regulatory guidelines.
qRT-PCR
Gene expression in clinical samples and siRNA-induced target gene silencing were validated with qRT-PCR. For the primary prostate cancer tissue samples (n = 33) obtained from the Department of Pathology at Turku University Hospital, frozen tissue blocks were sectioned and hematoxylin and eosin staining was used for confirmation and localization of cancerous tissue. Skin biopsy equipment was then used to collect cancer samples for RNA extractions. The histology of the three normal prostate tissue samples was also confirmed to be free of any pathological alteration. RNA samples extracted with an RNeasy mini kit (Qiagen, Valencia, CA) were reverse-transcribed to cDNA (high capacity cDNA reverse transcription kit; Applied Biosystems, Foster City, CA) and TaqMan qRT-PCR was performed with an Applied Biosystems 7900HT instrument (Finnish DNA Microarray Centre, Turku Centre For Biotechnology, University of Turku, Finland). The primers and probes used are listed in Table 1. The results were analyzed with the manufacturer's software packages (sequence detection system SDS 2.3 with RQ relative quantification software; Applied Biosystems). β-Actin was used as an endogenous control. In clinical samples, relative mRNA expression for each gene in the normal control tissue samples (n = 3) was set as 1 (mean relative expression).
Table 1.
TaqMan qRT-PCR Primers and Probes Designed Using Roche Universal ProbeLibrary Assay Design Center and Used to Validate Target Gene Silencing by siRNAs and mRNA Expression in Clinical Prostate Cancer Samples
| Gene | Forward primer | Reverse primer | Probe |
|---|---|---|---|
| ALOX15B | 5′-TGAGGTCTTCACCCTGGCTA-3′ | 5′-TTGATGTGCAGGGTGTATCG-3′ | 43 |
| AR | 5′-GCCTTGCTCTCTAGCCTCAA-3′ | 5′-GTCGTCCACGTGTAAGTTGC-3′ | 14 |
| CYP4F8 | 5′-CATCTTCAGCTTTGACAGCAA-3′ | 5′-TGAGCTCCATGATCGCAGTA-3′ | 2 |
| EPHX2 | 5′-TTCTGCTGGACACCCTGAA-3′ | 5′-TTCAGATTAGCCCCGATGTC-3′ | 45 |
| ERG | 5′-CAGGTGAATGGCTCAAGGA-3′ | 5′-AGTTCATCCCAACGGTGTCT-3′ | 44 |
| FAAH | 5′-CTCTGCTGCCAAGGCTGT-3′ | 5′-TGCAGTTCCCAGAGTTTTCC-3′ | 73 |
| HPGD | 5′-TGGTCAATAATGCTGGAGTGA-3′ | 5′-GGTTCCACTGATAACAGAAACCA-3′ | 48 |
| KIF11 | 5′-CATCCAGGTGGTGGTGAGAT-3′ | 5′-TATTGAATGGGCGCTAGCTT-3′ | 53 |
| MT1X | 5′-CTTCTCCTTGCCTCGAAATG-3′ | 5′-ACAGGCACAGGAGCCAAC-3′ | 15 |
| MT2A | 5′-CTAGCCGCCTCTTCAGCA-3′ | 5′-GCAGGTGCAGGAGTCACC-3′ | 68 |
| PLA2G2A | 5′-ACCTGCCCTGTCTCCAAAC-3′ | 5′-TTTGTTCTGCACTCCTGCTC-3′ | 32 |
| PLA2G7 | 5′-TGGCTCTACCTTAGAACCCTGA-3′ | 5′-TTTTGCTCTTTGCCGTACCT-3′ | 63 |
| PLK1 | 5′-CACAGTGTCAATGCCTCCA-3′ | 5′-TTGCTGACCCAGAAGATGG-3′ | 30 |
The results are presented as means ± SD. Statistical analyses were performed using Student's t-test (P < 0.05, P < 0.01, and P < 0.001) and Pearson's correlation coefficient, unless otherwise indicated.
Immunohistochemistry
Acetone-fixed frozen sections (6 μm) of primary prostate cancer samples were dried and endogenous peroxidase activity was blocked using 0.3% hydrogen peroxide (H2O2). Goat serum was used to prevent unspecific staining. The slides were incubated with primary antibodies affinity-purified IgG to human PLA2G7 protein (1:200; Cayman Chemical, Ann Arbor, MI) or to HPGD (1:400; PA005679; Sigma-Aldrich, St. Louis, MO) at 4°C overnight. After Tris-buffered saline washes, the slides were incubated with biotinylated goat anti-rabbit secondary antibody (1:200; Vectastain; Vector Laboratories, Burlingame, CA), followed by Vectastain ABC reagent incubation and diaminobenzidine staining. Mayer's hematoxylin was used in counterstaining.
The paraffin-mounted metastatic prostate cancer tissue microarrays and the paraffin-mounted tissue samples containing histologically normal (n = 14) and hyperplastic (n = 5) prostate were stained using a Lab Vision autostainer (Thermo Fisher Scientific, Fremont, CA) and PowerVision+ Poly-HRP immunohistochemistry detection system kit reagents (ImmunoVision Technologies, Burlingame, CA). Samples were deparaffinized and antigen retrieval was performed in Tris-buffered saline (Dako Target retrieval solution, pH 9; Dako, Glostrup, Denmark) using microwaving for 2 × 7 minutes. Endogenous peroxidase activity was blocked with 3% H2O2, and slides were incubated with dilution buffer (Dako) for 10 minutes before incubation with the primary antibodies (1:200–1:400) for 1 hour. After washing and 20 minutes of PowerVision post-blocking, slides were incubated with PowerVision Poly-HRP anti-rabbit IgG for 30 minutes and with diaminobenzidine for 10 minutes. Mayer's hematoxylin was used in counterstaining, and slides were coated with coverslips after ethanol series and xylene. Protein expression in each cancer sample was graded into four groups, based on staining intensity (−, +, ++, and +++).
An Olympus BX50 microscope (Olympus, Tokyo, Japan), Nikon ACT-1 software version 2.62 (Nikon, Tokyo, Japan), and a digital camera (DXM1200; A.G. Heinze, Lake Forest, CA) were used in the photographing.
Cell Culture
LNCaP androgen-sensitive human prostate adenocarcinoma cells26 were provided by Dr. Marco Cecchini (University of Bern, Switzerland) and were grown in T-Medium (Invitrogen, Carlsbad, CA). VCaP vertebral prostate cancer cells27 were received from Drs. Adrie van Bokhoven (University Medical Center, Nijmegen, Netherlands) and Kenneth Pienta (University of Michigan, MI) and were maintained in RPMI-1640.
RNAi Experiments
For the RNAi studies, siRNAs (HP GenomeWide; Qiagen) were plated onto 384-well plate (Greiner Bio-One, Frickenhausen, Germany), followed by addition of the transfection agent (siLentFect lipid reagent; Bio-Rad Laboratories, Hercules, CA) in Opti-MEM medium (Invitrogen) and an appropriate quantity of cells (1500–2000 per well). The final siRNA concentration was 13 nmol/L. After 48 hours (VCaP) or 72 hours (LNCaP) incubation, cells were assayed with CellTiter-Blue (Promega, Madison WI) for viability and ApoONE (Promega) for apoptosis according to the manufacturer's instructions. The EnVision multilabel plate reader (Perkin-Elmer, Waltham, MA) was used for signal quantification. The raw results were normalized using B-score,28 and siRNAs reducing cell viability by ≥2 SD from the median of the controls (corresponding to P < 0.05) were considered putative antiproliferative siRNAs.
AllStars negative control (scrambled siRNA; Qiagen) and lipid only were used as negative controls; siRNAs against KIF11 (kinesin family member 11) and PLK1 (polo-like kinase 1) were used as positive controls.
Oxidative Stress Response Analysis
VCaP cells were transfected with siRNAs as described above. At 48 hours after transfection, H2O2 (0, 100, 200, and 400 μmol/L) was added to the cells for 6 hours. Cells were washed and incubated for an additional 24 hours in normal media. Cell viability was determined with CellTiter-Glo (Promega) according to the manufacturer's instructions.
Western Blot Analysis
Western blot analysis was performed to validate the antibodies used in immunohistochemical analysis (see Supplemental Figure S1 at http://ajp.amjpathol.org) and to visualize the possible changes in AR and prostate-specific antigen (PSA) expression in response to target inhibition. Cells were lysed and proteins were separated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane. Antibodies against AR (1:1000, NeoMarkers, Fremont, CA), PSA (1:1000, Dako), platelet activating factor (PAF) acetylhydrolase (encoded by PLA2G7) (1:500, Cayman Chemical), 15-PGDH (encoded by HPGD) (1:500, Sigma-Aldrich), and β-actin (1:5000, mouse-monoclonal, Becton Dickinson, Franklin Lakes, NJ) were used. Signal was detected with 1:4000 dilution of horseradish peroxidase-conjugated secondary antibodies (Invitrogen Molecular Probes, Carlsbad, CA), followed by visualization with an enhanced chemiluminescence reagent (Amersham Biosciences, Piscataway, NJ).
Flutamide Treatment
Anti-androgen flutamide was purchased from Sigma-Aldrich and diluted in ethanol. LNCaP cells were plated and transfected with siRNAs as described above 24 hours before addition of 10 μmol/L flutamide or ethanol control. Cell viability was determined with CellTiter-Glo (Promega) after 48 hours treatment.
Results
Expression Levels of Arachidonic Acid Pathway Genes in Prostate Cancer
We first performed a detailed bioinformatic analysis of the mRNA expression levels for all 36 central AA pathway genes in both prostate cancer and healthy prostate, as well as in all normal tissues present in the GeneSapiens database (Figure 1A and Supplemental Table S2 at http://ajp.amjpathol.org). Six genes (ALOX15B, CYP4F8, EPHX2, FAAH, PLA2G2A, and PLA2G7) were highly expressed in prostate cancer samples, compared with expression levels in the normal tissues studied. ALOX15B, CYP4F8, EPHX2, FAAH, and PLA2G2A showed more prostate-specific than prostate cancer-specific expression, whereas PLA2G7 mRNA levels were clearly elevated in prostate cancer, compared with normal prostate. In addition, HPGD mRNA expression was significantly elevated in a subset of prostate cancer samples, compared with normal prostate, even though the mean expression level was not significantly different in prostate cancer in comparison to all normal tissue types. These seven genes were selected for further studies. The expression profiles for these genes across all 43 healthy and 68 malignant tissue types, as well as in prostate cancer samples compared with normal prostate samples, are presented in Supplemental Figure S2 (available at http://ajp.amjpathol.org). Interestingly, high HPGD expression was associated with metastatic prostate cancer, a finding supported also by independent datasets in Oncomine29 (see Supplemental Figure S3 at http://ajp.amjpathol.org); no such enrichment in advanced disease was found for the other six genes (data not shown).
Figure 1.
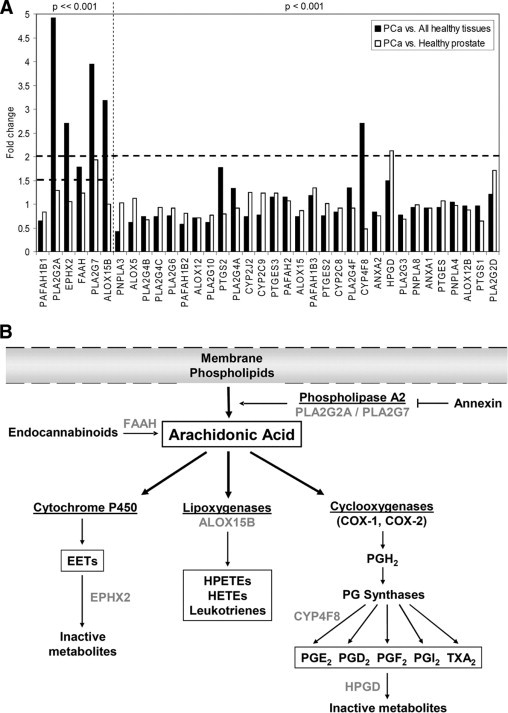
Arachidonic acid (AA) pathway. A:In silico gene expression analysis of 36 central AA pathway genes showing fold changes between mean mRNA expression in prostate cancer (n = 329) and all normal tissue samples (n = 1626) or prostate cancer and healthy prostate tissue samples (n = 147). Sorted in ascending order of the P value. Horizontal dashed lines indicate the fold change limits in the selection criteria. Vertical dashed line separates the different P value limits in the prostate cancer vs. all healthy tissues comparison. B: AA is released from plasma membranes by PLA2 and is converted to various eicosanoids and signaling molecules by COXs, LOXs, or P450 cytochromes. Alternatively, AA can also be hydrolyzed from endocannabinoids by FAAH. The annexins are membrane-binding proteins, some of which are known to inhibit PLA2. AA may be metabolized by various P450 cytochromes to epoxy fatty acids, which are inactivated by EPHX2. Lipoxygenase family consists of three main enzymes differing in their expression profiles and hydroxyeicosatetraenoic acids (HETEs) produced from AA. COXs form thromboxane A2 and different prostaglandins. HPGD is the key enzyme inactivating prostaglandins. Modified from a review article by Patel et al.7 Genes included in the functional experiments are indicated with gray. EET, epoxyeicosatrienoic acid, HPETEs, hydroperoxyeicosatetraenoic acids.
An overview of the roles of the seven genes in the AA cascade is presented in Figure 1B. Three of the genes (PLA2G2A, FAAH, and ALOX15B) have previously been reported to regulate prostate cancer growth. The PLA2G2A protein (group IIA phospholipase A2, a member of the secretory phospholipase A2 family) is a gatekeeper for the pathway releasing AA from the membrane phospholipids. The FAAH protein, fatty-acid amide hydrolase 1, increases AA concentrations by hydrolyzing endocannabinoids. Inhibition of either one of these enzymes is known to decrease prostate cancer cell growth.20,22,31 In contrast, The ALOX15B protein (arachidonate 15-lipoxygenase B) metabolizes AA to 15-hydroxyeicosatetraenoic acid (HETE) and has been described as a functional tumor suppressor in prostate cancer.32
Four other genes (ie, PLA2G7, CYP4F8, EPHX2, and HPGD) have not previously been shown to be functionally involved in prostate cancer and were therefore of special interest. The PLA2G7 protein (platelet-activating factor acetylhydrolase, or PAF-acetylhydrolase; also known as LDL-associated phospholipase 2), is a potent pro- and antiinflammatory enzyme with the ability to degrade PAF and truncated membrane phospholipids generated by oxidative stress.33 Based on enzymatic assays, the CYP4F8 protein (cytochrome P450 4F8) has been proposed to oxygenate and hydroxylate COX-derived products to 19-hydroxy-PGE2 (prostaglandin E2).34 The EPHX2 protein (epoxide hydrolase 2) degrades AA-derived and CYP-produced bioactive epoxy fatty acids and has been suggested as a potential metastasis suppressor gene in breast cancer.35 On the other hand, the HPGD protein [15-hydroxyprostaglandin dehydrogenase (NAD+), or 15-PGDH] inactivates eicosanoids, mainly prostaglandins, and it has also been suggested to be a tumor suppressor.36–39
In Vivo Validation of the Expression of Arachidonic Acid Pathway Genes in Clinical Prostate Cancer Samples
To confirm the expression of PLA2G7, CYP4F8, EPHX2, and HPGD in prostate cancer, qRT-PCR analysis was performed with 33 primary prostate tumors compared with normal prostate (n = 3). For comparison, mRNA levels for the key prostate cancer oncogenes AR and ERG were also determined (Figure 2).
Figure 2.
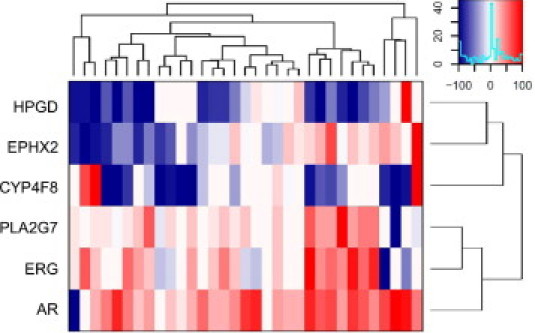
Heatmap visualization of the gene-wise scaled relative mRNA expression values for CYP4F8, EPHX2, HPGD, PLA2G7, ERG, and AR in 33 primary prostate cancer tissues. The heatmap is drawn based on unsupervised hierarchical clustering of the expression values. Relative mean expression level in normal control samples (n = 3) was set as 0.
PLA2G7 mRNA was highly expressed (>3-fold increase) in 24 of 33 (73%) primary tumors, compared with normal prostate (P < 0.001). In addition, PLA2G7 mRNA expression correlated positively (r = 0.66, P < 0.001) with ERG expression (Figure 3A). Immunohistochemical staining of eight primary tumor samples (four ERG-negative and four ERG-positive) confirmed the elevated PLA2G7 protein expression in ERG-positive prostate cancer samples (Figure 3B). Immunohistochemical staining of histologically normal (n = 14) and hyperplastic (n = 5) samples confirmed low PLA2G7 expression in nonmalignant prostate. Weak expression (+) was detected in 68% (n = 13) of samples, and one normal sample showed strong (++) staining intensity, others had no staining. No significant differences were detected between the histologically normal and hyperplastic tissues (Figure 3C). Results from immunohistochemical staining of tissue microarrays containing 80 metastatic prostate cancer samples from 47 patients showed PLA2G7 expression in 33 of 47 (70%) patients and in 56 of 80 (70%) samples. A total of 36 of the 80 (45%) metastatic samples, collected from 20 of the 47 (43%) of the patients, showed strong (++/+++) PLA2G7 protein staining (Figure 3D).
Figure 3.
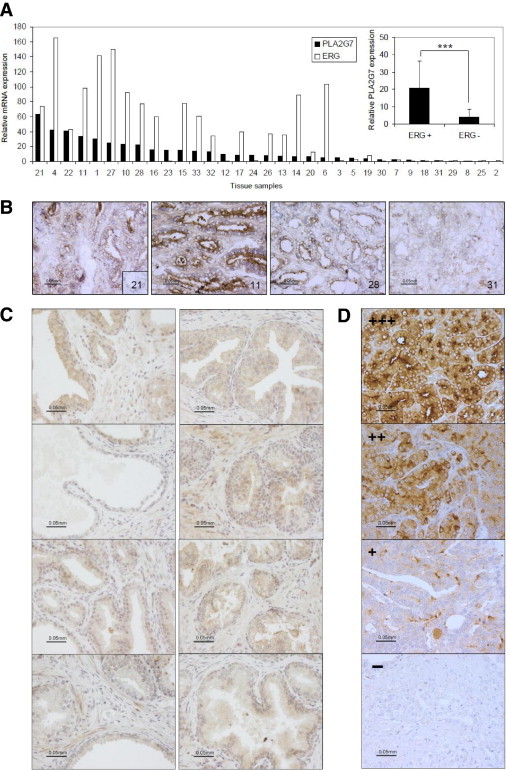
PLA2G7 expression in prostate tissue samples. A: Relative ERG and PLA2G7 mRNA expression in individual primary prostate cancer samples, sorted based on PLA2G7 expression. Inset: PLA2G7 mRNA expression in ERG-positive and -negative tumors, ***P < 0.001. For detailed sample information, see Supplemental Table S1 (http://ajp.amjpathol.org). B: Immunohistochemical results from frozen primary cancer sections show PLA2G7 expression in the cytoplasm and membrane (especially the luminal side) of epithelial prostate cancer cells. The sample number is given on each image. Inset: Negative control staining (no first antibody) of sample 21. C: Immunohistochemical staining of PLA2G7 in histologically normal (left) and hyperplastic (right) prostate tissue samples. D: Immunohistochemical staining of paraffin-mounted tissue microarray sections containing 80 samples from 47 patients with metastatic prostate cancer show PLA2G7 expression in 70% (33/47) of the patients and 70% of samples (56/80). Staining of four individual samples is shown, ranging in intensity from − to +++. The magnification scale is identical for all 16 images.
According to qRT-PCR analysis, CYP4F8 expression was elevated in 10 of the 33 (30%) primary cancer samples. Immunohistochemical staining of CYP4F8 was not performed, for lack of functional and specific antibodies. EPHX2 mRNA was expressed at the same level in both primary cancer and normal prostate samples, in accordance with immunohistochemical staining results published previously.40,41 In addition, EPHX2 mRNA expression was found to correlate significantly (r = 0.43, P = 0.001) with AR expression in our data set.
HPGD was highly expressed in 15% (n = 5) of the primary cancer samples, compared with normal prostate. Interestingly, the prostate tumor sample with highest HPGD expression had high AR mRNA expression (Figures 2 and 4A) and was derived from a patient who had experienced tumor progression (based on PSA elevation) after radical prostatectomy (see Supplemental Table S1 at http://ajp.amjpathol.org). Results from histological analysis indicated that this sample was undifferentiated, and immunohistochemical HPGD staining showed cytoplasmic HPGD expression in the epithelial cancer cells (Figure 4A). We then analyzed a set of 19 advanced prostate cancer samples for HPGD mRNA expression. The results showed high expression in 5 of the 19 (26%) advanced cancer samples, and the statistical analysis confirmed significant difference in HPGD expression between normal tissue and all cancers (P = 0.046), as well as between primary and advanced tumors (P = 0.01; Mann-Whitney U-test) (Figure 4B). Immunohistochemical staining of histologically normal (n = 14) and hyperplastic (n = 5) samples confirmed low HPGD expression in nonmalignant prostate. Weak expression (+) was detected in 7 of the 19 (37%) samples, and one benign prostatic hyperplasia sample showed strong (++) staining intensity. No significant differences were detected between normal and hyperplastic tissues (Figure 4C). Interestingly, immunohistochemical staining results from metastatic prostate cancer samples (77 samples from 47 patients) indicated HPGD expression in as many as 62% of the patients (29/47) and 52% of the samples (40/77). In accordance with the qRT-PCR findings, 11 of 47 (23%) patients and 15 of 77 (19%) samples showed strong (++/+++) HPGD protein staining (Figure 4D).
Figure 4.
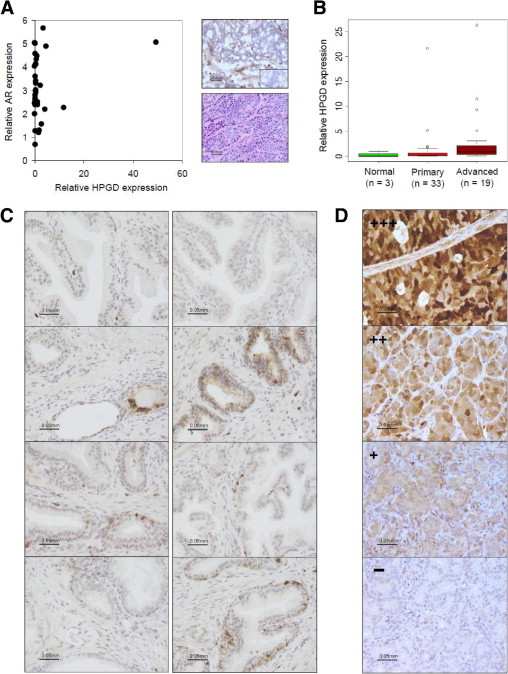
HPGD expression in prostate cancer samples. A: Relative HPGD and AR mRNA expression graphed for primary cancer samples (left). The sample with highest HPGD mRNA expression has undifferentiated histology (right, bottom) and shows cytoplasmic HPGD staining in the cancer cells and weak staining in the stroma (right, top). Inset: Negative control staining (no first antibody). B: Box plot analysis of relative HPGD mRNA expression levels in normal prostate and in primary and advanced prostate cancer samples. The box refers to the quartile distribution range (25% to 75%), with the median indicated by a vertical line. The whiskers indicate the 95% range, and individual outlier samples are also shown. C: Immunohistochemical staining of HPGD in histologically normal (left column) and hyperplastic (right column) prostate tissue samples. D: Immunohistochemical staining of paraffin-mounted tissue microarray sections containing metastatic prostate cancer samples (77 samples from 47 patients) indicate HPGD expression in 62% of the patients (29/47) and 52% of the samples (40/77). Staining shows diffuse and variable cytosolic and nuclear localization of HPGD in the epithelial prostate cancer cells. Staining of four individual samples is shown. Staining of four individual samples is shown, ranging in intensity from − to +++. The magnification scale is identical for all 16 images.
Taken together, the clinical validation showed that PLA2G7, CYP4F8, EPHX2, and HPGD are expressed in clinical prostate cancer samples, thereby confirming the results of the bioinformatic surveys. Furthermore, the analysis points to important differences in their involvement in different types of tumors, with PLA2G7 being highly expressed and correlating with ERG in prostate cancer, whereas HPGD was expressed in a subset of AR-overexpressing advanced and metastatic prostate cancers.
Gene Silencing Revealed a Significant Role for Arachidonic Acid Pathway in Prostate Cancer Cell Growth in Vitro
We conducted gene silencing studies of the seven most promising AA pathway members in prostate cancer cells. siRNA molecules were transfected in VCaP and LNCaP cells, and cell viability and apoptosis were measured as endpoints. The results indicated that all seven genes reduce prostate cancer cell viability in one or both model systems used (Figure 5). The siRNAs for two genes (CYP4F8 and PLA2G2A) reduced cell viability in both cell lines; for four genes (ALOX15B, EPHX2, FAAH, and HPGD), the siRNAs reduced cell viability only in LNCaP cells; for the seventh gene, PLA2G7, siRNA-induced inhibition was seen only in the ERG oncogene-positive VCaP cells (Figure 5A). In addition, a moderate but significant induction of apoptosis was detected in LNCaP cells after ALOX15B, EPHX2, and FAAH silencing and in VCaP cells after PLA2G7 silencing (Figure 5B). The induction of apoptosis in response to PLA2G7 silencing in VCaP cells and EPHX2 silencing in LNCaP cells at the 72-hour time point was confirmed with Western blot analysis of cleaved poly(ADP-ribose) polymerase (cPARP). The results confirmed that both PLA2G7 and EPHX2 silencing increased PARP cleavage, confirming the induction of apoptosis (data not shown). The efficacy of target gene silencing by the siRNAs was validated by qRT-PCR (Figure 5C).
Figure 5.
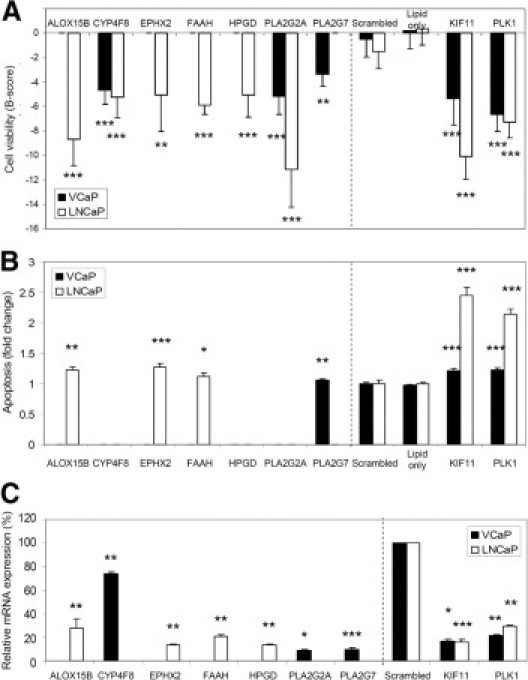
RNAi results. A: The effect of target gene silencing on VCaP and LNCaP cell viability. Mean B-score values from three separate cell viability assays given. Only results exceeding −2 SD from the median are presented. B: Induction of apoptosis after target gene silencing. Data are presented as mean fold change from at least two separate experiments, compared with the scrambled control. Only significant results (P < 0.05) are presented. C: Validation of target gene silencing with qRT-PCR. Relative mRNA expression level in the scrambled control was set as 100%. For the LNCaP-selective antiproliferative siRNAs, results are shown in LNCaP cells; for others, in VCaP cells. The results for the control experiments are shown in both cell lines. The vertical dashed lines separate the results from the selected AA pathway members from the control experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
PLA2G7 Silencing Sensitizes Prostate Cancer Cells to Oxidative Stress
One of the main functions of PLA2G7 is to hydrolyze truncated phospholipids generated by oxidative attack and to participate in the maintenance of membrane integrity.33 A yeast PLA2G7 ortholog has been shown to suppress oxidative death,42 and PLA2G7 expression has been associated with elevated risk of cardiac disease and with high expression of oxidative stress markers.43 We therefore studied the effect of PLA2G7 silencing on the sensitivity to oxidative stress in VCaP cells. PLA2G7 siRNA and H2O2 cotreatment induced a 30% (100 μmol/L, P = 0.01) to 40% (200 μmol/L, P = 0.007) reduction in cell viability, compared with scrambled siRNA and H2O2 cotreated controls, when cell viability was normalized to siRNA-transfected but not H2O2-treated samples (Figure 6A).
Figure 6.
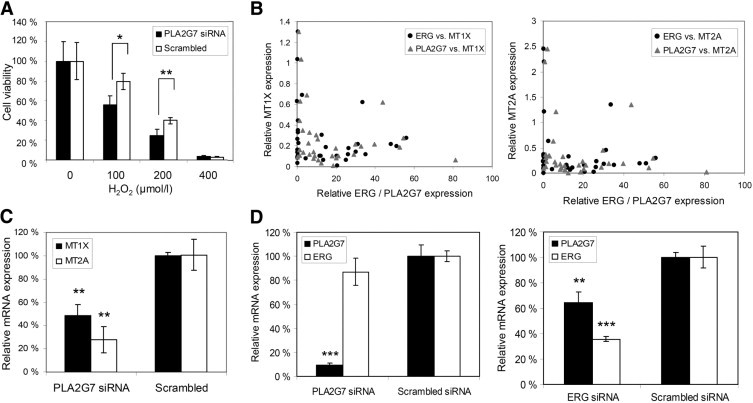
Mechanistic exploration of PLA2G7 inhibition. A: Combinatorial effect of PLA2G7 silencing and oxidative stress on VCaP cell viability. Cell viability in samples not H2O2-treated but siRNA transfected was set as 100%. P values are compared with the scrambled control. B: Relative MT1X/MT2A and PLA2G7/ERG mRNA expression levels in primary prostate cancer samples. The relative mRNA expression for each gene in normal control sample was set as 1. C: Relative MT1X and MT2A mRNA expression levels after PLA2G7 silencing in VCaP cells. Relative mRNA expression level in the scrambled control was set as 100%. D: Relative PLA2G7 and ERG mRNA expression levels in VCaP cells after PLA2G7 and ERG silencing. Relative mRNA expression level in the scrambled control was set as 100%. *P < 0.05, **P < 0.01, ***P < 0.001.
Metallothioneins are known to protect cells from oxidative stress.44 Recent high-throughput compound screening results from our research group showed that ERG-positive VCaP cells are vulnerable to compounds inducing oxidative stress accompanied with induction of metallothionein expression.45 Bioinformatic analysis indicated low metallothionein 1X (MT1X) and 2A (MT2A) expression in ERG- and PLA2G7-positive prostate tumors (data not shown). This finding was confirmed with qRT-PCR in primary prostate cancer samples (Figure 6B). In addition, PLA2G7 silencing in vitro significantly reduced MT1X and MT2A mRNA levels (51.4%, P = 0.007 and 72.1%, P = 0.002 respectively) (Figure 6C). These results indicate that PLA2G7 silencing sensitizes prostate cancer cells to the damage caused by oxidative stress, leading to reduced cell viability.
Because PLA2G7 expression was found to correlate with ERG mRNA expression in clinical prostate cancers and to sensitize ERG-positive prostate cancer cells to oxidative stress, we analyzed whether there is a functional link between these two genes. The qRT-PCR results showed that silencing of the ERG oncogene reduces PLA2G7 mRNA expression significantly (P = 0.007) in ERG-positive VCaP cells, but PLA2G7 silencing does not affect ERG mRNA expression (Figure 6D).
Inhibition of EPHX2 Decreases Androgen Receptor Signaling and Potentiates the Growth Inhibitory Effect of Flutamide in Prostate Cancer Cells
The EPHX2 protein has been recently suggested to regulate testosterone levels in mice.46 Because EPHX2 mRNA was found to significantly correlate with AR expression in clinical prostate cancers, Western blot analysis was performed to analyze the possible effects of EPHX2 silencing on AR and PSA levels. EPHX2 siRNA transfection reduced PSA protein levels to 42%, compared with scrambled siRNA control transfected LNCaP cells, whereas no changes were detected in AR protein expression (Figure 7A). EPHX2 inhibition by RNAi was then tested alone and in combination with the antiandrogen flutamide in LNCaP cells. EPHX2 siRNA transfection alone reduced cell viability by 22.2% (P < 0.001) compared with the scrambled siRNA transfected control cells. Combinatorial treatment with EPHX2 siRNA and flutamide reduced cell viability by 35.0% (P < 0.001) compared with the respective control samples treated with scrambled siRNA and flutamide (Figure 7B). These results indicate that EPHX2 silencing potentiates (P = 0.002) the growth-inhibitory effect of flutamide in prostate cancer cells.
Figure 7.
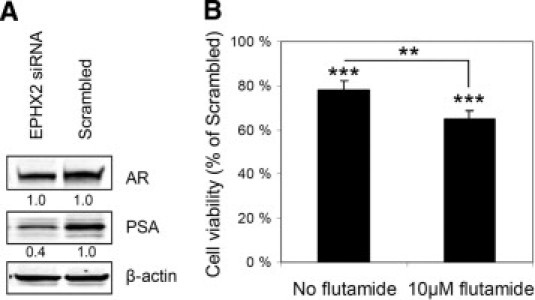
EPHX2 and AR signaling. A: Western blot analysis of AR and PSA protein levels after siRNA-induced EPHX2 gene silencing. β-Actin was used as a loading control. Relative value for each blot is indicated; relative expression in control sample was set as 1. B: Combinatorial treatment of LNCaP cells with EPHX2 siRNA and flutamide. Cell viability of EPHX2 siRNA-treated cells is shown. Cell viability in the scrambled control sample was set as 100% in both treatments (no flutamide or 10 μmol/L flutamide). P values for individual treatments are given, compared with the scrambled control. P value for the difference in cell viability in EPHX2 siRNA treated samples after different treatments is also indicated. **P < 0.01, ***P < 0.001.
Discussion
We decided to systematically explore the role of the arachidonic acid pathway in prostate cancer by combining gene expression data from clinical specimens with RNAi knockdown studies in cultured prostate cancer cells. Combination of the results from in vivo and in vitro studies implicated PLA2G7, CYP4F8, EPHX2, and HPGD as putative novel therapeutic targets for prostate cancer. Even though all four genes are critical for prostate cancer cell viability, we found that their expression profiles in clinical specimens and the consequences of in vitro silencing were highly different.
The enzyme encoded by PLA2G7 is a member of the arachidonic acid releasing PLA2 family. High PLA2G7 expression has been previously associated with ERG-positive prostate tumors in silico.25 Here we confirm PLA2G7 overexpression in a majority of clinical prostate tumors, especially in ERG oncogene-positive prostate cancers, compared with histologically normal and hyperplastic prostate tissues. In addition, silencing of ERG reduced PLA2G7 mRNA expression in ERG-positive prostate cancer cells, supporting a functional link between these two genes. Furthermore, PLA2G7 expression was required for ERG-positive prostate cancer cell viability. Interestingly, ERG oncogene-positive prostate cancer samples were found to express low levels of metallothioneins, known to protect cells against oxidative stress. This supports recent high-throughput compound screening results from our research group,45 suggesting that ERG-positive prostate cancers are vulnerable to inducers of oxidative stress. As noted above, PLA2G7 ortholog has been shown to protect yeast cells from oxidative stress.42 The present results indicated that PLA2G7 silencing further reduced the expression of the protective metallothionein genes and also sensitized ERG-positive prostate cancer cells to oxidative stress. Taken together, these results suggest that PLA2G7 could have utility as a biomarker and therapeutic target, especially in ERG-positive prostate cancers.
CYP4F8 silencing significantly reduced cell viability in prostate cancer cells, even with a partial knockdown of CYP4F8 expression. The protein encoded by CYP4F8 is known to oxygenate and hydroxylate COX-derived products to 19-hydroxy-PGE2.34 In contrast to PGE2, which may indiscriminately stimulate a variety of prostanoid receptor subtypes, 19-hydroxy-PGE2 has been found to exhibit selectivity for the EP2 receptor subtype.47 In prostate cancer, the EP2 receptor has been shown to induce vascular endothelial growth factor (VEGF) secretion, cell motility, growth, and angiogenesis on binding to PGE2.23,48 Even though the mechanism of CYP4F8 inhibition-induced growth reduction in prostate cancer requires further study, the prostate-specific expression profile of CYP4F8 and EP2 receptor activation by 19-hydroxy-PGE2 suggests that CYP4F8 inhibition could be an attractive therapeutic alternative to COX-2 inhibition (which is associated with cardiovascular adverse effects).
The EPHX2 protein is a bifunctional enzyme harboring epoxide hydrolase and phosphatase domains, both with different biological functions.49 In the AA pathway, this protein degrades bioactive epoxy fatty acids derived from arachidonic acid by CYP metabolism. EPHX2 has also been associated with androgen signaling,50 and recently it was suggested to regulate testosterone levels in mice through cholesterol biosynthesis and metabolism.46 In the present study, EPHX2 expression was shown to correlate with androgen receptor mRNA in clinical primary prostate tumor samples. In addition, EPHX2 silencing induced apoptosis in prostate cancer cells, as well as reducing cell viability and androgen receptor signaling. Furthermore, EPHX2 siRNA transfection potentiated the antiproliferative effect of antiandrogen flutamide, indicating a putative additive therapeutic value in prostate cancer treatment and also confirming a functional role of EPHX2 in androgen receptor signaling. These results are in accordance with the previous literature describing EPHX2 as a regulator of testosterone metabolism. Interestingly, EPHX2-null mice are reported to be fertile and healthy,46 further suggesting that EPHX2 is a safe and attractive drug target.
The HPGD protein is the key enzyme inactivating eicosanoids, mainly prostaglandins. In our study, HPGD was highly expressed in a subset of androgen receptor-overexpressing advanced and metastatic prostate tumors, indicating potential relevance in this subset of typically incurable prostate cancer. Previous results have indicated that HPGD protein is induced by androgens in LNCaP cells.51 We confirmed a clear increase in HPGD protein expression in LNCaP cells in response to androgen stimulation (data not shown). In addition, our results showed a clear dependency of LNCaP cell growth and survival on HPGD expression. HPGD has been reported to function as a potential tumor suppressor in breast cancer, modulating estrogen receptor signaling,36 and decreased expression has been reported also in other cancers.37–39 The results from our bioinformatic exploration of gene expression data support these findings, because the mean HPGD expression in many cancer types was lower than in the corresponding normal tissues. However, multiple cancer types had a small subset of samples expressing very high levels of HPGD, compared with the levels in normal tissues (see Supplemental Figure S2 at http://ajp.amjpathol.org). Our results confirmed a similar pattern of HPGD expression in prostate tissues. Based on the present results, we conclude that high HPGD expression is associated with advanced metastatic disease and that HPGD inhibition may provide therapeutic opportunities, especially in advanced prostate cancer.
As expected, known regulators of prostate cancer growth in the AA pathway were found to affect proliferation. In accordance with earlier findings, the present results support the potential involvement of FAAH in prostate tumorigenesis.22 PLA2G2A expression did not differ much between cancerous and healthy prostate, but the results from RNAi studies support its role in regulating prostate cancer cell growth.20,31,52 ALOX15B has been suggested as a potential tumor suppressor whose function may be lost in prostate cancer.32 It has been reported to regulate human prostate epithelial cell differentiation, senescence, and growth in vitro and tumor development in vivo.32,53,54 Our bioinformatic analyses indicated that ALOX15B mRNA is highly expressed in both healthy and malignant prostates, and qRT-PCR results confirmed expression also in LNCaP prostate cancer cells. ALOX15B silencing clearly decreased cell viability and increased apoptosis in LNCaP cells.
Taken together, the present results highlight the significance of the arachidonic acid pathway in prostate cancer cell growth regulation. Although the mechanisms inducing the changes observed after target gene silencing are most likely diverse, inhibition of this metabolic signaling cascade, or the balance between different branches of the pathway, appears to affect the growth and survival of prostate cancer cells. We identified four novel putative therapeutic targets with biomarker potential for different subtypes of prostate cancer. PLA2G7 appears to be the most relevant in ERG oncogene-positive prostate tumors, whereas HPGD is associated with advanced metastatic disease. Although further validation is needed to confirm the frequency of expression of these novel targets in prostate cancer, the results from our clinical data suggest that targeting only PLA2G7 and HPGD would cover the majority of prostate cancers. Finally, as shown by our in vitro results, inhibition of EPHX2 and PLA2G7 may reduce prostate cancer cell viability even more effectively when combined with other treatments, such as androgen deprivation or induction of oxidative stress.
Acknowledgments
We thank Niko Sahlberg, Arttu Heinonen, Jouni Lahoniitty, and Rami Mäkelä for technical assistance with high-throughput screening. Special thanks to Elmar Bucher, Marcin Magnus, Tommi Pisto, and Henri Sara for excellent help with the bioinformatics, to Katariina Lassila for assistance in performing qRT-PCR, and to Sinikka Kollanus for immunohistochemical stainings.
Footnotes
Supported by Marie Curie Canceromics (MEXT-CT-2003-2728), EU-PRIMA project (contract no. LSHC-CT-204-504587), TIME collaborative project, Academy of Finland, Cancer Organizations of Finland, and Sigrid Juselius Foundation.
Supplemental material for this article can be found on http://ajp.amjpathol.org and at doi:10.1016/j.ajpath.2010.10.002.
Supplementary data
Validation of antibodies used in immunohistochemical staining. A:PLA2G7 antibody. PLA2G7 siRNA treated cells compared with the scrambled control. B:HPGD antibody. HPGD siRNA treated cells compared with the scrambled control. β-Actin was used as a loading control.
Normalized expression levels of the selected arachidonic acid pathway genes in prostate tissue samples, as well as in all available tissue samples in silico. Box plot analysis of normalized expression levels in healthy and malignant prostate tissue samples in silico is shown in the upper figures. The box refers to the quartile distribution (25 to 75%) range, with the median shown as a vertical line (green in normal samples, red in malignant). In addition, the 95% range and individual outlier samples are shown. Tissue- and disease-wide in silico expression profiles are shown in the lower figures. All samples are represented along the x axis in a predetermined fixed order, first the normal tissues, then cancers and other diseases. The y axis reflects the relative level of gene expression after normalization. Each dot represents the expression of indicated gene in one sample. Anatomical origin of the samples is marked with colored bars and indicated in the end of the figure. Sample types having higher than average expression or an outlier expression profile are additionally colored in each gene plot (legend at the top left corner of each figure).
In silico analysis of HPGD mRNA expression. A.In silico data anlysis of prostate tissue in GeneSapiens database indicate high HPGD expression in metastatic disease. B. Yu et al. study30 in Oncomine29 shows HPGD to be highly expressed in metastatic prostate cancer.
References
- 1.Tannock I.F., de Wit R., Berry W.R., Horti J., Pluzanska A., Chi K.N., Oudard S., Théodore C., James N.D., Turesson I., Rosenthal M.A., Eisenberger M.A., TAX 327 Investigators Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351:1502–1512. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- 2.Holzbeierlein J., Lal P., LaTulippe E., Smith A., Satagopan J., Zhang L., Ryan C., Smith S., Scher H., Scardino P., Reuter V., Gerald W.L. Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen-responsive genes and mechanisms of therapy resistance. Am J Pathol. 2004;164:217–227. doi: 10.1016/S0002-9440(10)63112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pienta K.J., Bradley D. Mechanisms underlying the development of androgen-independent prostate cancer. Clin Cancer Res. 2006;12:1665–1671. doi: 10.1158/1078-0432.CCR-06-0067. [DOI] [PubMed] [Google Scholar]
- 4.Taichman R.S., Loberg R.D., Mehra R., Pienta K.J. The evolving biology and treatment of prostate cancer. J Clin Invest. 2007;117:2351–2361. doi: 10.1172/JCI31791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dillard P.R., Lin M.F., Khan S.A. Androgen-independent prostate cancer cells acquire the complete steroidogenic potential of synthesizing testosterone from cholesterol. Mol Cell Endocrinol. 2008;295:115–120. doi: 10.1016/j.mce.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Attard G., Reid A.H., Olmos D., de Bono J.S. Antitumor activity with CYP17 blockade indicates that castration-resistant prostate cancer frequently remains hormone driven. Cancer Res. 2009;69:4937–4940. doi: 10.1158/0008-5472.CAN-08-4531. [DOI] [PubMed] [Google Scholar]
- 7.Patel M.I., Kurek C., Dong Q. The arachidonic acid pathway and its role in prostate cancer development and progression. J Urol. 2008;179:1668–1675. doi: 10.1016/j.juro.2007.12.037. [DOI] [PubMed] [Google Scholar]
- 8.Chaudry A., McClinton S., Moffat L.E., Wahle K.W. Essential fatty acid distribution in the plasma and tissue phospholipids of patients with benign and malignant prostatic disease. Br J Cancer. 1991;64:1157–1160. doi: 10.1038/bjc.1991.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., Corr J.G., Thaler H.T., Tao Y., Fair W.R., Heston W.D. Decreased growth of established human prostate LNCaP tumors in nude mice fed a low-fat diet. J Natl Cancer Inst. 1995;87:1456–1462. doi: 10.1093/jnci/87.19.1456. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh J., Myers C.E. Arachidonic acid stimulates prostate cancer cell growth: critical role of 5-lipoxygenase. Biochem Biophys Res Commun. 1997;235:418–423. doi: 10.1006/bbrc.1997.6799. [DOI] [PubMed] [Google Scholar]
- 11.Locke J.A., Guns E.S., Lehman M.L., Ettinger S., Zoubeidi A., Lubik A., Margiotti K., Fazli L., Adomat H., Wasan K.M., Gleave M.E., Nelson C.C. Arachidonic acid activation of intratumoral steroid synthesis during prostate cancer progression to castration resistance. Prostate. 2010;70:239–251. doi: 10.1002/pros.21057. [DOI] [PubMed] [Google Scholar]
- 12.Fürstenberger G., Krieg P., Müller-Decker K., Habenicht A.J. What are cyclooxygenases and lipoxygenases doing in the driver's seat of carcinogenesis? Int J Cancer. 2006;119:2247–2254. doi: 10.1002/ijc.22153. [DOI] [PubMed] [Google Scholar]
- 13.Matsuyama M., Yoshimura R. The target of arachidonic acid pathway is a new anticancer strategy for human prostate cancer. Biologics. 2008;2:725–732. doi: 10.2147/btt.s3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D., Dubois R.N. Eicosanoids and cancer. Nat Rev Cancer. 2010;10:181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu A.L., Ching T.T., Wang D.S., Song X., Rangnekar V.M., Chen C.S. The cyclooxygenase-2 inhibitor celecoxib induces apoptosis by blocking Akt activation in human prostate cancer cells independently of Bcl-2. J Biol Chem. 2000;275:11397–11403. doi: 10.1074/jbc.275.15.11397. [DOI] [PubMed] [Google Scholar]
- 16.Narayanan B.A., Narayanan N.K., Pittman B., Reddy B.S. Regression of mouse prostatic intraepithelial neoplasia by nonsteroidal anti-inflammatory drugs in the transgenic adenocarcinoma mouse prostate model. Clin Cancer Res. 2004;10:7727–7737. doi: 10.1158/1078-0432.CCR-04-0732. [DOI] [PubMed] [Google Scholar]
- 17.Patel M.I., Subbaramaiah K., Du B., Chang M., Yang P., Newman R.A., Cordon-Cardo C., Thaler H.T., Dannenberg A.J. Celecoxib inhibits prostate cancer growth: evidence of a cyclooxygenase-2-independent mechanism. Clin Cancer Res. 2005;11:1999–2007. doi: 10.1158/1078-0432.CCR-04-1877. [DOI] [PubMed] [Google Scholar]
- 18.Kearney P.M., Baigent C., Godwin J., Halls H., Emberson J.R., Patrono C. Do selective cyclo-oxygenase-2 inhibitors and traditional non-steroidal anti-inflammatory drugs increase the risk of atherothrombosis?: Meta-analysis of randomised trials. BMJ. 2006;332:1302–1308. doi: 10.1136/bmj.332.7553.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moretti R.M., Montagnani Marelli M., Sala A., Motta M., Limonta P. Activation of the orphan nuclear receptor RORalpha counteracts the proliferative effect of fatty acids on prostate cancer cells: crucial role of 5-lipoxygenase. Int J Cancer. 2004;112:87–93. doi: 10.1002/ijc.20387. [DOI] [PubMed] [Google Scholar]
- 20.Sved P., Scott K.F., McLeod D., King N.J., Singh J., Tsatralis T., Nikolov B., Boulas J., Nallan L., Gelb M.H., Sajinovic M., Graham G.G., Russell P.J., Dong Q. Oncogenic action of secreted phospholipase A2 in prostate cancer. Cancer Res. 2004;64:6934–6940. doi: 10.1158/0008-5472.CAN-03-3018. [DOI] [PubMed] [Google Scholar]
- 21.Chaudry A.A., Wahle K.W., McClinton S., Moffat L.E. Arachidonic acid metabolism in benign and malignant prostatic tissue in vitro: effects of fatty acids and cyclooxygenase inhibitors. Int J Cancer. 1994;57:176–180. doi: 10.1002/ijc.2910570208. [DOI] [PubMed] [Google Scholar]
- 22.Endsley M.P., Thill R., Choudhry I., Williams C.L., Kajdacsy-Balla A., Campbell W.B., Nithipatikom K. Expression and function of fatty acid amide hydrolase in prostate cancer. Int J Cancer. 2008;123:1318–1326. doi: 10.1002/ijc.23674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain S., Chakraborty G., Raja R., Kale S., Kundu G.C. Prostaglandin E2 regulates tumor angiogenesis in prostate cancer. Cancer Res. 2008;68:7750–7759. doi: 10.1158/0008-5472.CAN-07-6689. [DOI] [PubMed] [Google Scholar]
- 24.Kilpinen S., Autio R., Ojala K., Iljin K., Bucher E., Sara H., Pisto T., Saarela M., Skotheim R.I., Björkman M., Mpindi J.P., Haapa-Paananen S., Vainio P., Edgren H., Wolf M., Astola J., Nees M., Hautaniemi S., Kallioniemi O. Systematic bioinformatic analysis of expression levels of 17,330 human genes across 9,783 samples from 175 types of healthy and pathological tissues. Genome Biol. 2008;9:R139. doi: 10.1186/gb-2008-9-9-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iljin K., Wolf M., Edgren H., Gupta S., Kilpinen S., Skotheim R.I., Peltola M., Smit F., Verhaegh G., Schalken J., Nees M., Kallioniemi O. TMPRSS2 fusions with oncogenic ETS factors in prostate cancer involve unbalanced genomic rearrangements and are associated with HDAC1 and epigenetic reprogramming. Cancer Res. 2006;66:10242–10246. doi: 10.1158/0008-5472.CAN-06-1986. [DOI] [PubMed] [Google Scholar]
- 26.Harris S.E., Rong Z., Harris M.A., Lubahn D.B. Androgen receptor in human prostate carcinoma LNCaP/ADEP cells contains a mutation which alters the specificity of the steroid dependent transcriptional activation region. Program and Abstracts, 72nd Annual Meeting of the Endocrine Society, Atlanta, GA. 1990:93. [Google Scholar]
- 27.Korenchuk S., Lehr J.E., MClean L., Lee Y.G., Whitney S., Vessella R., Lin D.L., Pienta K.J. VCaP, a cell-based model system of human prostate cancer. In Vivo. 2001;15:163–168. [PubMed] [Google Scholar]
- 28.Brideau C., Gunter B., Pikounis B., Liaw A. Improved statistical methods for hit selection in high-throughput screening. J Biomol Screen. 2003;6:634–647. doi: 10.1177/1087057103258285. [DOI] [PubMed] [Google Scholar]
- 29.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Y.P., Landsittel D., Jing L., Nelson J., Ren B., Liu L., McDonald C., Thomas R., Dhir R., Finkelstein S., Michalopoulos G., Becich M., Luo J.H. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22(14):2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 31.Dong Q., Patel M., Scott K.F., Graham G.G., Russell P.J., Sved P. Oncogenic action of phospholipase A2 in prostate cancer. Cancer Lett. 2006;240:9–16. doi: 10.1016/j.canlet.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 32.Tang D.G., Bhatia B., Tang S., Schneider-Broussard R. 15-Lipoxygenase 2 (15-LOX2) is a functional tumor suppressor that regulates human prostate epithelial cell differentiation, senescence, and growth (size) Prostaglandins Other Lipid Mediat. 2007;82:135–146. doi: 10.1016/j.prostaglandins.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 33.Stafforini D.M. Biology of platelet-activating factor acetylhydrolase (PAF-AH, lipoprotein associated phospholipase A2) Cardiovasc Drugs Ther. 2009;23:73–83. doi: 10.1007/s10557-008-6133-8. [DOI] [PubMed] [Google Scholar]
- 34.Bylund J., Hidestrand M., Ingelman-Sundberg M., Oliw E.H. Identification of CYP4F8 in human seminal vesicles as a prominent 19-hydroxylase of prostaglandin endoperoxides. J Biol Chem. 2000;275:21844–21849. doi: 10.1074/jbc.M001712200. [DOI] [PubMed] [Google Scholar]
- 35.Thomassen M., Tan Q., Kruse T.A. Gene expression meta-analysis identifies chromosomal regions and candidate genes involved in breast cancer metastasis [Erratum appeared in Breast Cancer Res Treat 2009, 113:251–252] Cancer Res Treat. 2009;113:239–249. doi: 10.1007/s10549-008-9927-2. [DOI] [PubMed] [Google Scholar]
- 36.Wolf I., O'Kelly J., Rubinek T., Tong M., Nguyen A., Lin B.T., Tai H.H., Karlan B.Y., Koeffler H.P. 15-Hydroxyprostaglandin dehydrogenase is a tumor suppressor of human breast cancer. Cancer Res. 2006;66:7818–7823. doi: 10.1158/0008-5472.CAN-05-4368. [DOI] [PubMed] [Google Scholar]
- 37.Huang G., Eisenberg R., Yan M., Monti S., Lawrence E., Fu P., Walbroehl J., Löwenberg E., Golub T., Merchan J., Tenen D.G., Markowitz S.D., Halmos B. 15-Hydroxyprostaglandin dehydrogenase is a target of hepatocyte nuclear factor 3beta and a tumor suppressor in lung cancer. Cancer Res. 2008;68:5040–5048. doi: 10.1158/0008-5472.CAN-07-6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thiel A., Ganesan A., Mrena J., Junnila S., Nykänen A., Hemmes A., Tai H.H., Monni O., Kokkola A., Haglund C., Petrova T.V., Ristimäki A. 15-Hydroxyprostaglandin dehydrogenase is down-regulated in gastric cancer. Clin Cancer Res. 2009;15:4572–4580. doi: 10.1158/1078-0432.CCR-08-2518. [DOI] [PubMed] [Google Scholar]
- 39.Tseng-Rogenski S., Gee J., Ignatoski K.W., Kunju L.P., Bucheit A., Kintner H.J., Morris D., Tallman C., Evron J., Wood C.G., Grossman H.B., Lee C.T., Liebert M. Loss of 15-hydroxyprostaglandin dehydrogenase expression contributes to bladder cancer progression. Am J Pathol. 2010;176:1462–1468. doi: 10.2353/ajpath.2010.090875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Enayetallah A.E., French R.A., Barber M., Grant D.F. Cell-specific subcellular localization of soluble epoxide hydrolase in human tissues. J Histochem Cytochem. 2006;54:329–335. doi: 10.1369/jhc.5A6808.2005. [DOI] [PubMed] [Google Scholar]
- 41.Enayetallah A.E., French R.A., Grant D.F. Distribution of soluble epoxide hydrolase, cytochrome P450 2C8, 2C9 and 2J2 in human malignant neoplasms. J Mol Histol. 2006;37:133–141. doi: 10.1007/s10735-006-9050-9. [DOI] [PubMed] [Google Scholar]
- 42.Foulks J.M., Weyrich A.S., Zimmerman G.A., McIntyre T.M. A yeast PAF acetylhydrolase ortholog suppresses oxidative death. Free Radic Biol Med. 2008;45:434–442. doi: 10.1016/j.freeradbiomed.2008.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J.Y., Hyun Y.J., Jang Y., Lee B.K., Chae J.S., Kim S.E., Yeo H.Y., Jeong T.S., Jeon D.W., Lee J.H. Lipoprotein-associated phospholipase A2 activity is associated with coronary artery disease and markers of oxidative stress: a case-control study. Am J Clin Nutr. 2008;88:630–637. doi: 10.1093/ajcn/88.3.630. [DOI] [PubMed] [Google Scholar]
- 44.Kang Y.J. Metallothionein redox cycle and function. Exp Biol Med (Maywood) 2006;231:1459–1467. doi: 10.1177/153537020623100903. [DOI] [PubMed] [Google Scholar]
- 45.Iljin K., Ketola K., Vainio P., Halonen P., Kohonen P., Fey V., GrafströM R.C., Perälä M., Kallioniemi O. High-throughput cell-based screening of 4910 known drugs and drug-like small molecules identifies disulfiram as an inhibitor of prostate cancer cell growth. Clin Cancer Res. 2009;15:6070–6078. doi: 10.1158/1078-0432.CCR-09-1035. [DOI] [PubMed] [Google Scholar]
- 46.Luria A., Morisseau C., Tsai H.J., Yang J., Inceoglu B., De Taeye B., Watkins S.M., Wiest M.M., German J.B., Hammock B.D. Alteration in plasma testosterone levels in male mice lacking soluble epoxide hydrolase. Am J Physiol Endocrinol Metab. 2009;297:E375–E383. doi: 10.1152/ajpendo.00131.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woodward D.F., Protzman C.E., Krauss A.H., Williams L.S. Identification of 19 (R)-OH prostaglandin E2 as a selective prostanoid EP2-receptor agonist. Prostaglandins. 1993;46:371–383. doi: 10.1016/0090-6980(93)90102-d. [DOI] [PubMed] [Google Scholar]
- 48.Wang X., Klein R.D. Prostaglandin E2 induces vascular endothelial growth factor secretion in prostate cancer cells through EP2 receptor-mediated cAMP pathway. Mol Carcinog. 2007;46:912–923. doi: 10.1002/mc.20320. [DOI] [PubMed] [Google Scholar]
- 49.Newman J.W., Morisseau C., Harris T.R., Hammock B.D. The soluble epoxide hydrolase encoded by EPXH2 is a bifunctional enzyme with novel lipid phosphate phosphatase activity. Proc Natl Acad Sci USA. 2003;100:1558–1563. doi: 10.1073/pnas.0437724100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinot F., Grant D.F., Spearow J.L., Parker A.G., Hammock B.D. Differential regulation of soluble epoxide hydrolase by clofibrate and sexual hormones in the liver and kidneys of mice. Biochem Pharmacol. 1995;50:501–508. doi: 10.1016/0006-2952(95)00167-x. [DOI] [PubMed] [Google Scholar]
- 51.Tong M., Tai H.H. Induction of NAD(+)-linked 15-hydroxyprostaglandin dehydrogenase expression by androgens in human prostate cancer cells. Biochem Biophys Res Commun. 2000;276:77–81. doi: 10.1006/bbrc.2000.3437. [DOI] [PubMed] [Google Scholar]
- 52.Mirtti T., Laine V.J., Hiekkanen H., Hurme S., Rowe O., Nevalainen T.J., Kallajoki M., Alanen K. Group IIA phospholipase A as a prognostic marker in prostate cancer: relevance to clinicopathological variables and disease-specific mortality. APMIS. 2009;117:151–161. doi: 10.1111/j.1600-0463.2008.00002.x. [DOI] [PubMed] [Google Scholar]
- 53.Tang Y., Wang M.T., Chen Y., Yang D., Che M., Honn K.V., Akers G.D., Johnson S.R., Nie D. Downregulation of vascular endothelial growth factor and induction of tumor dormancy by 15-lipoxygenase-2 in prostate cancer. Int J Cancer. 2009;124:1545–1551. doi: 10.1002/ijc.24118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhatia B., Maldonado C.J., Tang S., Chandra D., Klein R.D., Chopra D., Shappell S.B., Yang P., Newman R.A., Tang D.G. Subcellular localization and tumor-suppressive functions of 15-lipoxygenase 2 (15-LOX2) and its splice variants. J Biol Chem. 2003;278:25091–25100. doi: 10.1074/jbc.M301920200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Validation of antibodies used in immunohistochemical staining. A:PLA2G7 antibody. PLA2G7 siRNA treated cells compared with the scrambled control. B:HPGD antibody. HPGD siRNA treated cells compared with the scrambled control. β-Actin was used as a loading control.
Normalized expression levels of the selected arachidonic acid pathway genes in prostate tissue samples, as well as in all available tissue samples in silico. Box plot analysis of normalized expression levels in healthy and malignant prostate tissue samples in silico is shown in the upper figures. The box refers to the quartile distribution (25 to 75%) range, with the median shown as a vertical line (green in normal samples, red in malignant). In addition, the 95% range and individual outlier samples are shown. Tissue- and disease-wide in silico expression profiles are shown in the lower figures. All samples are represented along the x axis in a predetermined fixed order, first the normal tissues, then cancers and other diseases. The y axis reflects the relative level of gene expression after normalization. Each dot represents the expression of indicated gene in one sample. Anatomical origin of the samples is marked with colored bars and indicated in the end of the figure. Sample types having higher than average expression or an outlier expression profile are additionally colored in each gene plot (legend at the top left corner of each figure).
In silico analysis of HPGD mRNA expression. A.In silico data anlysis of prostate tissue in GeneSapiens database indicate high HPGD expression in metastatic disease. B. Yu et al. study30 in Oncomine29 shows HPGD to be highly expressed in metastatic prostate cancer.


