Abstract
Transcriptional activation of Wnt/Wg-responsive genes requires the stabilization and nuclear accumulation of β-catenin, a dedicated coactivator of LEF/TCF enhancer-binding proteins. Here we report that recombinant β-catenin strongly enhances binding and transactivation by LEF-1 on chromatin templates in vitro. Interestingly, different LEF-1 isoforms vary in their ability to bind nucleosomal templates in the absence of β-catenin, owing to N-terminal residues that repress binding to chromatin, but not nonchromatin, templates. Transcriptional activation in vitro requires both the armadillo (ARM) repeats and the C terminus of β-catenin, whereas the phosphorylated N terminus is inhibitory to transcription. A fragment spanning the C terminus (CT) and ARM repeats 11 and 12 (CT–ARM), but not the CT alone, functions as a dominant negative inhibitor of LEF-1–β-cat activity in vitro and can block ATP-dependent binding of the complex to chromatin. LEF-1–β-cat transactivation in vitro was also repressed by inhibitor of β-catenin and Tcf-4 (ICAT), a physiological inhibitor of Wnt/Wg signaling that interacts with ARM repeats 11 and 12, and by the nonsteroidal anti-inflammatory compound, sulindac. None of these transcription inhibitors (CT–ARM, ICAT, or sulindac) could disrupt the LEF-1–β-cat complex after it was stably bound to chromatin. We conclude that the CT–ARM region of β-catenin functions as a chromatin-specific activation domain, and that several inhibitors of the Wnt/Wg pathway directly modulate LEF-1–β-cat activity on chromatin.
Keywords: Wnt signaling, β-catenin, LEF-1, chromatin, transcription regulation
The Wnt/Wingless signaling pathway specifies cell fate, segment polarity, and tissue and organ identity in many organisms through activating the armadillo-related coactivator, β-catenin (for reviews, see Nusse 1999; Bienz and Clevers 2000; Hecht and Kemler 2000; Polakis 2000; Huelsken and Birchmeier 2001). Most cellular β-catenin localizes to cellular membrane adherens junctions to promote E-cadherin-dependent cell adhesion. However, a small fraction is bound in cytoplasmic complexes with the adenomatous polyposis coli (APC) tumor suppressor, Axin/Conductin, and glycogen synthase kinase-3β (GSK3β), and it is this pool that mediates the transcriptional response to Wnt signaling. In the absence of Wnt ligands, β-catenin becomes phosphorylated by GSK3β within the APC complex, marking it for SCF-directed ubiquitination and proteosomal degradation. Wnt signaling ultimately inactivates GSK3β and disrupts the APC regulatory complex, and nonphosphorylated β-catenin enters the nucleus to bind LEF-1/TCF enhancer factors (Behrens et al. 1996; Molenaar et al. 1996; Brunner et al. 1997). Another Wnt-responsive transcriptional coactivator, γ-catenin (plakoglobin), is phosphorylated similarly to β-catenin although its stability is not affected by Wnt signaling (Kolligs et al. 2000). Interestingly, these Wnt-induced coactivators differentially regulate downstream target genes (Zhurinsky et al. 2000), i.e., β-catenin preferentially activates synthetic enhancers with tandem LEF/TCF binding sites, whereas γ-catenin more strongly induces the native c-myc gene (Kolligs et al. 2000).
Defects in the Wnt pathway contribute to several human cancers, most notably colon cancers and melanomas (Korinek et al. 1997; Morin et al. 1997; for review, see Bienz and Clevers 2000; Polakis 2000). Most human colorectal tumors either lack the APC tumor suppressor or express a severely truncated form that is unable to regulate GSK3β-mediated phosphorylation of β-catenin (Korinek et al. 1997; Morin et al. 1997). A small number of cancers arise from N-terminal mutations in β-catenin that prevent phosphorylation (Morin et al. 1997), which enhance β-catenin activity in transcription and transformation assays (Gat et al. 1998; Kolligs et al. 1999). Interestingly, the LEF/TCF proteins are also important targets of Wnt signaling in transformed cells (Roose et al. 1999; Hovanes et al. 2001). Activation of the Wnt pathway up-regulates transcription of full-length LEF-1 without affecting the expression of an N-terminal truncated form of LEF-1 (ΔNLEF-1) that is unable to bind β-catenin and functions as a feedback inhibitor of Wnt signaling in vivo (Hovanes et al. 2001). Thus the balance between active and dominant-interfering forms of LEF-1 changes on Wnt signaling in colon epithelial cells. Disruption of this balance, for example in mice lacking the TCF1 gene, results in enhanced activity of other LEF/TCF proteins and an increased susceptibility towards developing intestinal and colon neoplasms (Roose et al. 1999). Other targets of Wnt signaling in transformed cells include the genes encoding cyclin D1 (Shtutman et al. 1999; Tetsu and McCormick 1999) and PPARδ (He et al. 1999).
The LEF/TCF family members (LEF-1, TCF1, TCF3, TCF4) are monomeric high mobility group (HMG) proteins that contact the minor groove, bending the DNA strongly (for review, see Eastman and Grosschedl 1999). Binding to β-catenin is mediated through a conserved N-terminal motif, and certain family members, such as LEF-1, also contain a context-dependent activation domain (CAD) that participates in Wnt-independent activation of the T-cell receptor α-chain (TCRα) gene. The C terminus (CT) of β-catenin harbors a strong activation domain (van de Wetering et al. 1997; Hsu et al. 1998; Hecht et al. 1999). Different regions of β-catenin also interact with CBP/p300 (Hecht et al. 2000; Miyagishi et al. 2000; Sun et al. 2000; Takemaru and Moon 2000), the Brg-1-containing SWI/SNF chromatin remodeling complex (Barker et al. 2001), pontin52 (Bauer et al. 1998), and the TATA-binding protein, TBP (Hecht et al. 1999). The central core of β-catenin contains 12 armadillo (ARM) repeats that mediate mutually-exclusive binding interactions with LEF/TCF, APC, and other proteins required for Wnt signaling. Crystallographic studies have shown that the first eight ARM repeats form a flat and compacted superhelical structure that gradually bends through repeats 8 and 9, changing the orientation of ARM repeats 11 and 12 relative to the rest of the molecule (Huber et al. 1997). Recent analysis of a Xtcf3–ARM cocrystal further revealed that the charged amino terminus of LEF/TCF proteins forms an extended antiparallel structure with ARM repeats 3–8, without disturbing the overall conformation of β-catenin (Graham et al. 2000).
Many Wnt-responsive genes are strongly repressed by LEF/TCF proteins in the absence of Wnt signaling. Genetic studies in Drosophila indicate that repression is mediated through Groucho corepressors that interact with histone deacetylases to modulate chromatin structure (Cavallo et al. 1998; Levanon et al. 1998; Roose et al. 1998; Chen et al. 1999), and Osa, a component of the Brahma-containing SWI/SNF chromatin remodeling complex (Collins and Treisman 2000). Although little is known about the process that converts or replaces repressive complexes with active ones, it is clear that different LEF/TCF proteins vary in their relative ability to activate or repress transcription in vivo. For example, the TCF3 protein is a potent repressor of Wnt signaling in zebrafish (Kim et al. 2000) and in mouse epidermal stem cells (Merrill et al. 2001), even though it retains the ability to bind β-catenin. A variety of inhibitory pathways further restrict β-catenin activity in the nucleus, including small polypeptide inhibitors such as ICAT (inhibitor of β-catenin and Tcf-4; Tago et al. 2000) and I-mfa (Snider et al. 2001).
We have previously used a chromatin-based cell-free transcription system to examine context-dependent activation of the HIV-1 and TCRα enhancers by LEF-1 (Sheridan et al. 1995, 1997; Mayall et al. 1997). These studies showed that LEF-1 has a low intrinsic affinity for chromatin templates but can bind and function cooperatively with other enhancer-binding proteins to regulate TCRα and HIV-1 transcription in a CAD- and chromatin-dependent manner. We show that LEF-1 also binds and activates transcription cooperatively with β-catenin on a Wnt-responsive enhancer in vitro. Cooperative binding results from an inhibitory effect of the N terminus of LEF/TCF proteins that is exhibited on binding to chromatin, but not nonchromatin, templates. β-Catenin activity in vitro is enhanced by p300 and chromatin remodeling activities, and requires the C-terminal activation domain and inhibited by the N terminus. We also find that LEF-1–β-cat transactivation is selectively inhibited by ICAT and by a dominant-negative fragment of β-catenin, and is sensitive to the nonsteroidal anti-inflammatory drug (NSAID) sulindac. Thus this system provides a useful new approach to explore the mechanism of LEF/TCF–β-cat-mediated transcription of chromatin-assembled genes.
Results
LEF-1–β-cat activates transcription in a chromatin-dependent manner in vitro
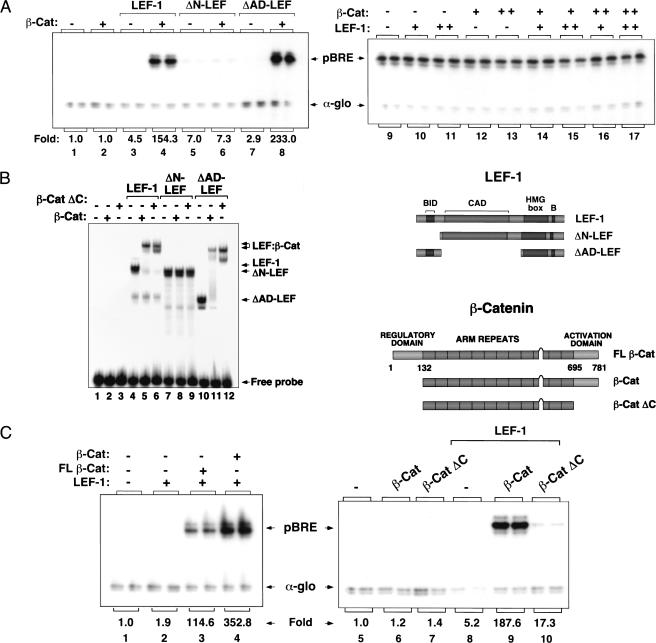
To assess whether β-catenin is sufficient to activate transcription when bound with LEF-1 to chromatin, we purified wild-type and mutant LEF-1 and β-catenin proteins and examined their ability to activate the pBRE (β-catenin response element) plasmid, which contains four LEF-1-binding sites positioned upstream of a TATA-containing core promoter. In vivo, LEF-1 is unable to activate pBRE or the related TOPFlash reporter gene in the absence of β-catenin (Korinek et al. 1997; data not shown). The various LEF-1 and β-catenin proteins we tested are indicated schematically in Figure 1. For the initial experiments, we used an N-terminal truncated form of β-catenin (β-cat) that resists proteolysis and functions as a strong constitutive inducer of Wnt signaling in vivo (Gat et al. 1998). Chromatin assembly was carried out as described by Bulger and Kadonaga (1994) using a Drosophila embryo S190 extract and purified core histones, and RNA was analyzed by primer extension following incubation of the pBRE chromatin template with a HeLa nuclear extract. Because it was unclear whether LEF-1 would require the activity of chromatin remodeling complexes to bind a nucleosomal template, the purified His-tagged LEF-1 and GST-tagged β-catenin proteins were allowed to bind to the pBRE enhancer during nucleosome assembly. Under these conditions, neither LEF-1 nor β-catenin activated transcription alone (Fig. 1A, lanes 2,3), whereas together the two proteins strongly induced pBRE transcription (Fig. 1A, lane 4). β-catenin failed to activate a truncated LEF-1 protein (ΔN-LEF) that lacks the β-catenin interaction domain (Fig. 1A, lane 6), but was a potent activator when complexed with a LEF-1 mutant lacking the CAD (ΔAD-LEF; Fig. 1A, cf. lanes 7 and 8). β-Catenin was unable to activate pBRE transcription on naked DNA, either alone or together with LEF-1 (Fig. 1A, lanes 9–17), indicating that chromatin structure is essential for LEF-1–β-cat transactivation in vitro.
Figure 1.
Chromatin-specific activation of transcription by purified recombinant LEF-1 and β-catenin. (A) Primer-extension analysis of pBRE transcription in the presence of wild-type or mutant LEF-1 proteins on chromatin (left panel) or nonchromatin (right panel) templates in vitro. Reactions either lacked enhancer factors (lane 1), or contained β-cat (120 nM, lanes 2,4,6,8), full-length LEF-1 (120 nM, lanes 3,4), ΔN-LEF (120 nM, lanes 5,6), or ΔAD-LEF (120 nM, lanes 7,8). (Right panel) Transcription reactions either lacked enhancer factors (lane 9), or contained LEF-1 (250 nM, lanes 10,14,16; 750 nM, lanes 11,15,17), β-cat (250 nM, lanes 12,14,15; 750 nM, lanes 13,16,17). Arrows designate transcription from the pBRE template (pBRE) or the alpha-globin promoter (α-glo), which was added as a nonchromatin template to the HeLa transcription extract as a positive control for RNA recovery. (B) pBRE chromatin transcription reactions with full-length (FLβ-cat) and truncated β-cat proteins (β-cat and β-catΔC). Reactions either lacked enhancer factors (lanes 1,5) or contained LEF-1 (120 nM, lanes 2–4 and 8–10), FLβ-cat (120 nM, lane 3), β-cat (120nM, lanes 4,6,9), or β-cat ΔC (120 nM, lanes 7,10). (C) Western blot analysis of β-cat and FLβ-cat proteins before (t = 0) and after (t = 5 h) chromatin assembly. At the right is a schematic of the different mutant LEF-1 and β-catenin proteins examined in this study.
Transient expression studies have shown that β-catenin carries a strong C-terminal transcription activation domain and may also contain a second N-terminal activation domain (Hsu et al. 1998; Hecht et al. 1999). However, it has been difficult to assess the contribution of the amino terminus of β-catenin to transactivation in vivo because mutants lacking this domain are much more stable than the full-length protein and show increased activity in transcription and transformation assays (Hsu et al. 1998; Hecht et al. 1999; Kolligs et al. 1999). Interestingly, we find that the N-terminal truncated β-catenin protein (β-cat) is more active than the full-length protein (FLβ-cat) in vitro (Fig. 1B, cf. lanes 3 and 4). Western blot experiments indicate that the two proteins are equally stable during chromatin assembly and transcription (Fig. 1C), even though the FLβ-cat protein undergoes N-terminal phosphorylation by protein kinases in the chromatin assembly extract (R. Landry and K. Jones, unpubl.). We conclude that the N-terminal truncated form of β-catenin is an intrinsically stronger coactivator than the full-length protein. In contrast, LEF-1 was only weakly activated by a β-catenin mutant lacking the C-terminal activation domain (β-catΔC; Fig. 1B, cf. lanes 9 and 10), even though β-catΔC readily formed a stable complex with LEF-1 on DNA (data not shown). The residual low-level transcriptional activity of β-catΔC may reflect the ability of the ARM repeats to contribute to transactivation or to regulate binding of LEF-1 to chromatin (see below). We conclude that β-catenin functions as a powerful, selective, and chromatin-specific transcriptional coactivator of LEF-1 in vitro.
β-Catenin enhances binding of LEF-1 to chromatin templates in vitro
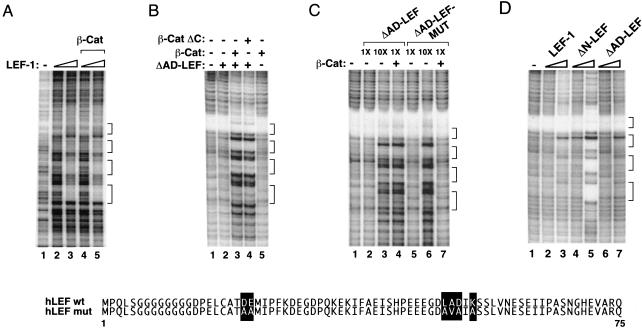
We have previously reported that LEF-1 binds weakly on its own to chromatin templates but can interact cooperatively with other enhancer-binding factors to regulate T-cell enhancer activity in vitro (Mayall et al. 1997). From these observations we inferred that β-catenin might also promote the interaction of LEF-1 with chromatin. However, as reported in earlier studies, β-catenin does not affect binding of LEF-1 to naked DNA in gel shift or DNase I footprint experiments (Fig. 2A, cf. lanes 2 and 4, or lanes 3 and 5). In contrast, β-catenin strongly enhanced binding of full-length LEF-1 or ΔAD-LEF when the factors were incubated with the pBRE template during nucleosome assembly (Fig. 2B, cf. lanes 2 and 3; other data not shown). Beta-catenin did not bind to the pBRE enhancer in the absence of LEF-1 (Fig. 2B, lane 5), nor did it stimulate transcription by unrelated enhancer factors such as TFE3 (data not shown). Cooperative binding was also observed with β-catΔC (Fig. 2B, lane 4), which lacks the C-terminal activation domain. In all cases, the pattern of footprint protection observed with the different LEF-1–β-cat complexes was identical to that obtained with high levels of LEF-1 alone (Fig. 2C, cf. lanes 3 and 4). In contrast, a LEF-1 mutant containing multiple substitutions in the β-catenin interaction domain (ΔAD-LEF-MUT) failed to bind cooperatively with β-catenin to the template (Fig. 2C, lane 7), even though it bound the pBRE enhancer independently of β-catenin at higher levels (Fig. 2C, lane 6). We conclude (1) that β-catenin and LEF-1 bind cooperatively to the pBRE enhancer when incubated with the template during chromatin assembly, but (2) do not bind in a cooperative manner to naked DNA, and (3) that cooperative binding does not require the C-terminal activation domain of β-cat.
Figure 2.
β-catenin strongly enhances the binding of LEF-1 to chromatin, but not nonchromatin templates. (A) DNase I footprint analysis of binding of LEF-1 and LEF-1–β-cat to (nonchromatin) pBRE DNA. Binding reactions either lacked enhancer factors (lane 1) or contained LEF-1 (112 nM, lanes 2,4; 560 nM, lanes 3,5) and β-cat (560 nM, lanes 4,5). The four LEF-1 binding sites in the pBRE enhancer are indicated with brackets. (B) DNase I footprint analysis of pBRE chromatin assembled in the absence of enhancer factors (lane 1) or in the presence of ΔAD-LEF (120 nM, lanes 2–4), β-cat (120 nM, lanes 3,5) or β-cat ΔC (120 nM, lane 4). (C) DNase I footprint analysis of pBRE chromatin assembled in the absence of enhancer factors (lane 1) or in the presence of β-cat (120 nM, lanes 4,7), ΔAD-LEF (112 nM, lanes 2,4; 1 μM, lane 3), or ΔAD-LEF-MUT (112 nM, lanes 5,7; 1 μM, lane 6). Substitutions in the amino terminus of ΔAD-LEF-MUT that abrogate binding to β-cat are indicated with bold type. (D) The N terminus of LEF-1 inhibits binding to chromatin, but not nonchromatin, templates. DNase I footprint analysis of the binding of LEF-1 and mutant LEF-1 proteins to pBRE chromatin. Chromatin assembly reactions either lacked enhancer factors (lane 1) or contained full-length LEF-1 (120 nM, lane 2; 1 μM, lane 3), ΔN-LEF (120 nM, lane 4; 1 μM, lane 5), or ΔAD-LEF (120 nM, lane 6; 1 μM, lane 7). Brackets indicate the four LEF/TCF binding sites in the pBRE enhancer.
The N terminus of LEF-1 inhibits binding to chromatin, but not nonchromatin, templates
The observation that β-catenin can modulate binding of LEF-1 to chromatin without itself contacting DNA raised the possibility that the conformation or structure of native LEF-1 might impede binding to chromatin in the absence of β-catenin. Consequently, we asked whether mutant LEF-1 proteins differed in their ability to bind to the nucleosomal pBRE template in the absence of β-cat. Interestingly, although LEF-1, ΔN-LEF, and ΔAD-LEF bound with similar affinities to the nonchromatin pBRE template in DNase I footprint and gel mobility shift experiments (data not shown), we observed striking differences in the ability of these proteins to interact with the nucleosomal pBRE enhancer (Fig. 2D). In particular, the ΔN-LEF protein (Fig. 2D, lane 5) bound more avidly than wild-type LEF-1 (Fig. 2D, lane 3) to pBRE chromatin templates. In contrast, removal of the LEF-1 CAD did not enhance binding to chromatin (ΔAD-LEF; Fig. 2D, lane 7). Thus the LEF-1 N terminus strongly inhibits binding to chromatin in the absence of β-catenin. Because the N-terminal substitutions in ΔAD-LEF-MUT failed to derepress binding to chromatin (Fig. 2D, cf. lanes 4 and 7), we conclude that the region that inhibits LEF-1 binding overlaps, but is not identical to, the β-catenin interaction motif.
The β-catenin CT–ARM fragment is a potent and selective inhibitor of LEF-1–β-cat transcription in vitro
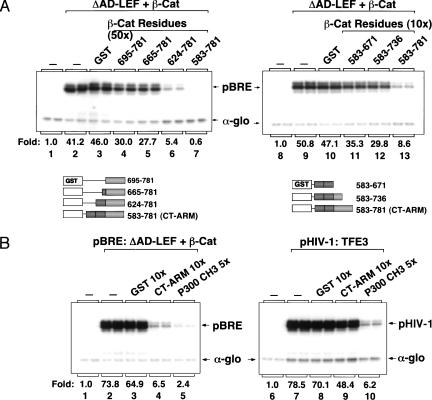
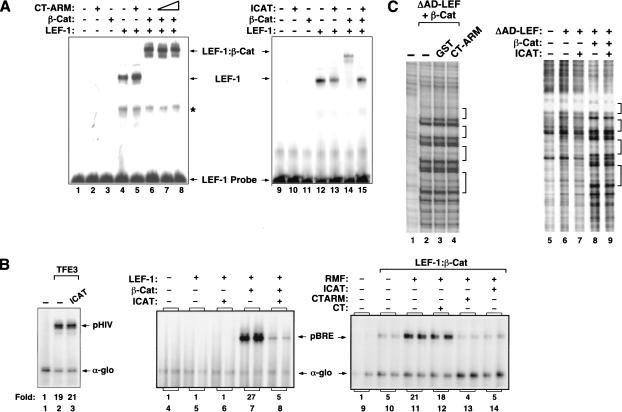
To better characterize the chromatin-dependent activation domain at the C terminus of β-catenin, we sought to identify fragments from this region that interfere with transcription by sequestering targeted coactivators from the complex bound to the pBRE enhancer. Unexpectedly, LEF-1–β-cat transcription was unaffected by incubation with a 50-fold molar excess (relative to β-cat) of a β-cat fragment (CT, amino acids 695–781) that spans the entire C terminus (Fig. 3A, cf. lanes 2 and 4). Reasoning that the activation domain required for transcription on chromatin might extend into the C-terminal ARM repeats, we asked if larger fragments would inhibit transcription in vitro. Interestingly, LEF-1–β-cat activity was inhibited by β-cat (amino acids 624–781), which includes ARM repeat 12 (Fig. 3A, lane 6), and was even more effectively blocked by a larger fragment that includes ARM repeat 11 (CT–ARM; Fig. 3A, lane 7). The CT–ARM fragment also inhibited transcription at a 10-fold excess to β-catenin (Fig. 3A, lane 13), whereas no inhibition was observed with GST alone (Fig. 3A, lane 10) or with an ARM 11/12 fragment that lacks the C terminus (Fig. 3A, lane 11). The CT–ARM fragment did not affect global chromatin assembly as assessed by micrococcal nuclease digestion of pBRE chromatin, nor did it affect transcription of nonchromatin pBRE or α-globin templates (data not shown). The CT–ARM fragment failed to disrupt both TFE3-mediated activation of the HIV-1 enhancer (Fig. 3B, cf. lanes 7 and 9), and Notch-regulated transcription in vitro (C. Fryer and K. Jones, unpubl.), indicating that the inhibition is specific for LEF-1–β-cat transcription. These results suggest that ARM repeats 11 and 12 function synergistically with the C terminus to activate transcription on chromatin.
Figure 3.
The β-cat CT–ARM domain fragment selectively inhibits LEF-1–β-cat transcription on chromatin in vitro. (A) (Left panel) Chromatin was assembled in absence of enhancer factors (lane 1) or with 120 nM ΔAD-LEF and 120 nM β-cat (lanes 2–7) in the presence of a 50-fold molar excess (relative to β-cat) of either GST alone (lane 3), GST–β-cat aa695–781 (lane 4), GST–β-cat aa665–781 (lane 5), GST–β-cat aa624–781 (lane 6), or GST–β-cat aa583–781 (CT–ARM, lane 7). (Right panel) Chromatin was assembled in the absence of enhancer factors (lane 8) or with 120 nM ΔAD-LEF and 120 nM β-cat (lanes 9–13) and a 10-fold molar excess (relative to β-cat) of either GST alone (lane 10), GST–β-cat amino acids 583–671 (lane 11), GST–β-cat amino acids 583–736 (lane 12), or GST–β-cat amino acids 583–781 (lane 13). The GST–β-cat fragments are represented schematically below the figure. (B) Comparison of the transcription inhibitory effects of the β-cat CT–ARM and p300 CH3 domains on pBRE (left panel) and HIV-1 (right panel) chromatin templates. (Left panel) Transcription reactions either lacked enhancer factors (lane 1) or contained 120 nM ΔAD-LEF and 120 nM β-cat (lanes 2–5), along with a 10-foldmolar excess (relative to β-cat) of GST (lane 3), GST–CT–ARM (lane 4) or the GST–p300/CH3 fragment (lane 5). (Right panel) Transcription reactions either lacked enhancer factors (lane 6) or contained GST–TFE3 (30 nM, lanes 7–10) and a 10-fold molar excess (relative to TFE3) of GST (lane 8), GST–CT–ARM (lane 9) or the GST–p300/CH3 fragment (lane 10). Arrows indicate pBRE, pHIV-1, and α-globin transcripts.
Previous studies have indicated that the CT–ARM region of β-catenin can interact with the CH3 domain of the p300 coactivator (Hecht et al. 2000; Takemaru and Moon 2000). We therefore asked if the p300 CH3 domain would also selectively block LEF-1–β-cat activity in vitro. As shown in Figure 3B, the CH3 fragment strongly inhibited LEF-1–β-cat transactivation of the pBRE template (lane 5). However, the CH3 domain fragment also strongly inhibited TFE3-mediated activation of the HIV-1 enhancer (Fig. 3B, lane 10) as well as Notch-regulated transcription in vitro (C. Fryer and K. Jones, unpubl.). The CH3 inhibitor did not disrupt α-globin or pBRE core promoter activity on naked DNA (data not shown). We conclude that whereas the CT–ARM fragment is a selective inhibitor of β-catenin activity in vitro, the p300 CH3 domain acts in a more general manner to disrupt enhancer-dependent transcription on chromatin. In addition, these findings strongly suggested that p300 is required for LEF-1–β-cat activity in vitro.
p300 and a chromatin remodeling fraction facilitate LEF-1–β-cat transcription in vitro
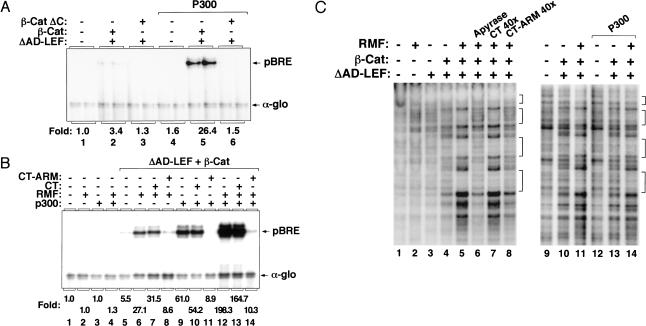
Although some DNA-binding proteins recognize their binding sites in chromatin efficiently in vitro (Pazin et al. 1998), others must be incubated with specific chromatin remodeling complexes or chromatin-modifying enzymes to activate transcription from fully-assembled chromatin templates (Armstrong et al. 1998; Kadam et al. 2000). Therefore, it was important to assess whether the recombinant LEF-1–β-cat complex can activate transcription from a preassembled nucleosomal template. LEF-1–β-cat transactivation was very inefficient when the complex was incubated with the pBRE template after nucleosome assembly (Fig. 4A, lane 2), but transcription was enhanced significantly in the presence of purified recombinant p300 (Fig. 4A, lane 5). Activation by p300 was specific because it did not enhance transcription without enhancer factors (Fig. 4A, lane 4), or when incubated with LEF-1–β-catΔC (Fig. 4A, lane 6). We also asked whether LEF-1–β-cat activity could be enhanced by ATP-dependent chromatin remodeling complexes. Although we were unable to activate the complex with a purified SWI/SNF fraction (data not shown), LEF-1–β-cat activity was stimulated by a partially-purified chromatin remodeling fraction (RMF), which contains the hSWI/SNF and hACF/ISWI remodeling complexes and is devoid of p300 (Fig. 4B, lane 6). The effect of RMF was similar to that observed with recombinant p300 (Fig. 4B, lane 9), and in combination the two fractions functioned synergistically (Fig. 4B, lane 12). LEF-1–β-cat activation under these conditions could still be repressed selectively by CT–ARM (Fig. 4B, lane 14), and not by the CT fragment of β-catenin (Fig. 4B, lane 13). Enhanced binding in the presence of the RMF fraction was more pronounced with LEF-1–β-cat than with LEF-1 alone (data not shown). Thus LEF-1–β-cat can strongly activate transcription from fully assembled chromatin templates when incubated with p300 and chromatin remodeling enzymes.
Figure 4.
p300 and RMF enhance transcription from fully assembled chromatin templates by LEF-1 and β-catenin in vitro. (A) Chromatin transcription reactions either lacked enhancer factors (lane 1), or contained ΔAD-LEF (120 nM, lanes 2,3,5,6), β-cat (120 nM, lanes 2,5), or β-cat ΔC (120 nM, lanes 3,6). Where indicated, purified recombinant p300 (60 nM, lanes 4–6) was incubated together with LEF-1 and β-cat after completion of pBRE chromatin assembly. (B) Transcription reactions either lacked enhancer factors (lane 1), or contained 120 nM ΔAD-LEF and 120 nM β-cat (lanes 5–14) added after pBRE chromatin assembly. Where indicated, reactions also contained p300 (60 nM, lanes 3,4,9–14), RMF (1 μg, lanes 2,4,6–8,12–14), GST–CT (4.2 μg, lanes 7,10,13), or GST–CT–ARM (4.8 μg, lanes 8,11,14), added simultaneously with LEF-1 and β-cat. (C) Binding of LEF-1–β-cat to assembled pBRE chromatin templates in the presence or absence of p300, RMF, and the CT–ARM inhibitor. Binding reactions either lacked enhancer factors (lanes 1,9) or contained ΔAD-LEF (120 nM, lanes 3–8,10–11,13–14), β-cat (120 nM, lanes 4–8,10–11,13–14), p300 (60 nM, lanes 12–14), RMF (1 μg, lanes 2,5–8,11,14), or a 10-fold excess of GST–CT (lane 7), GST–CT–ARM (lane 8), and apyrase (0.5 unit; lane 6), added with the LEF-1–β-cat complex after chromatin assembly.
DNase I footprint analysis of these transcription reactions revealed that the LEF-1–β-cat complex also binds very poorly on its own to the pBRE enhancer when added to the template after the chromatin template has been fully assembled (Fig. 4C, lane 4), and under these conditions binding of the complex was enhanced considerably by the addition of RMF (Fig. 4C, lane 5). Enhanced binding of LEF-1–β-cat to chromatin in the presence of RMF was completely inhibited by apyrase (Fig. 4C, lane 6), indicating that an ATP-dependent chromatin remodeling step is required. Interestingly, RMF-enhanced binding could also be competed by the CT–ARM fragment (Fig. 4C, lane 8) and not by the CT fragment (Fig. 4C, lane 7). In these experiments the CT and CT–ARM inhibitors, or apyrase, were added together with the LEF-1–β-cat complex and the RMF fraction after the completion of nucleosome assembly. In contrast, purified recombinant p300 did not affect the binding of LEF-1–β-cat to chromatin (Fig. 4C, cf. lanes 10 and 13). Taken together, these data indicate that LEF-1–β-cat transactivation requires p300 and chromatin remodeling, and that the CT–ARM fragment can block both ATP-dependent binding of the LEF-1–β-cat complex to preassembled chromatin, as well as the transcriptional activity of the complex after it has bound stably to chromatin.
ICAT, a physiological inhibitor of Wnt signaling, selectively represses LEF-1–β-cat transactivation in vitro
To further test the specificity of LEF-1–β-cat transactivation in this system, we asked whether activation was sensitive to ICAT, a physiological inhibitor of Wnt signaling in Xenopus oocytes (Tago et al. 2000). ICAT was of particular interest because it binds to β-cat ARM repeats 11 and 12, spanning the portion of the ARM repeats that is necessary for CT–ARM inhibition in vitro. Thus binding of ICAT to β-cat might disrupt interactions with chromatin-specific co-activators. However, it has also been shown that ICAT can block the interaction between LEF-1 and β-catenin, even though its binding site does not directly overlap the region of the ARM repeats (3–8) that bind LEF-1. We therefore asked whether ICAT would block LEF-1–β-cat activity in vitro and, if so, how inhibition by ICAT would compare with that we observe for the β-cat CT–ARM fragment.
As reported previously (Tago et al. 2000), high levels of purified recombinant ICAT can block the formation of the LEF-1–β-cat complex in electrophoretic mobility shift (EMSA) experiments (Fig. 5A, cf. lanes 14 and 15). In contrast, the LEF-1–β-cat complex was not affected by the CT–ARM inhibitor (Fig. 5A, cf. lane 6 with lanes 7,8). In addition, ICAT efficiently repressed LEF-1–β-cat transcription in vitro when incubated with the complex during chromatin assembly (Fig. 5B, cf. lanes 5 and 6). The inhibition was specific because ICAT did not affect TFE3-directed transcription from the HIV-1 enhancer (Fig. 5B, cf. lanes 2 and 3) or Notch-dependent transcription in vitro (C. Fryer and K. Jones, unpubl.). Both CT–ARM and ICAT also inhibited LEF-1–β-cat activation of preassembled pBRE chromatin templates when added to the template in the presence of RMF and p300 (Fig. 5B, cf. lane 8 with lanes 10,11). Unexpectedly, however, ICAT failed to block the cooperative binding observed when LEF-1 and β-cat were incubated with the pBRE enhancer during nucleosome assembly, even when ICAT was present at levels 10-fold higher than that required to block transcription (Fig. 5C, cf. lanes 6 and 7). Therefore, the interaction between LEF-1 and β-catenin in chromatin footprint reactions is not appreciably weakened by ICAT, under conditions where transcription is efficiently repressed. We conclude that ICAT and CT–ARM display a similar specificity of transcription inhibition but function through different mechanisms. Moreover, these experiments suggest that, in addition to its effects on complex formation, ICAT may be able to recognize and block the activity of LEF-1–β-cat complexes that have previously bound to chromatin.
Figure 5.
ICAT is a potent and selective inhibitor of LEF-1–β-cat transcription on chromatin. (A) Analysis of the effects of CT–ARM and ICAT on the formation of the LEF-1–β-cat–DNA ternary complex in gel mobility shift experiments. Binding reactions either lacked enhancer factors (lanes 1,9) or contained 110 nM LEF-1 (lanes 4–8,12–15), 110 nM β-cat (lanes 3,6–8,11,14,15), GST–CT–ARM (500 nM, lane 7; 2.5 μM, lanes 2,5,8) or GST–ICAT (2.5 μM, lanes 10,13,15). Arrows indicate the position of LEF-1–DNA and LEF-1–β-cat–DNA complexes. Asterisk indicates nonspecific band. (B) Analysis of the effect of ICAT on HIV-1 (lanes 1–3) and pBRE transcription (lanes 4–14) in vitro. Chromatin was assembled in the absence of enhancer factors (lanes 1,4,9) or in the presence of 120 nM GST–TFE3 (lanes 2,3), or 120 nm ΔAD-LEF and 120 nM β-cat (lanes 5–8,10–14). Where indicated, reactions also contained 1.2 μM of ICAT (lanes 3,6,8,14). The enhancer factors were either incubated with the template during chromatin assembly (lanes 4–8) or incubated after chromatin assembly together with 1.0 μg RMF (lanes 11–14). Arrows indicate the pHIV, pBRE, and α-globin transcripts. (C) DNase I footprint analysis of the effects of CT–ARM and ICAT on the β-catenin-enhanced binding of LEF-1 to chromatin. Binding reactions either lacked enhancer factors (lanes 1,5) or contained 120 nM ΔAD-LEF and 120 nM β-cat (lanes 2–4,6,7). Where indicated, reactions also contained 1 μM each of GST (lane 3), GST–CT–ARM (lane 4), or GST–ICAT (lane 7).
LEF-1–β-cat transcription in vitro is also blocked by the NSAID compound, sulindac
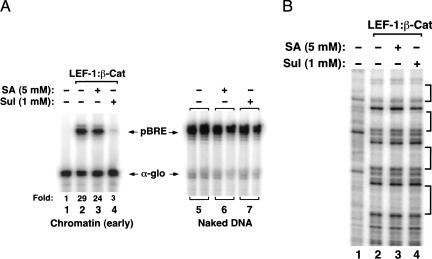
Finally, it was also of interest to examine whether LEF-1–β-cat activity might be influenced directly by NSAIDs. NSAIDs are anti-tumorigenic compounds that disrupt PPARδ activity and block induction of Wnt target genes (He et al. 1999). These compounds interfere with LEF/TCF–β-cat transactivation in cell lines (Dihlmann et al. 2000) and reduce intestinal polyp formation in APC(Min) mice, which express a truncated form of APC (for review, see Potter 1999). For these experiments, we asked whether the NSAIDs sulindac or salicylate would affect LEF-1–β-cat activity when incubated with the pBRE template during nucleosome assembly. We tested the NSAIDs at levels comparable to those shown to block IKK-β and other enzymes in vitro (Yin et al. 1998). Interestingly, we found that LEF-1–β-cat transcription in vitro was inhibited 10-fold by sulindac (1 mM) (Fig. 6A, cf. lanes 2 and 4), but was unaffected by salicylate (5 mM; Fig. 6A, lane 3). Sulindac did not block cooperative binding of LEF-1 and β-catenin to the nucleosomal pBRE enhancer (Fig. 6B), and also failed to disrupt the interaction between the two proteins in EMSA experiments (data not shown). Neither sulindac nor salicylate inhibited pBRE transcription on naked DNA (Fig. 6A, lanes 6,7), suggesting that sulindac may target a chromatin-specific enzyme required for LEF-1–β-cat activity. Thus certain NSAIDs, such as sulindac, have the potential to affect LEF-1–β-cat transcription directly.
Figure 6.
LEF-1–β-cat transactivation on chromatin in vitro is inhibited by the NSAID, sulindac. (A) (Left panel) Analysis of the effects of NSAID on LEF-1–β-cat transcription of pBRE chromatin templates in vitro. Chromatin was assembled in the absence of enhancer factors (lane 1) or in the presence of 120 nM ΔAD-LEF and 120 nM β-cat (lanes 2–4), in the presence of 5 mM salicylic acid (lane 3) or 1 mM sulindac (lane 4). (Right panel) Analysis of the effects of NSAIDs on nonchromatin pBRE DNA templates. Reactions either lacked NSAIDs (lane 5) or contained either 5 mM salicylic acid (lane 6) or 1 mM sulindac (lane 7). Arrows indicate pBRE (chromatin) and α-globin (nonchromatin) transcripts. (B) DNase I footprint analysis of the effects of NSAIDs on binding of the LEF-1–β-cat complex to chromatin. Chromatin was assembled in the absence of enhancer factors (lane 1) or in the presence of 120 nM each of ΔAD-LEF and β-cat (lanes 2–4), incubated with either 5 mM salicylic acid (lane 3) or 1 mM sulindac (lane 4). Brackets indicate the four LEF/TCF binding sites in the pBRE enhancer.
Discussion
Many developmental signaling pathways that control cell fate and proliferation, such as the Wnt/Wg, Smad, and Notch pathways, regulate the activity or availability of dedicated, factor-specific transcriptional coactivators. In each case, the induced coactivator interacts with one or a small subset of enhancer-binding proteins to convert a highly repressed gene to an active state and to reconfigure local chromatin structure. Although many studies address the mechanisms that mobilize these coactivators, relatively little is known about how these intermediary proteins function on chromatin to induce transcription in a rapid but transient manner. The chromatin-based cell-free transcription system used here provides a useful approach to assess the mechanism of LEF/TCF–β-cat activation of Wnt-responsive genes.
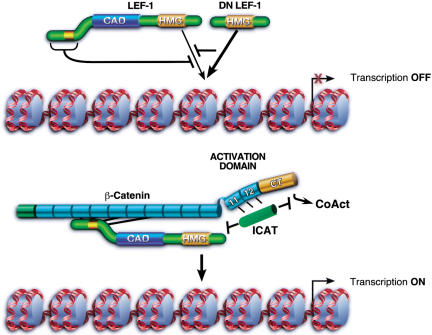
Our findings in this system support and extend current models of LEF-1–β-cat transactivation in several important ways (Fig. 7). First, we find that the activation domain of β-catenin is required and sufficient for LEF-1 to initiate transcription on chromatin in vitro. Although LEF-1 contains a CAD that is essential for T-cell enhancer regulation, this region of LEF-1 is dispensable for LEF-1–β-cat activity in vitro (Fig. 1) and in vivo. Second, transactivation is accompanied by cooperative binding of LEF-1 and β-catenin to chromatin, which results from an inhibitory effect of the LEF-1 N terminus on binding to nucleosomal templates. Interestingly, neither of these effects can be observed on naked DNA. Thus different LEF/TCF isoforms will vary in their intrinsic affinity for chromatin, even though all of these isoforms contain identical DNA-binding domains. Third, we show that LEF-1–β-cat transcription in vitro is sensitive to Wnt pathway inhibitors, including the CT–ARM fragment of β-catenin, ICAT, and the NSAID, sulindac. Taken together, these data suggest that β-catenin targets full-length LEF-1 proteins to nucleosomal sites and directs transcription through a C-terminal regulatory domain that extends into the adjacent ARM repeats. Because fusion of the β-catenin C-terminal domain to LEF-1 is sufficient to enable LEF-1 to activate transcription and to enhance transformation in transient expression assays (Aoki et al. 1999; Hecht et al. 1999; Galceran et al. 2001), the contribution of the ARM repeats may be evident only on chromatin templates.
Figure 7.
Chromatin-specfic effects of β-catenin on binding and transcriptional activation by LEF-1 in vitro. In the absence of β-catenin, Wnt-responsive genes are repressed by LEF/TCF–Groucho complexes, and binding of uncomplexed LEF-1 to chromatin is inhibited through amino-terminal residues. LEF-1 isoforms that lack the inhibitory amino-terminal domain bind chromatin more avidly and function as potent feedback inhibitors of Wnt signaling in vivo (dominant negative DN LEF-1). The interaction of the central ARM repeats of β-catenin with the N terminus of LEF-1 alleviates this inhibition, and LEF-1 binds cooperatively with its coactivator to chromatin. Optimal transactivation requires an activation domain located at the C terminus that may also include C-terminal ARM repeats 11 and 12. LEF-1–β-cat transcription can be selectively blocked in vitro by a dominant-negative fragment of β-catenin that spans the carboxyl terminus and ARM repeats 11/12, as well as by the physiological Wnt inhibitor, ICAT. ICAT, but not CT–ARM, can disrupt the interaction between LEF-1 and β-catenin. Moreover, ICAT can block transcription from complexes bound stably to chromatin, indicating that it may also disrupt interactions between β-catenin and downstream transcriptional coactivators (CoAct).
These observations have interesting biological implications for the regulation of Wnt/Wg-responsive genes with different isoforms of the LEF/TCF proteins. Cooperative binding to chromatin may ensure that full-length LEF-1 is not targeted to enhancers unless previously complexed with β-catenin. In contrast, ΔN-LEF, which is unable to bind β-catenin and functions in vivo as a feedback inhibitor of Wnt signaling, lacks the inhibitory region and binds chromatin independently of β-catenin. It is unclear why LEF/TCF proteins recognize their binding site in chromatin in a qualitatively distinct manner than DNA, although it could be related to the unique abilities of HMG domain proteins to recognize distorted and bent DNA structures (Travers 2000). One possibility is that nucleosomal histone tails repel the highly charged N terminus of LEF-1. Binding of LEF-1 to β-catenin effectively buries the N terminus along the central ARM repeats (Graham et al. 2000), which may neutralize its inhibitory effect on chromatin recognition.
The cooperative binding interactions we have characterized are observed when LEF-1 and β-catenin are incubated with the pBRE template during chromatin assembly, as would occur in vivo with actively replicating templates. However, we show that binding of the LEF-1–β-cat complex to fully assembled chromatin also requires an ATP-dependent chromatin remodeling step (Fig. 5). The observation that the CT–ARM fragment can block ATP-dependent binding of the complex in extracts (Fig. 5) implies that this region of β-catenin targets a chromatin remodeling complex. Our preliminary data indicate that the CT–ARM fragment interacts directly with a remodeling complex and likely binds to other transcriptional coactivators as well (A.V. Tutter and K. Jones, unpubl.). In contrast, a recent report indicates that a BRG-1 containing SWI/SNF remodeling complex can bind the central ARM repeats of β-catenin (Barker et al. 2001), and therefore it is possible that the CT–ARM fragment might block the recruitment or subsequent action of this complex. Regardless of the mechanism, these data together strongly suggest that one essential function of LEF–β-cat is to recruit chromatin remodeling complex. It is likely that the relevant remodeling complex does more than enhance binding of LEF-1–β-cat to chromatin, because we show that the CT–ARM fragment also inhibits transcription when the factors are allowed to bind the template during chromatin assembly (Fig. 3). Therefore a chromatin remodeling activity recruited through LEF–β-cat may be needed to open adjacent regions of the promoter for access by other transcription factors.
Genetic studies indicate that the repression of Wg target genes in Drosophila is mediated by Osa- and BRM-containing Swi/SNF remodeling complexes (Collins and Treisman 2000), and by extension the ability of ΔNLEF-1 and other dominant-negative LEF/TCF isoforms to assemble into enhancer complexes may also require chromatin remodeling in vivo. It will be important to assess whether similar or distinct remodeling complexes are used for the purposes of gene activation and repression by the various LEF/TCF proteins. Cooperative binding interactions facilitated by specific remodeling complexes on chromatin may help explain how the distinct LEF/TCF family members that mediate activation or repression of Wnt target genes become differentially assembled into enhancer complexes in vivo.
As has been observed previously in vivo, we find that p300 is a positive coactivator of LEF–β-cat transcription in vitro. Different studies have suggested that either the CH1 (Sun et al. 2000), CH3 (Hecht et al. 2000; Miyagishi et al. 2000), or KIX (Takemaru and Moon 2000) domains of CBP/p300 interact with either the N terminus (Sun et al. 2000), ARM repeats (Miyagishi et al. 2000), or CT–ARM (Hecht et al. 2000; Takemaru and Moon 2000) regions of β-catenin. We have not detected CBP/p300 among the nuclear proteins that interact with the CT–ARM fragment in HeLa extracts (A. Tutter, C. Fryer, and K. Jones, unpubl.), and we show here that the CH3 domain of p300 not only blocks LEF-1–β-cat activity, but can also inhibit the activity of unrelated enhancer factors in a chromatin-specific manner (Fig. 4). This latter observation is consistent with a previous report that the CH3 region of p300 plays a general role in enhancer activation in vitro (Kraus et al. 1999). Experiments are underway to examine how p300 is recruited to the LEF-1–β-cat complex in vitro.
Although previous studies have suggested that the N terminus of β-catenin may provide an auxiliary activation domain (Hsu et al. 1998; Hecht et al. 1999), we find that this region is inhibitory to LEF-1–β-cat transactivation in vitro (Fig. 1). One possibility is that the N terminus may fold back over the ARM repeats to weaken the interaction with LEF-1, and indeed we find that LEF-1–FLβ-cat complexes bind relatively weakly to DNA (R. Landry and K. Jones, unpubl.). However, phosphorylation of the N terminus may also contribute to the inhibition we observe in the chromatin assembly extract. Because a subset of colon cancers arise from mutations in β-catenin that prevent N-terminal phosphorylation (Kolligs et al. 1999), it will be interesting to learn whether these unmodified proteins can function as constitutive activators of LEF-1 in vivo.
LEF-1–β-cat activity in vitro can also be selectively inhibited by ICAT, a small nuclear and cytoplasmic protein that binds to β-cat ARM repeats 11 and 12 (Tago et al. 2000). The ability of ICAT to mask the binding of β-cat to LEF-1 (Tago et al. 2000) may be its primary function in the cytoplasm. However, ICAT was unable to disrupt cooperative binding of LEF-1 and β-catenin to the nucleosomal pBRE enhancer (Fig. 5), and therefore it may also be able to recognize LEF-1–β-cat complexes bound to chromatin and block transcription at a later step (Fig. 7). If so, then ICAT may be able to block all β-catenin-mediated transcription in cells, regardless of whether it is mediated through LEF/TCF proteins.
Considerable efforts have been extended to identify inhibitors of the Wnt pathway due to its central role in several important human cancers. The NSAID sulindac was shown to block both the induction and the activity of PPARδ, an important target of Wnt signaling, and to inhibit the DNA-binding activity of PPARδ in vitro (He et al. 1999). In addition, sulindac can block Wnt signaling in colon epithelial cells without affecting β-catenin stability or disrupting the LEF-1–β-cat complex in vivo (Dihlmann et al. 2000). In vitro, we find that sulindac inhibits LEF-1–β-cat transcription without affecting binding to chromatin, whereas salicylic acid failed to block transcription. However, sulindac is also a weak inhibitor of other chromatin-dependent enhancer factors in vitro (C. Fryer and K. Jones, unpubl.), and therefore its specificity of inhibition is lower than that of the ICAT and CT–ARM inhibitors. Salicylate and sulindac at the levels tested here also block ATP binding to IKK-β and enzymatic activity in vitro (Yin et al. 1998). By extension, sulindac may target an ATP-dependent enzyme required to remodel or modify chromatin, although we find that it does not block p300-mediated acetylation of core histones (C. Fryer and K. Jones, unpubl.). Differential effects of NSAIDs on tumor formation have also been reported in APC(Min) mice, which respond to sulindac (Boolbol et al. 1996) but not salicylate (Ritland et al. 1999). Therefore it may be useful to compare sulindac with other inhibitors of chromatin modifying enzymes, as such compounds may be useful inhibitors of cellular transformation.
In summary, the chromatin-transcription system used here recapitulates important aspects of Wnt-regulated transcription in cells and can be used to test additional aspects of the mechanism of LEF-1–β-cat transactivation. This system is responsive to selective inhibitors of the Wnt pathway, and could also be used to explore how different LEF/TCF isoforms and associated corepressors establish repressive complexes that shut down transcription in the absence of Wnt signaling.
Materials and methods
Protein expression and purification
Recombinant His-tagged LEF-1 and GST-tagged β-catenin (β-cat and FLβ-cat) proteins were affinity-purified from bacterial lysates for use in chromatin transcription and binding reactions. The full-length human LEF-1 and various mutants described in the text were subcloned into pET28a+ (Novagen) by standard PCR methods to yield constructs encoding in-frame fusions with a C-terminal six-His affinity tag. PCR fragments encoding ΔN-LEF were inserted into the NcoI and HindIII sites of pET28a+. ΔAD-LEF was subcloned by inserting LEF-1 amino acids 1–75 into the NcoI and PmlI sites of pET–LEF-1, replacing sequences up-stream of the HMG domain. Protein expression was induced by addition of IPTG to a final concentration of 2 mM and incubation at 37°C for 2 h. Protein pellets from 500 mL induced cultures were resuspended in 8 mL of His–Lysis buffer (PBS with NaCl added to a final concentration of 250 mM, 1% Triton X-100, 10 mM imidazole, 4 mM β-mercaptoethanol, 0.1 mM PMSF, 2 mg/mL benzamidine, 1 μg/mL pepstatin A, 4 μg/mL leupeptin, 10 μg/mL aprotinin, 20 μg/mL soybean trypsin inhibitor). Cleared lysates were incubated for 2 h at 4°C with 600 μL of Ni–NTA Superflow resin (QIAGEN). Bound proteins were eluted in His-elution buffer (HEG 0.1 M KCl, 300 mM imidazole, 4 mM β-mercaptoethanol, 0.1 mM PMSF), and further purified by chromatography on a Mono-S HR 5/5 column (Pharmacia). All purified protein preparations were dialyzed against HEG 0.1 M KCl supplemented with 2 mM DTT and 0.1 mM PMSF and frozen in small aliquots before use.
The full-length human β-catenin and truncated mutants were subcloned into pGEX–KG and expressed as fusion proteins with GST. A PCR fragment encoding the CH3 domain of p300 (amino acids 1723–1815) was inserted into the NcoI and HindIII sites of pGEX–KG. Transformed bacterial pellets were resuspended in 8 mL of GST–Lysis buffer (50 mM Tris at pH 7.9, 100 mM KCl, 1% Triton X-100, 2 mM DTT. 0.1mM PMSF, 2 mg/mL benzamidine, 1 μg/mL pepstatin A, 4 μg/mL leupeptin, 10 μg/mL aprotinin, 20 μg/mL soybean trypsin inhibitor). Cleared lysates were incubated for 2 h at 4°C with 600 μL of glutathione sepharose 4B (Pharmacia). Bound proteins were eluted three times with 0.75 mL of GST-elution buffer (50 mM Tris-HCl at pH 7.9, 100 mM KCl, 15 mM reduced glutathione, 0.2 mM EDTA, 10% glycerol, 2 mM DTT, 0.1 mM PMSF). Eluted proteins were further purified by loading onto a Mono-Q HR 5/5 column (Pharmacia) and eluted with a 120–300 mM KCl gradient. Purified recombinant proteins were dialyzed against HEG 0.1 M KCl with 2 mM DTT and 0.1 mM PMSF and frozen in small aliquots before use.
Chromatin assembly and transcription
Chromatin assembly was performed as described previously (Bulger and Kadonaga 1994). For chromatin reconstitution of pBRE or pHIV, 1.25 μg of supercoiled plasmid DNA was used in each 250 μL of chromatin assembly reaction. For the experiments shown in Figures 1–4, 6 and 7, the enhancer factors and, where indicated, the inhibitors or NSAIDs, were incubated with the pBRE template during chromatin assembly, whereas the enhancer factors were incubated with the pBRE chromatin template after nucleosome assembly for the experiments shown in Figures 5 and 6. The chromatin template (1.25 μg of DNA in 250 μL) was incubated with the enhancer factors for 30 min at 30°C, and 20-μL aliquots were incubated with 10 μL of HeLa cell nuclear extract (8–12 mg/mL) and 25 ng of nonchromatin α-globin DNA for 30 min at 30°C. Transcription was analyzed by primer extension as described by Mayall et al. (1997), and RNA levels were quantitated by PhosphorImager scanning.
DNA- and chromatin-binding experiments
EMSA were carried out with the high-affinity LEF-1 binding site (5′-TCTCAGCAGTCTTTGTAGTACAGCAGTCATAGTAG TA-3′) in a final reaction volume of 15 μL containing 20 mM HEPES (pH 8.0), 50 mM KCl, 2.5 mM EDTA, 8 mM MgCl2, 5 mM spermidine, 30–35 μg/mL poly(dIdC), 250 μg/mL BSA, 0.025% NP-40 and 15% glycerol. DNase I footprint reactions on chromatin were carried out as described previously (Mayall et al. 1997).
Acknowledgments
We thank Tsutomu Nakamura and T. Akiyama (Tokyo, Japan) for the GST–ICAT plasmid; Glenn McAlpine, Eric Suess, and Nathan Gomes for their contributions to the analysis of dominant negative inhibitors; and the members of our laboratory for their comments on the paper. This work was funded by grants to K.A.J. from the National Institutes of Health. A.V.T. carried out these studies as part of his doctoral research in the Graduate Program in Biology at the University of California, San Diego, and also received support from the Chapman Foundation. C.J.F. is supported by the Cancer Research fund of the Damon Runyan-Walter Winchell Foundation Fellowship (DRG-1610).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jones@salk.edu; FAX (858) 535-8194.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.946501.
References
- Aoki M, Hecht A, Kruse U, Kemler R, Vogt PK. Nuclear endpoint of Wnt signaling: Neoplastic transformation induced by transactivating lymphoid-enhancing factor 1. Proc Natl Acad Sci. 1999;96:139–144. doi: 10.1073/pnas.96.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong JA, Bieker JJ, Emerson BM. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H. The chromatin remodeling factor Brg-1 interacts with β-catenin to promote target gene activation. EMBO J. 2001;20:4935–4943. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A, Huber O, Kemler R. Pontin52, an interaction partner of beta-catenin, binds to the TATA box binding protein. Proc Natl Acad Sci. 1998;95:14787–14792. doi: 10.1073/pnas.95.25.14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- Boolbol S, Dannenberg A, Chadburn A, Martucci C, Guo X, Ramonetti J, Abreu-Goris M, Newmark H, Lipkin M, DeCosse J, Bertagnolli M. Cyclooxygenase-2 overexpression and tumor formation are blocked by sulindac in a murine model of familial adenomatous polyposis. Cancer Res. 1996;56:2556–2560. [PubMed] [Google Scholar]
- Brunner E, Peter O, Schweizer L, Basler K. pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature. 1997;385:829–833. doi: 10.1038/385829a0. [DOI] [PubMed] [Google Scholar]
- Bulger M, Kadonaga J. Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol Genet. 1994;5:242–262. [Google Scholar]
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor Groucho in Drosophila development. Genes & Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R, Treisman J. Osa-containing Brahma chromatin remodeling complexes are required for the repression of Wingless target genes. Genes & Dev. 2000;14:3140–3152. doi: 10.1101/gad.854300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dihlmann S, Siermann A, von Knebel Doeberitz M. The nonsteroidal anti-inflammatory drugs aspirin and indomethacin attenuate beta-catenin/TCF-4 signaling. Oncogene. 2000;20:645–653. doi: 10.1038/sj.onc.1204123. [DOI] [PubMed] [Google Scholar]
- Eastman Q, Grosschedl R. Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr Opin Cell Biol. 1999;11:233–240. doi: 10.1016/s0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- Galceran J, Hsu SC, Grosschedl R. Rescue of a Wnt mutation by an activated form of LEF-1: Regulation of maintenance but not initiation of Brachyury expression. Proc Natl Acad Sci. 2001;98:8668–8673. doi: 10.1073/pnas.151258098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gat U, DasGupta R, Degenstein L, Fuchs E. De novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- Graham TA, Weaver C, Mao F, Kimelman D, Xu W. Crystal structure of a beta-catenin/Tcf complex. Cell. 2000;103:885–896. doi: 10.1016/s0092-8674(00)00192-6. [DOI] [PubMed] [Google Scholar]
- He TC, Chan TA, Vogelstein B, Kinzler KW. PPARdelta is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell. 1999;99:335–345. doi: 10.1016/s0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Kemler R. Curbing the nuclear activities of β-catenin. EMBO Reports. 2000;1:24–28. doi: 10.1093/embo-reports/kvd012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Litterst CM, Huber O, Kemler R. Functional characterization of multiple transactivating elements in beta-catenin, some of which interact with the TATA-binding protein in vitro. J Biol Chem. 1999;274:18017–18025. doi: 10.1074/jbc.274.25.18017. [DOI] [PubMed] [Google Scholar]
- Hecht A, Vleminckx K, Stemmler M, van Roy F, Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 2000;19:1839–1850. doi: 10.1093/emboj/19.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovanes K, Li T, Munguia J, Truong T, Milovanovic T, Lawrence Marsh J, Holcombe R, Waterman M. Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat Genet. 2001;28:53–57. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- Hsu SC, Galceran J, Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- Huelsken J, Birchmeier W. New aspects of Wnt signaling pathways in higher vertebrates. Curr Opin Genet Dev. 2001;11:547–553. doi: 10.1016/s0959-437x(00)00231-8. [DOI] [PubMed] [Google Scholar]
- Kadam S, McAlpine GS, Phelan ML, Kingston RE, Jones KA, Emerson BM. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes & Dev. 2000;14:2441–2451. doi: 10.1101/gad.828000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Oda T, Itoh M, Jiang D, Artinger KB, Chandrasekharappa SC, Driever W, Chitnis AB. Repressor activity of Headless/Tcf3 is essential for vertebrate head formation. Nature. 2000;407:913–916. doi: 10.1038/35038097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolligs F, Hu G, Dang CV, Fearon E. Neoplastic transformation of RK3E by mutant beta-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol Cell Biol. 1999;19:5696–5706. doi: 10.1128/mcb.19.8.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolligs F, Kolligs B, Hajra K, Hu G, Tani M, Cho K, Fearon E. gamma-catenin is regulated by the APC tumor suppressor and its oncogenic activity is distinct from that of beta-catenin. Genes & Dev. 2000;14:1319–1331. [PMC free article] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin–Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Kraus WL, Manning ET, Kadonaga JT. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol Cell Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D, Goldstein RE, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayall TP, Sheridan PL, Montminy MR, Jones KA. Distinct roles for P-CREB and LEF-1 in TCR alpha enhancer assembly and activation on chromatin templates in vitro. Genes & Dev. 1997;11:887–899. doi: 10.1101/gad.11.7.887. [DOI] [PubMed] [Google Scholar]
- Merrill BJ, Gat U, DasGupta R, Fuchs E. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes & Dev. 2001;15:1688–1705. doi: 10.1101/gad.891401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishi M, Fujii R, Hatta M, Yoshida E, Araya N, Nagafuchi A, Ishihara S, Nakajima T, Fukamizu A. Regulation of Lef-mediated transcription and p53-dependent pathway by associating beta-catenin with CBP/p300. J Biol Chem. 2000;275:35170–35175. doi: 10.1074/jbc.C000258200. [DOI] [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Nusse R. WNT targets: Repression and activation. Trends Genet. 1999;15:1–3. doi: 10.1016/s0168-9525(98)01634-5. [DOI] [PubMed] [Google Scholar]
- Pazin MJ, Hermann JW, Kadonaga JT. Promoter structure and transcriptional activation with chromatin templates assembled in vitro. A single Gal4–VP16 dimer binds to chromatin or to DNA with comparable affinity. J Biol Chem. 1998;273:34653–34660. doi: 10.1074/jbc.273.51.34653. [DOI] [PubMed] [Google Scholar]
- Polakis P. Wnt signaling and cancer. Genes & Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- Potter JD. Colorectal cancer: Molecules and populations. J Natl Cancer Inst. 1999;91:916–932. doi: 10.1093/jnci/91.11.916. [DOI] [PubMed] [Google Scholar]
- Ritland S, Leighton J, Hirsch R, Morrow J, Weaver A, Gendler S. Evaluation of 5-aminosalicylic acid (5-ASA) for cancer chemoprevention: Lack of efficacy against nascent adenomatous polyps in the Apc(Min) mouse. Clin Cancer Res. 1999;5:855–863. [PubMed] [Google Scholar]
- Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destree O, Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- Roose J, Huls G, van Beest M, Moerer P, van der Horn K, Goldschmeding R, Logtenberg T, Clevers H. Synergy between tumor suppressor APC and the beta-catenin-Tcf4 target Tcf1. Science. 1999;285:1923–1926. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- Sheridan PL, Sheline CT, Cannon K, Voz ML, Pazin MJ, Kadonaga JT, Jones KA. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes & Dev. 1995;9:2090–2104. doi: 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- Sheridan PL, Mayall TP, Verdin E, Jones KA. Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes & Dev. 1997;11:3327–3340. doi: 10.1101/gad.11.24.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider L, Thirlwell H, Miller J, Moon R, Groudine M, Tapscott S. Inhibition of Tcf3 binding by I-mfa domain proteins. Mol Cell Biol. 2001;21:1866–1873. doi: 10.1128/MCB.21.5.1866-1873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Kolligs F, Hottiger M, Mosavin R, Fearon E, Nabel G. Regulation of beta-catenin transformation by the p300 transcriptional coactivator. Proc Natl Acad Sci. 2000;97:12613–12618. doi: 10.1073/pnas.220158597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tago K, Nakamura T, Nishita M, Hyodo J, Nagai S, Murata Y, Adachi S, Ohwada S, Morishita Y, Shibuya H, Akiyama T. Inhibition of Wnt signaling by ICAT, a novel beta-catenin-interacting protein. Genes & Dev. 2000;14:1741–1749. [PMC free article] [PubMed] [Google Scholar]
- Takemaru K, Moon R. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J Cell Biol. 2000;149:249–254. doi: 10.1083/jcb.149.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- Travers A. Recognition of distorted DNA structures by HMG domains. Curr Opin Struct Biol. 2000;10:102–109. doi: 10.1016/s0959-440x(99)00056-1. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, et al. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- Yin MJ, Yamamoto Y, Gaynor RB. The anti-inflammatory agents aspirin and salicylate inhibit the activity of I(kappa)B kinase-beta. Nature. 1998;396:77–80. doi: 10.1038/23948. [DOI] [PubMed] [Google Scholar]
- Zhurinsky J, Shtutman M, Ben-Ze'ev A. Differential mechanisms of LEF/TCF family-dependent transcriptional activation by beta-catenin and plakoglobin. Mol Cell Biol. 2000;20:4238–4252. doi: 10.1128/mcb.20.12.4238-4252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]