An increasing number of mitochondrial preproteins are sequentially processed upon import by the presequence mitochondrial processing peptidase (MPP) and the intermediate peptidase octapeptidyl aminopeptidase 1 (Oct1). We show that Oct1 removes destabilizing residues from import intermediates generated by MPP. Oct1 therefore acts as a quality control system, preventing premature substrate degradation.
Abstract
Most mitochondrial proteins are encoded in the nucleus as precursor proteins and carry N-terminal presequences for import into the organelle. The vast majority of presequences are proteolytically removed by the mitochondrial processing peptidase (MPP) localized in the matrix. A subset of precursors with a characteristic amino acid motif is additionally processed by the mitochondrial intermediate peptidase (MIP) octapeptidyl aminopeptidase 1 (Oct1), which removes an octapeptide from the N-terminus of the precursor intermediate. However, the function of this second cleavage step is elusive. In this paper, we report the identification of a novel Oct1 substrate protein with an unusual cleavage motif. Inspection of the Oct1 substrates revealed that the N-termini of the intermediates typically carry a destabilizing amino acid residue according to the N-end rule of protein degradation, whereas mature proteins carry stabilizing N-terminal residues. We compared the stability of intermediate and mature forms of Oct1 substrate proteins in organello and in vivo and found that Oct1 cleavage increases the half-life of its substrate proteins, most likely by removing destabilizing amino acids at the intermediate's N-terminus. Thus Oct1 converts unstable precursor intermediates generated by MPP into stable mature proteins.
INTRODUCTION
Virtually all mitochondrial proteins are synthesized on cytosolic ribosomes and are imported into the organelle in a posttranslational manner. Approximately 70% of these mitochondrial preproteins use the presequence pathway for import into mitochondria, involving the translocase of the outer membrane (TOM) and the presequence translocase of the inner membrane (TIM23) complexes ( Neupert and Herrmann, 2007; Endo and Yamano, 2009; Vögtle et al., 2009; van der Laan et al., 2010; Schmidt et al., 2010). Those preproteins possess N-terminal positively charged extensions that form amphipathic α-helices required for targeting and import into the organelle. Most presequences are removed upon import by the mitochondrial processing peptidase (MPP) located in the matrix ( Witte et al., 1988; Gavel and von Heijne, 1990; Glaser et al., 1998; Gakh et al., 2002; Habib et al., 2007; Huang et al., 2009; Yogev and Pines, 2011).
Some preproteins can undergo a second processing event after MPP cleavage. This can be performed, for example, by the inner membrane peptidase (IMP) or the rhomboid protease processing of cytochrome c peroxidase (Pcp1), which act to regulate submitochondrial sorting of preproteins ( Esser et al., 2002; Koppen and Langer, 2007). A further intermediate cleaving peptidase, Icp55, which cleaves preprotein intermediates after initial processing by MPP has recently been identified ( Naamati et al., 2009; Vögtle et al., 2009). Icp55 typically removes one amino acid residue from preprotein intermediates that carry a destabilizing amino acid residue at their N-terminus according to the N-end rule for bacterial proteins ( Mogk et al., 2007; Varshavsky, 2008). This leads to the conversion of a destabilizing N-terminus into a stable one, thereby preventing premature degradation of substrate proteins ( Vögtle et al., 2009).
Another protease, which cleaves precursor intermediates generated by MPP, is the matrix-located mitochondrial intermediate peptidase (MIP), octapeptidyl aminopeptidase 1 (Oct1). Oct1 cleaves an octapeptide from the N-terminus of the preprotein intermediate, thus generating the mature protein ( Gakh et al., 2002). Oct1-dependent substrate proteins were found in the mitochondrial matrix and inner membrane, and their functions are quite diverse. They play a role in the respiratory chain, the citric acid cycle, genome maintenance and translation, the urea cycle, protein folding, and stress response. Based on the presequences of the Oct1 substrates known so far, the cleavage motif RX(↓)(F/L/I)XX(T/S/G)XXXX(↓) has been described (arrows indicate MPP and Oct1 cleavage sites, respectively) ( Sztul et al., 1987; Kalousek et al., 1988; Hendrick et al., 1989; Branda and Isaya, 1995; Gakh et al., 2002; Vögtle et al., 2009). However, the functional role of the cleavage of an octapeptide by Oct1 is still not understood. It was speculated that the mature N-termini of dual-processed precursors would be incompatible with MPP processing and that the octapeptide is required to separate the MPP cleavage site from the mature part of the protein ( Gavel and von Heijne, 1990; Isaya et al., 1991). In the cases of the Oct1 substrates Cox4 and Rip1, it was proposed that their intermediate forms could be either missorted or involved in regulation of the assembly rate of respiratory complexes ( Isaya et al., 1994; Nett et al., 1997). However, it was also shown that the presequence of Rip1 can be cleaved by MPP in one step when the Oct1 cleavage site is replaced by an MPP motif ( Nett et al., 1997) and that the Rip1 preprotein can assemble and function also in its intermediate form ( Nett and Trumpower, 1999). To date, no explanation for the two-step cleavage by MPP/Oct1 has been found that would be applicable to general features of Oct1 substrate proteins.
We aimed to identify common characteristics of Oct1 substrate proteins that could lead to the uncovering of a functional role of Oct1. A recent proteomic approach led to the identification of five further substrates in Saccharomyces cerevisiae mitochondria ( Vögtle et al., 2009) that possess the largest number of Oct1 substrates known. Systematic inspection of their sequences revealed that the most common characteristic is a destabilizing N-terminal amino acid in the intermediate forms. This led to the identification of 1) a novel Oct1 substrate with an unusual sequence motif and 2) a novel function of Oct1 in converting destabilizing N-termini of intermediates into stable N-termini of the mature proteins. Thus Oct1 acts as a quality control system for MPP-processed preproteins.
RESULTS
Ygr031w/Imo32 encodes a novel Oct1 substrate protein with an unusual sequence motif
For the Oct1 substrate proteins identified to date, a motif including three characteristic amino acid residues, RX(↓)(F/L/I)XX(T/S/G)XXXX(↓), has been identified ( Hendrick et al., 1989; Gakh et al., 2002). The first arrow represents cleavage by MPP, whereas the second arrow indicates processing by Oct1. The most frequent amino acids within this motif are arginine at position −10 and phenylalanine at position −8 relative to the mature N-termini ( Vögtle et al., 2009). While arginine has been considered as strictly required in position −10 leucine and isoleucine were also tolerated in position −8 ( Hendrick et al., 1989; Gakh et al., 2002; Vögtle et al., 2009). Screening a large collection of experimentally determined mitochondrial presequences (N-proteome) from yeast for arginine in a −10 position led to the identification of five novel Oct1 substrate proteins ( Vögtle et al., 2009). We now screened the N-proteome for presequences that possess a phenylalanine in position −8. Those candidates were then tested by in organello import of the radiolabeled preproteins into isolated mitochondria from wild-type and oct1Δ yeast cells. For the as yet uncharacterized protein encoded by the open reading frame YGR031W, we found that the membrane potential–dependent processing from an intermediate form into the mature form is completely prevented in oct1Δ mitochondria ( Figure 1A), indicating that Ygr031w is a substrate of Oct1. We therefore termed the protein Imo32 for “intermediate cleaved by mitochondrial octapeptidyl aminopeptidase.” Imo32 is highly conserved from bacteria to humans, however, the functional role of this protein is still unclear. To test whether the first processing step of the Imo32 preprotein is mediated by MPP, we used a conditional mutant, which expresses one of the two MPP subunits (mas1), from a temperature-sensitive allele ( Witte et al., 1988). In this strain, MPP function can be impaired by incubating isolated mitochondria at 37°C during in organello import ( Vögtle et al., 2009). When radiolabeled Imo32 was imported into mas1 mitochondria at nonpermissive temperatures, the full-length precursor accumulated without cleavage of the presequence, indicating that MPP processing of Imo32 preprotein precedes processing by Oct1 ( Figure 1B). The N-terminus of mature Imo32 has been determined as glutamine 47 ( Vögtle et al., 2009). This implies that the predicted MPP cleavage site contains a cysteine at position −2 (position −10 relative to the mature protein) instead of arginine, which would not fit to the characteristic three amino acid motif of Oct1 substrate proteins ( Hendrick et al., 1989; Gakh et al., 2002). (To exclude any sequencing error, we also confirmed the DNA sequence coding for the Imo32 presequence including cysteine −10.) In yeast, only one further variation at this position has been found, with Prx1 allowing a lysine instead of arginine at position −10 ( Vögtle et al., 2009). To assess the intermediate N-terminus of Imo32, we synthesized radiolabeled truncated versions lacking either the complete presequence (Δ1–46) or the MPP-processed part (Δ1–38), and compared their migration on high-resolution SDS–PAGE with the processed forms after in organello import ( Figure 1C). In wild-type mitochondria truncated Imo32Δ1–46 migrated like the imported and fully processed protein ( Figure 2C, lanes 1–4; Vögtle et al., 2009), while in oct1Δ mitochondria, the truncated Imo32Δ1–38 migrated like the intermediate form ( Figure 2C, lanes 5–8), confirming that the actual cleavage by MPP indeed occurs very close to the predicted cleavage site (the next positively charged residue of Imo32, lysine, is located at position −14 and thus too far away to function as MPP recognition motif). We mutated the cysteine in position −2 (with regard to the predicted MPP cleavage site) to either alanine or aspartate and found that precursors with these residues are also processed by MPP upon in organello import, suggesting the presence of an unusual MPP cleavage site in the presequence of Imo32 ( Figure 1D). It must be considered that the peptide removed by Oct1 is shorter or longer than eight residues, and thus a residue different from cysteine −10 would be critical for cleavage by MPP. However, since the next positively charged residue in this case is also too far away, an unusual MPP recognition motif of an Oct1 substrate would be used.
FIGURE 1:
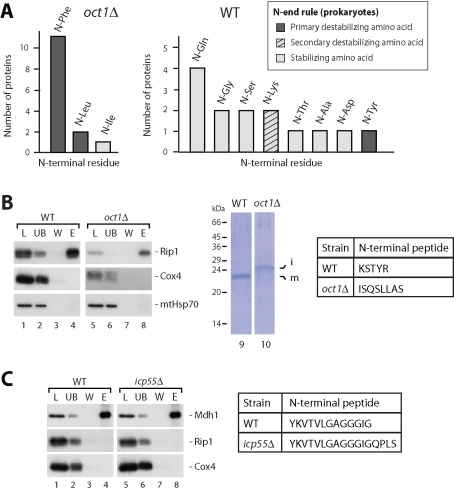
Ygr031w/Imo32 is a novel Oct1 substrate with an unusual cleavage site motif. (A) Radiolabeled Imo32 precursor was incubated with mitochondria isolated from wild-type (WT) or oct1Δ yeast cells, treated with proteinase K, and analyzed by SDS–PAGE and digital autoradiography. Where indicated, the membrane potential (Δψ) was dissipated prior to the import reaction. prec. and p, precursor; i, intermediate; m, mature. (B) Import of [35S]Imo32 into wild-type and mas1 mutant mitochondria. Samples were treated as in (A). (C) Radiolabeled Imo32 preprotein was incubated with isolated yeast mitochondria, and then treated with proteinase K. The samples, as well as 35S-labeled truncated forms of Imo32, were analyzed by SDS–PAGE and digital autoradiography. (D) Import of [35S]Imo32 with altered presequences into wild-type mitochondria. Cys-37 (corresponding to position −2 with regard to the proposed MPP cleavage site) was replaced by either alanine/A or aspartate/D. (E) Alignment of the cleavage motif of the 14 Oct1 substrate proteins from yeast. Characteristic three amino acid motif is highlighted in dark gray. Light gray denotes first amino acid of mature protein. Arrows indicate cleavage by MPP and Oct1, respectively. (F) Amino acid blot of relative frequency of the presequences (10 residues of the C-terminal segment) and first mature amino acids of the Oct1 substrate proteins identified in yeast (see Figure 1E). Arrows show cleavage sites of MPP and Oct1.
FIGURE 2:
Oct1-processing intermediates possess destabilizing N-terminal amino acids. (A) N-terminal amino acids of the 14 Oct1 substrate proteins in oct1Δ (left panel) and wild-type (WT; right panel) mitochondria. Amino acids are classified according to the N-end rule for Escherichia coli. (B) Immunoprecipitation of Rip1 from wild-type (WT) and oct1Δ mitochondria. Samples were analyzed by SDS–PAGE and immunoblotting (left panel) or protein bands of the elution fraction were visualized by Coomassie Blue staining (right panel). L, load (5%); UB, unbound (5%); W, wash (5%); E, elution (100%). The table shows the amino acid sequence of the Rip1 N-terminus from wild-type (WT) and oct1Δ mitochondria obtained by Edman degradation. (C) Immunoprecipitation of Mdh1 from wild-type (WT) and icp55Δ mitochondria ( Vögtle et al., 2009). Analysis was performed as in (B).
We also noticed that the amino acid leucine at position −5 does not correspond to the published Oct1 motif, which would require threonine, serine, or glycine. This position, however, seems to be generally more variable, as shown by an alignment of the 10 N-terminal amino acids (relative to the mature form) from all experimentally verified Oct1 substrate proteins in S. cerevisiae ( Figure 1E). An amino acid frequency profile of this region is shown in Figure 1F.
Thus Imo32 represents a novel Oct1 substrate protein, which matches only in one amino acid (phenylalanine −8) the characteristic three amino acid motif known to date.
Oct1-processing intermediates possess destabilizing N-terminal amino acids
Inspection of the N-terminal amino acid residues from intermediate and mature forms of the Oct1 substrate proteins revealed that most of the intermediates carry a destabilizing amino acid at the N-terminus, while most of the mature forms carry a stabilizing amino acid according to the N-end rule for degradation of bacterial proteins ( Figure 2A; Mogk et al., 2007; Varshavsky, 2008). The intermediate forms of 11 substrate proteins start with phenylalanine and 2 with leucine. Both phenylalanine and leucine are primary destabilizing amino acids ( Mogk et al., 2007; Varshavsky, 2008). However, in the case of Rip1, the intermediate form carries an isoleucine representing a stabilizing amino acid. Regarding the mature N-termini, we found 10 proteins with a stabilizing amino acid, 2 with secondary destabilizing amino acids, and in the case of Mdh1, the primary destabilizing amino acid was determined to be tyrosine. For Mdh1, it is important to note that within the group of destabilizing amino acids N-tyrosine was found to be more stable than N-phenylalanine ( Bachmair et al., 1986). In the case of Rip1, we observed that the presequence contains a destabilizing leucine in position −9. It has been speculated, that Oct1 could also remove intermediate peptides with 9 amino acids ( Gavel and von Heijne, 1990). The N-terminus of the yeast Rip1 intermediate in oct1Δ mitochondria has not yet been determined experimentally. We therefore isolated Rip1 by immunoprecipitation from wild-type and oct1Δ mitochondria, and determined the corresponding N-terminal amino acids by Edman degradation. We confirmed the mature N-terminal amino acid lysine ( Beckmann et al., 1987), and found that the intermediate indeed starts with isoleucine ( Figure 2B). Regarding Mdh1, we wondered if the N-terminal tyrosine could be additionally cleaved by Icp55, thus generating a stabilizing N-terminus of the mature protein. We isolated Mdh1 from wild-type and icp55Δ mitochondria, and found by Edman sequencing that in both cases the N-terminal residue is a tyrosine ( Figure 2C; Thompson et al., 1988).
Our results indicate that, with the exception of Rip1 and Mdh1, Oct1 substrate proteins possess destabilizing N-termini at their preprotein intermediates and stabilizing residues at their mature N-terminus.
Oct1 processing of preprotein intermediates in mitochondria increases protein stability
To test whether processing of preprotein intermediates by Oct1 indeed plays a role in regulation of protein turnover, we planned to compare the stability of substrate proteins in wild-type and oct1Δ mitochondria. It is known that deletion of OCT1 results in loss of mitochondrial DNA and, therefore, respiratory deficiency ( Isaya et al., 1994). When we compared the import efficiency of the matrix-targeted precursor protein Atp2 into oct1Δ and wild-type (Rho+) mitochondria, we found a reduced import rate in oct1Δ mitochondria ( Figure 3A, Top), which is probably due to a reduced membrane potential in the mutant. However, import capability seemed to be enhanced in oct1Δ mitochondria when compared with wild-type mitochondria from a Rho0 yeast strain ( Meisinger et al., 2004) that lacks mitochondrial DNA ( Figure 3A, Bottom). Therefore we decided to reexpress Oct1 under its endogenous promoter and terminator in the oct1Δ background and to use it as a control strain. Indeed, the import efficiency for Atp2 was now similar in mitochondria from the oct1Δ strain, which reexpressed Oct1 from a plasmid (oct1Δ OCT1) and the oct1Δ strain transformed with an empty control vector ( Figure 3B, Top). Recovery of Oct1 function after reexpression was controlled by full processing of the Cox4 precursor intermediate upon import into isolated mitochondria ( Figure 3B, Bottom).
FIGURE 3:
Reexpression of Oct1 in oct1Δ yeast cells. (A) Radiolabeled Atp2 precursor was incubated with mitochondria from WTRho+, oct1Δ, or WTRho0 cells over increasing periods of time (3, 6, 12 min). Samples were treated with proteinase K and analyzed by SDS–PAGE and digital autoradiography. For the longest time point, the membrane potential (Δψ) was dissipated prior to the import reaction. p, precursor; m, mature. (B) Mitochondria isolated from oct1Δ yeast cells transformed with the empty control plasmid and oct1Δ OCT1 yeast reexpressing Oct1 from its endogenous promoter were incubated with 35S-Atp2 or 35S-Cox4. Samples were analyzed as described in (A) and Materials and Methods. i, intermediate.
To monitor protein stability, isolated mitochondria from oct1Δ and oct1Δ OCT1 yeast cells were incubated over an increasing period of time. Samples were taken at regular time points and analyzed by SDS–PAGE and immunoblotting. We chose three different Oct1 substrates for analysis: Cox4 as a typical substrate with respect to the N-terminal amino acids in the intermediate and mature forms, Mdh1 with a destabilizing amino acid at both N-termini, and Rip1 with a stabilizing amino acid at the N-terminus of the intermediate form. For Mdh1 and Cox4, we observed a faster degradation in oct1Δ mitochondria compared with mitochondria from the control strain, while Rip1 and the control protein Atp2 were degraded with similar kinetics in both samples ( Figure 4A, lanes 1–14). Due to the faster turnover rate of Rip1 and Cox4, a second degradation kinetic with shorter time intervals was performed ( Figure 4A, lanes 15–28). Again, Cox4 was found to be less stable in oct1Δ mitochondria, while Rip1 stability was unaffected, as were the control proteins Atp2, mtHsp70, Atp3, and Nfs1. Atp3 and Nfs1 were recently identified as substrate proteins of the intermediate cleaving peptidase, Icp55 ( Naamati et al., 2009; Vögtle et al., 2009). Quantification revealed a reduction of approximately 50% for Mdh1 (after 166 h) and 70% for Cox4 (after 76 h) in oct1Δ mitochondria compared with control samples. Stability of Rip1, however, was unaffected or even slightly higher after 76 h in oct1Δ ( Figure 4B). We conclude that processing of preprotein intermediates by Oct1 leads to a stabilization of proteins due to the removal of destabilizing N-terminal amino acids that appear after initial cleavage by MPP. Stability of Rip1, which represents the only Oct1 substrate in yeast with a stabilizing N-terminal amino acid in the intermediate form, is not affected, thus fully confirming our finding.
FIGURE 4:
The half-life of Oct1 substrate proteins is prolonged upon full processing of the intermediate forms in organello. (A) Mitochondria from oct1Δ and oct1Δ OCT1 (reexpressing the OCT1 gene) yeast cells were incubated at 37°C for the indicated time. Mitochondria were reisolated and analyzed by SDS–PAGE and immunoblotting. N-terminal amino acids of Oct1 intermediate (Int.) and mature (Mat.) forms of the proteins tested are indicated. (B) Quantification of protein turnover: graphs indicate averaged protein levels of the last incubation time points (166, 76 h) in comparison to the level at 0 h. Values of oct1Δ OCT1 mitochondria were set to 100% (control). Error bars represent SD from three different experiments.
Accelerated degradation of Oct1-dependent precursor intermediates in vivo
We next compared the stability of the intermediate and mature forms of Oct1 substrates in vivo. For this purpose, oct1Δ and oct1Δ OCT1 cells were treated with cycloheximide to block translation, and samples were taken after different time spans. Whole cell extracts were separated by SDS–PAGE and protein degradation profiles were analyzed by immunoblotting ( Vögtle et al., 2009; Claypool et al., 2011). For Mdh1 we observed a fast degradation of the intermediate form, while the fully processed mature form was stable over the whole time frame tested ( Figure 5A). Quantification revealed that the level of the Mdh1 intermediate form decreased to < 40% after 46 h compared with the mature form, while the stability of the control proteins Atp2 and mtHsp70 was not dependent on Oct1 ( Figure 5A). The cytosolic Pgk1 was used as a loading control. Due to their faster degradation kinetic, the stability of Cox4 and Rip1 was tested in a shorter time frame ( Figure 5B) (the half-lives of different mitochondrial proteins have been found to cover a broad range from hours to several days; Kambacheld et al., 2005; Belle et al., 2006; Major et al., 2006; Yen et al., 2008; Shen et al., 2008; Vögtle et al., 2009; Claypool et al., 2011). Cox4 intermediate form decreased to approx. 50% compared with the mature form after 9 h, while the degradation kinetic of Rip1 was not affected by the absence of Oct1 ( Figure 5B). Our results also show that in vivo Oct1-dependent precursor intermediates with destabilizing N-terminal amino acid residues are much more rapidly degraded than their fully processed mature forms.
FIGURE 5:
Oct1 processing leads to stabilization of its substrate proteins in vivo. (A) and (B) In vivo degradation of proteins analyzed in oct1Δ and oct1Δ OCT1 yeast cells. Protein translation was blocked by addition of cycloheximide. Samples were obtained at various time points, subjected to postalkaline protein extraction, and analyzed by SDS–PAGE and immunoblotting. N-terminal amino acids of Oct1 intermediate (Int.) and mature (Mat.) forms of the proteins tested are indicated. Quantification was performed as in Figure 4.
DISCUSSION
Since its discovery 20 yr ago, several different functions have been assigned to Oct1 and its homologues from other organisms. It was proposed that the removal of an octapeptide after initial processing by MPP separates the MPP cleavage site from mature parts of the N-terminus or regulates submitochondrial protein sorting or is involved in protein complex assembly ( Gavel and von Heijne, 1990; Isaya et al., 1991, 1994; Nett et al., 1997). However, so far no satisfying experimental evidence corroborating these different theories has been shown, as all proposed functions were presented only for a single substrate and could never be applied to general features in the sequence motif of Oct1 substrates. The recent identification of novel substrate proteins by a proteomic approach in yeast enabled a more comprehensive analysis of common characteristics of Oct1 substrates. We found two interesting new features of the cleavage site motif in Oct1-dependent preproteins. First, MPP does not strictly require an arginine in position −10 of Oct1 substrate proteins, but can cleave precursors with lysine (Prx1) or cysteine, alanine, or aspartate (Imo32) residues at this position. This raises the possibility that many more Oct1 substrates with less characteristic sequence motifs await identification. Second, the N-termini of the Oct1 intermediates can be classified as destabilizing amino acids according to the N-end rule, whereas processing by Oct1 converts those N-termini into more stabilizing ones. Mutational analysis of the Oct1 cleavage site motif has shown that the amino acid in position −8 is the crucial one for recognition and cleavage by Oct1 ( Isaya et al., 1992). Using in organello and in vivo degradation assays we could show that processing by Oct1 indeed leads to a prolonged half-life of its substrate proteins. A first indication that Oct1-dependent precursor intermediates are degraded faster than the mature proteins was reported by Isaya et al. (1994); however, this rapid degradation was explained by a possible missorting of the precursor protein and not connected to a destabilizing N-terminus of the intermediate.
While we cannot exclude further functions of the octapeptide, like allowing a more efficient processing of precursors by MPP, the most common feature of the Oct1 substrates is the different classification of their intermediate and mature N-terminal amino acids according to the N-end rule. Only 2 of the 13 Oct1 substrates analyzed here are showing exceptions in the categorization of intermediate/destabilizing and mature/more stabilizing N-termini. 1) The intermediate form of the Rip1 protein has the stabilizing amino acid isoleucine at its N-terminus and is nevertheless processed by the Oct1 protease. Interestingly, the bovine homologue (Rieske iron–sulfur protein) is processed in a single step resulting in isoleucine at the mature N-terminus. It is likely that the two-step processing has been abolished during evolution, due to the already stabilizing N-terminus after the first cleavage by MPP. In yeast, intermediate Rip1 and mature-sized Rip1 are degraded at a similar rate. Interestingly the N-terminal amino acid of mature Rip1, lysine, represents a secondary destabilizing amino acid according to the N-end rule in prokaryotes ( Mogk et al., 2007; Varshavsky, 2008; Dougan et al., 2010). Lysine is also found at the mature N-terminus of another Oct1 substrate protein, Prx1. In the cytosol of eukaryotes and prokaryotes, such residues are typically converted by specific enzymes into primary destabilizing amino acid ( Mogk et al., 2007; Varshavsky, 2008; Dougan et al., 2010). However, no homologues of these enzymes have been identified in mitochondria so far. Therefore lysine could be considered as a stabilizing residue in line with our results. 2) Regarding Mdh1, we found that the mature form with an N-terminal tyrosine is more stable than the intermediate form, which carries a phenylalanine. Both residues are classified as primary destabilizing amino acids, but systematic analysis of fusion constructs exposing different residues at the N-termini revealed that proteins with N-terminal tyrosine are three times more stable than those starting with phenylalanine ( Bachmair et al., 1986), explaining the more rapid degradation of the Mdh1 intermediate form found in this study. The Mdh1 homologues in mouse and rat have maintained the destabilizing phenylalanine in position −8, but their mature N-termini possess the stabilizing amino acid alanine ( Hendrick et al., 1989).
When we compared verified Oct1 substrate proteins from other organisms, we found that they also contain primary destabilizing N-terminal amino acids at the intermediate form and stabilizing ones at the mature form in full agreement with our findings ( Hartl et al., 1986; Sztul et al., 1987; Grant et al., 1986; Tropschug et al., 1988; Hendrick et al., 1989). It is not determined which proteolytic systems mediate degradation of mitochondrial proteins according to the N-end rule pathway ( Vögtle et al., 2009; Dougan et al., 2010). Due to the absence of a ubiquitin–proteasome system in mitochondria, putative candidates comprise eukaryotic homologues of the ClpP family, which are involved in the bacterial N-end rule pathway, as well as Pim1 (Lon; Meinnel et al., 2006; Mogk et al., 2007; Dougan et al., 2010). (ClpP is present in mitochondria of higher eukaryotes but not in yeast.) Mitochondrial presequences have become essential upon transfer of mitochondrial genes to the nucleus during evolution in order to enable import of nuclear encoded mitochondrial proteins. Due to the presence of the highly conserved Oct1 and Icp55 peptidases in the mitochondrial matrix, an optimization of the presequence cleavage motif by MPP according to the N-end rule has not been necessary. Icp55 and Oct1 apparently recognize different subsets of precursor intermediates. Whereas N-terminal tyrosine is not found at intermediates recognized and cleaved by Oct1, this residue represents the main substrate of Icp55 ( Vögtle et al., 2009).
In summary, our results indicate that Oct1 and Icp55 together constitute a quality control system in the mitochondrial matrix, ensuring stabilization of precursor protein intermediates upon import into the organelle.
MATERIALS AND METHODS
Yeast strains
The S. cerevisiae strains used are derived from BY4741: wild-type strain (Mata, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) and deletion strain of Oct1 (Mata, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YKL134c::kanMX4). For reexpression of OCT1 in the oct1Δ strain, the open reading frame, which included the endogenous promoter and terminator region, was cloned into the pRS413 expression vector. Yeast cells were grown on minimal medium (6.7% [wt/vol] yeast nitrogen base without amino acids, 2% [wt/vol] glucose, 0.77% [wt/vol] Complete Supplement Mixture [BD Biosciences] minus histidine).
Isolation of mitochondria
Yeast cells were grown at 30°C on nonfermentable (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto Peptone, 3% [wt/vol] glycerol, pH 5.0) or fermentable (2% [wt/vol] sucrose instead of glycerol) medium. For yeast strains transformed with expression plasmids, cells were grown on minimal medium minus histidine, as described above. Cells were harvested at an OD600 of 0.7–1.5 and disrupted by homogenization, and mitochondria were isolated by differential centrifugation, as previously described ( Meisinger et al., 2006). Aliquots were stored in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS-KOH, pH 7.2) at −80°C.
Immunoprecipitation
Mitochondria (3 mg protein) were solubilized in lysis buffer (1% [wt/vol] SDS, 0.5% (vol/vol) Triton X-100, 150 mM NaCl, 10 mM Tris-HCl, pH 7.2) and incubated for 5 min at 95°C. Protein A Sepharose (GE Healthcare) loaded with antibodies against Mdh1 or Rip1 was equilibrated with lysis buffer without SDS and incubated with mitochondrial extract for 2 h at 4°C. After washing, proteins were eluted with 100 mM glycine, pH 2.5, precipitated with trichloroacetic acid (TCA), and analyzed by SDS–PAGE and Western blotting. Rip1 and Mdh1 protein bands were visualized by staining with Coomassie Brilliant Blue after transfer on PVDF membranes. Amino acid sequences were determined by Edman degradation (pulse liquid-phase sequencer, Procise 492 cLC; Applied Biosystems, Foster City, CA).
In vitro import of preproteins into mitochondria
Radiolabeled precursor proteins were generated by in vitro transcription/translation in the presence of [35S]methionine using a rabbit reticulocyte lysate system (Promega). Mitochondria (40 μg) and 3 μl radiolabeled precursor proteins were incubated in import buffer (10 mM MOPS-KOH, pH 7.2, 3% [wt/vol] bovine serum albumin [BSA], 250 mM sucrose, 5 mM MgCl2, 80 mM KCl, 5 mM KPi) supplemented with 2 mM ATP and 2 mM NADH. To abolish the membrane potential across the inner mitochondrial membrane, 1 μM valinomycin, 20 μM oligomycin, and 8 μM antimycin A (AVO mix) were added. The import reaction was stopped after the indicated time period by adding 0.01 volumes of AVO mix and putting the samples on ice. Samples were treated with 50 μg/ml proteinase K (PK) to remove nonimported precursor proteins. After PK digestion for 10 min on ice, the protease was inhibited by the addition of 2 mM phenylmethylsulfonyl fluoride (PMSF). Mitochondria were reisolated and washed with SEM buffer. For testing full Cox4 processing in oct1Δ OCT1 mitochondria, radiolabeled Cox4 was imported for 30 min into isolated mitochondria. After addition of AVO mix, mitochondria were reisolated and incubated for 60 min at 25°C. After SDS gel-electrophoresis, radiolabeled proteins were detected by digital autoradiography (PhosphorImager; GE Healthcare). For analysis of the truncated versions of Imo32, 40 μg mitochondrial extract per lane was loaded together with 0.2 μl of the radiolabeled protein onto SDS–PAGE.
For analysis of Imo32 presequence mutations, Imo32 was cloned into pRS415 and point mutations were introduced with the QuickChange II Site-Directed Mutagenesis Kit (Stratagene). All constructs were confirmed by sequencing.
Analysis of protein turnover
Mitochondrial protein turnover was analyzed as described ( Vögtle et al., 2009; Claypool et al., 2011). Isolated mitochondria were incubated in SEM buffer at 37°C. Samples were taken at various time points and analyzed by SDS–PAGE and immunoblotting. For analysis of protein turnover in vivo, yeast precultures were diluted in minimal medium (minus histidine) to an OD600 of 0.6 and grown at 37°C for 2 h. Cycloheximide (50 mg/ml stock in dimethyl sulfoxide [DMSO]) was added to a final concentration of 50 μg/ml to terminate protein translation. Cells were harvested at various time points and whole-cell extracts were obtained by postalkaline extraction. Samples were analyzed by SDS–PAGE and immunoblotting.
Miscellaneous
Western blots were scanned using Multi Gauge (http://multi-gauge.winsite.com) or ImageJ (http://rsbweb.nih.gov/ij/download.html) software. For quantification of protein turnover, the protein amount of the last time point was set in relation to the level of the first time point (0 h). Protein levels in oct1Δ were compared with levels of oct1Δ OCT1 as control (set to 100%). Standard deviation was calculated from at least three different experiments.
Acknowledgments
We thank C. Hunte and T. Langer for discussion and M. Yaffe for yeast strains. This work was supported by the Deutsche Forschungsgemeinschaft, Bundesministerium für Bildung und Forschung (Dynamo), Trinationales Graduiertenkolleg (GRK 1478), and the Excellence Initiative of the German federal and state governments (EXC 294 BIOSS).
Abbreviations used:
- BSA
bovine serum albumin
- DMSO
dimethyl sulfoxide
- Icp55
intermediate cleaving peptidase 55
- Imo32
intermediate cleaved by mitochondrial octapeptidyl aminopeptidase 1
- IMP
inner membrane peptidase
- MIP
mitochondrial intermediate peptidase
- MPP
mitochondrial processing peptidase
- Oct1
octapeptidyl aminopeptidase 1
- Pcp1
processing of cytochrome c peroxidase
- PK
proteinase K
- PMSF
phenylmethylsulfonyl fluoride
- TCA
trichloroacetic acid
- TIM
translocase of the inner membrane
- TOM
translocase of the outer membrane.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-02-0169) on April 27, 2011.
REFERENCES
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Beckmann JD, Ljungdahl PO, Lopez JL, Trumpower BL. Isolation and characterization of the nuclear gene encoding the Rieske iron-sulfur protein (RIP1) from Saccharomyces cerevisiae. J Biol Chem. 1987;262:8901–8909. [PubMed] [Google Scholar]
- Belle A, Tanay A, Bitincka L, Shamir R, O’Shea EK. Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci USA. 2006;103:13004–13009. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Isaya G. Prediction and identification of new natural substrates of the yeast mitochondrial intermediate peptidase. J Biol Chem. 1995;270:27366–27373. doi: 10.1074/jbc.270.45.27366. [DOI] [PubMed] [Google Scholar]
- Claypool SM, Whited K, Srijumnong S, Han X, Koehler CM. Barth syndrome mutations that cause tafazzin complex lability. J Cell Biol. 2011;192:447–462. doi: 10.1083/jcb.201008177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan DA, Truscott KN, Zeth K. The bacterial N-end rule pathway: expect the unexpected. Mol Microbiol. 2010;76:545–558. doi: 10.1111/j.1365-2958.2010.07120.x. [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K. Multiple pathways for mitchondrial protein traffic. Biol Chem. 2009;390:723–730. doi: 10.1515/BC.2009.087. [DOI] [PubMed] [Google Scholar]
- Esser K, Tursun B, Ingenhoven M, Michaelis G, Pratje E. A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease Pcp1. J Mol Biol. 2002;323:835–843. doi: 10.1016/s0022-2836(02)01000-8. [DOI] [PubMed] [Google Scholar]
- Gakh O, Cavadini P, Isaya G. Mitochondrial processing peptidases. Biochim Biophys Acta. 2002;1592:63–77. doi: 10.1016/s0167-4889(02)00265-3. [DOI] [PubMed] [Google Scholar]
- Gavel Y, von Heijne G. Cleavage-site motifs in mitochondrial targeting peptides. Protein Eng. 1990;4:33–37. doi: 10.1093/protein/4.1.33. [DOI] [PubMed] [Google Scholar]
- Glaser E, Sjöling S, Tanudji M, Whelan J. Mitochondrial protein import in plants: signals, sorting, targeting, processing and regulation. Plant Mol Biol. 1998;38:311–338. doi: 10.1023/a:1006020208140. [DOI] [PubMed] [Google Scholar]
- Grant PM, Tellam J, May VL, Strauss AW. Isolation and nucleotide sequence of a cDNA clone encoding rat mitochondrial malate dehydrogenase. Nucleic Acids Res. 1986;14:6053–6066. doi: 10.1093/nar/14.15.6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib SJ, Neupert W, Rapaport D. Analysis and prediction of mitochondrial targeting signals. Methods Cell Biol. 2007;80:761–781. doi: 10.1016/S0091-679X(06)80035-X. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Schmidt B, Wachter E, Weiss H, Neupert W. Transport into mitochondria and intramitochondrial sorting of the Fe/S protein of ubiquinol-cytochrome c reductase. Cell. 1986;47:939–951. doi: 10.1016/0092-8674(86)90809-3. [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Hodges PE, Rosenberg LE. Survey of amino-terminal proteolytic cleavage sites in mitochondrial precursor proteins: leader peptides cleaved by two matrix proteases share a three-amino acid motif. Proc Natl Acad Sci USA. 1989;86:4056–4060. doi: 10.1073/pnas.86.11.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Taylor NL, Whelan J, Millar AH. Refining the definition of plant mitochondrial presequences through analysis of sorting-signals, N-terminal modifications, and cleavage motifs. Plant Physiol. 2009;150:1272–1285. doi: 10.1104/pp.109.137885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaya G, Kalousek F, Fenton WA, Rosenberg LE. Cleavage of precursors by the mitochondrial processing peptidase requires a compatible mature protein or an intermediate octapeptide. J Cell Biol. 1991;113:65–76. doi: 10.1083/jcb.113.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaya G, Kalousek F, Rosenberg LE. Amino-terminal octapeptides function as recognition signals for the mitochondrial intermediate peptidase. J Biol Chem. 1992;267:7904–7910. [PubMed] [Google Scholar]
- Isaya G, Miklos D, Rollins RA. MIP1, a new yeast gene homologous to the rat mitochondrial intermediate peptidase gene, is required for oxidative metabolism in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:5603–5616. doi: 10.1128/mcb.14.8.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalousek F, Hendrick JP, Rosenberg LE. Two mitochondrial matrix proteases act sequentially in the processing of mammalian matrix enzymes. Proc Natl Acad Sci USA. 1988;85:7536–7540. doi: 10.1073/pnas.85.20.7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambacheld M, Augustin S, Tatsuta T, Müller S, Langer T. Role of the novel metallopeptidase MoP112 and Saccharolysin for the complete degradation of proteins residing in different subcompartments of mitochondria. J Biol Chem. 2005;280:20132–20139. doi: 10.1074/jbc.M500398200. [DOI] [PubMed] [Google Scholar]
- Koppen M, Langer T. Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit Rev Biochem Mol Biol. 2007;42:221–242. doi: 10.1080/10409230701380452. [DOI] [PubMed] [Google Scholar]
- Major T, von Janowsky B, Ruppert T, Mogk A, Voos W. Proteomic analysis of mitochondrial protein turnover: identification of novel substrate proteins of the matrix protease Pim1. Mol Cell Biol. 2006;26:762–776. doi: 10.1128/MCB.26.3.762-776.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T, Serero A, Giglione C. Impact of the N-terminal amino acid on targeted protein degradation. Biol Chem. 2006;387:839–851. doi: 10.1515/BC.2006.107. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Pfanner N, Truscott KN. Isolation of yeast mitochondria. Methods Mol Biol. 2006;313:33–39. doi: 10.1385/1-59259-958-3:033. [DOI] [PubMed] [Google Scholar]
- Meisinger C, et al. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev Cell. 2004;7:61–71. doi: 10.1016/j.devcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Mogk A, Schmidt R, Bukau B. The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol. 2007;17:165–172. doi: 10.1016/j.tcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Naamati A, Regev-Rudzki N, Galperin S, Lill R, Pines O. Dual targeting of Nfs1 and discovery of its novel processing enzyme, Icp55. J Biol Chem. 2009;284:30200–30208. doi: 10.1074/jbc.M109.034694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Nett JH, Denke E, Trumpower BL. Two-step processing is not essential for the import and assembly of functionally active iron-sulfur protein into the cytochrome bc1 complex in Saccharomyces cerevisiae. J Biol Chem. 1997;272:2212–2217. doi: 10.1074/jbc.272.4.2212. [DOI] [PubMed] [Google Scholar]
- Nett JH, Trumpower BL. Intermediate length Rieske iron-sulfur protein is present and functionally active in the cytochrome bc1 complex of Saccharomyces cerevisiae. J Biol Chem. 1999;274:9253–9257. doi: 10.1074/jbc.274.14.9253. [DOI] [PubMed] [Google Scholar]
- Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- Shen Y, Hixson KK, Tolic N, Camp DG, Purvine SO, Moore RJ, Smith RD. Mass spectrometry analysis of proteome-wide proteolytic post-translational degradation of proteins. Anal Chem. 2008;80:5819–5828. doi: 10.1021/ac800077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztul ES, Hendrick JP, Kraus JP, Wall D, Kalousek F, Rosenberg LE. Import of rat ornithine transcarbamylase precursor into mitochondria: two-step processing of the leader peptide. J Cell Biol. 1987;105:2631–2639. doi: 10.1083/jcb.105.6.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LM, Sutherland P, Steffan JS, McAlister-Henn L. Gene sequence and primary structure of mitochondrial malate dehydrogenase from Saccharomyces cerevisiae. Biochemistry. 1988;27:8393–8400. doi: 10.1021/bi00422a015. [DOI] [PubMed] [Google Scholar]
- Tropschug M, Nicholson DW, Hartl FU, Köhler H, Pfanner N, Wachter E, Neupert W. Cyclosporin A-binding protein (cyclophilin) of Neurospora crassa. One gene codes for both the cytosolic and mitochondrial forms. J Biol Chem. 1988;263:14433–14440. [PubMed] [Google Scholar]
- van der Laan M, Hutu DP, Rehling P. On the mechanism of preprotein import by the mitochondrial presequence translocase. Biochim Biophys Acta. 2010;1803:732–739. doi: 10.1016/j.bbamcr.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule at atomic resolution. Nat Struct Mol Biol. 2008;15:1238–1240. doi: 10.1038/nsmb1208-1238. [DOI] [PubMed] [Google Scholar]
- Vögtle FN, et al. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- Witte C, Jensen RE, Yaffe MP, Schatz G. MAS1, a gene essential for yeast mitochondrial assembly, encodes a subunit of the mitochondrial processing protease. EMBO J. 1988;7:1439–1447. doi: 10.1002/j.1460-2075.1988.tb02961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- Yogev O, Pines O. Dual targeting of mitochondrial proteins: mechanism, regulation and function. Biochim Biophys Acta. 2011;1808:1012–1020. doi: 10.1016/j.bbamem.2010.07.004. [DOI] [PubMed] [Google Scholar]