A new pathway that controls pseudopod extension is identified. Results support a model in which Rab1A is required for chemoattractant-mediated Roco2 activation that functions through the F-actin cross-linker filamin to promote pseudopod extension.
Abstract
We identify a new pathway that is required for proper pseudopod formation. We show that Roco2, a leucine-rich repeat kinase 2 (LRRK2)-related Roco kinase, is activated in response to chemoattractant stimulation and helps mediate cell polarization and chemotaxis by regulating cortical F-actin polymerization and pseudopod extension in a pathway that requires Rab1A. We found that Roco2 binds the small GTPase Rab1A as well as the F-actin cross-linking protein filamin (actin-binding protein 120, abp120) in vivo. We show that active Rab1A (Rab1A-GTP) is required for and regulates Roco2 kinase activity in vivo and that filamin lies downstream from Roco2 and controls pseudopod extension during chemotaxis and random cell motility. Therefore our study uncovered a new signaling pathway that involves Rab1A and controls the actin cytoskeleton and pseudopod extension, and thereby, cell polarity and motility. These findings also may have implications in the regulation of other Roco kinases, including possibly LRRK2, in metazoans.
INTRODUCTION
Dynamic reorganization of the cytoskeleton underlies a wide range of cellular processes that require changes in cell shape, including cytokinesis, random cell motility, and chemotaxis. Inherent in these major cytoskeletal reorganizations is the ability of the cell to polarize the cytoskeleton through an orchestrated redistribution and localized assembly/disassembly of F-actin and myosin II (MyoII). Leading-edge protrusion during chemotaxis of amoeboid cells is dependent, predominantly, on the localized polymerization of F-actin at the site on the cortex closest to the chemoattractant source. This requires not only new F-actin that pushes the membrane forward, but also F-actin cross-linking proteins that stabilize the mesh-like actin network and link it to the cortex (Stossel et al., 2001; Pollard and Borisy, 2003; Insall and Machesky, 2009; Pollard and Cooper, 2009). During the past decade, we have gained significant understanding of initiation of new F-actin polymerization and the signaling pathways and the machinery mediating the process of formation of the mesh-like network of F-actin that pushes the membrane forward (Kedrin et al., 2007; van Haastert and Veltman, 2007; Janetopoulos and Firtel, 2008; Insall and Machesky, 2009; Pollard and Cooper, 2009; Insall, 2010; Swaney et al., 2010). However, the pathways that control actin cross-linking have not been well defined.
Roco kinases are a novel family of highly conserved proteins, some of which regulate the cytoskeleton (Bosgraaf et al., 2002; Bosgraaf and van Haastert, 2003; Jaleel et al., 2007; van Egmond et al., 2008; Parisiadou et al., 2009; van Egmond and van Haastert, 2010; Zechel et al., 2010). In eukaryotes, Roco kinases share linked ROC (Ras of complex proteins), Ras-like GTP-binding/GTPase, and COR (C-terminal of Roc) domains, as well as a Ser/Thr kinase domain that shares sequence similarity with a mixed-lineage kinase (MLK) subfamily of MEKKs (mitogen-activated protein kinase kinase kinase) that is regulated by GTP binding to the Roc domain (Bosgraaf and van Haastert, 2003). Roco kinases often have distinct protein–protein interacting/regulatory domains, such as the leucine-rich repeats (LRRs), ankyrin, and WD40 repeats (Bosgraaf and van Haastert, 2003; Marin, 2006, 2008; van Egmond and van Haastert, 2010). In humans, mutations in LRRK2 (one of the two roco paralogous genes, LRRK1 and LRRK2), a leucine repeat–containing member of this family, are linked to both autosomal dominant and sporadic Parkinson's disease (PD; Paisan-Ruiz et al., 2004; Zimprich et al., 2004; Toft et al., 2005). LRRK2 has been proposed as a regulator of the cytoskeleton, neuronal outgrowth, and vesicular trafficking (Jaleel et al., 2007; Shin et al., 2008; Parisiadou et al., 2009; Xiong et al., 2010; Meixner et al., 2011). Dictyostelium encodes 11 Roco family members, and of these, 5 have been examined to various degrees (van Egmond and van Haastert, 2010). The first identified Roco kinase, Dictyostelium GbpC, is the sole effector of cGMP in Dictyostelium, an important regulator of MyoII in response to chemoattractant stimulation and osmotic stress (Goldberg et al., 2002; Bosgraaf and van Haastert, 2003; Bosgraaf et al., 2005). Roco2 regulates autocrine signaling associated with growth. Roco2-null cells grow more rapidly than wild-type cells, but also accumulate multinucleate cells, suggesting a possible cytokinesis defect; they have been reported to aggregate more slowly than wild-type cells and form larger aggregates (Abe et al., 2003; Phillips and Gomer, 2010; van Egmond and van Haastert, 2010). Another Roco kinase, Pats1, is required for proper cell cytokinesis (Abysalh et al., 2003), while roco4− and roco11− cells exhibit morphogenesis defects (van Egmond and van Haastert, 2010). Although several Roco kinases have been intensely studied, in part because of the linkage of LRRK2 to forms of PD, the mechanisms by which this family is regulated are not understood, with the exception of GbpC, which has a ligand-binding domain that regulates GTP binding and kinase activation (Bosgraaf et al., 2005; van Egmond et al., 2008).
In this article, we identify a regulatory pathway that is required for proper pseudopod extension in Dictyostelium. We determine that Dictyostelium Roco2 is activated by chemoattractant stimulation and is important for cortically localized F-actin polymerization and pseudopod extension during chemotaxis, and we present evidence that this pathway is mediated through the regulation of filamin by Roco2. We further demonstrate that Rab1A binds to Roco2 and regulates Roco2 kinase activity in vivo, thereby identifying a novel Rab1A effector and a new regulatory mechanism by which the cytoskeleton is controlled. Our studies may provide a model by which other Roco2 kinases, including possibly LRRK2, are regulated.
RESULTS
roco2− cells exhibit cell motility and cytokinesis defects
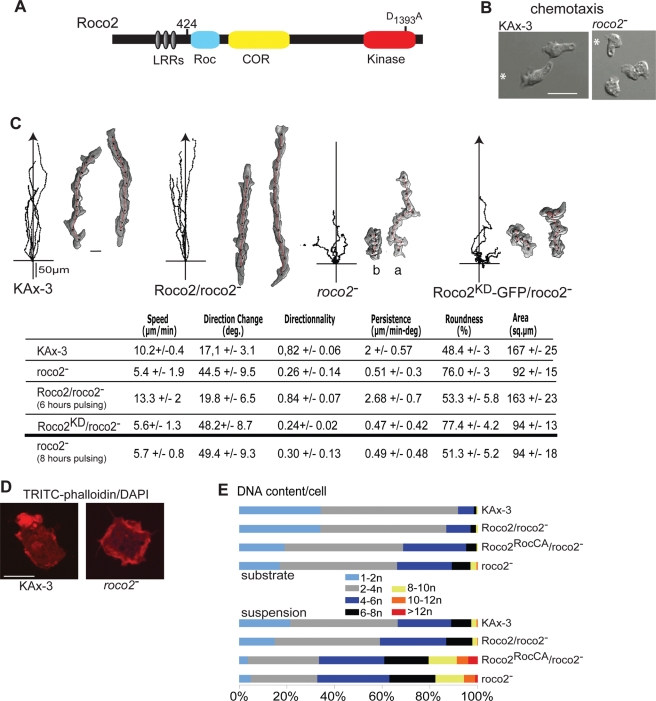
We identified Roco2 (QkgA), a member of the Roco family of protein kinases, as a potential regulator of the cytoskeleton by screening knockout strains of Dictyostelium Roco family members for defects in cell motility and chemotaxis. Roco2 contains an N-terminal LRR motif similar to that of LRRK2, in addition to the Roc (Ras-like), COR, and kinase domains present in eukaryotic Roco proteins (Figure 1A; Bosgraaf and van Haastert, 2003).
FIGURE 1:
(A) Domain structure of the Roco2 protein, the LRRs, a COR, and a ROC (Ras of complex protein) domain as defined by Bosgraaf and van Haastert (2003), and a kinase domain sharing similarity to MEKKs. The mutation created is indicated. (B) Images of chemotaxing wild-type and roco2− cells. (C) Trajectories of cells migrating to the chemoattractant cAMP emitted from a micropipette and images of chemotaxing cells taken at 1-min intervals obtained using DIAS software (Wessels et al., 1998). Wild-type, roco2−, GFP-Roco2/roco2−, and GFP-Roco2KD/roco2− cells are analyzed. For roco2− cells, two cells at different distances from the needle are shown: (a) a cell ∼400 μm from the needle; (b) a cell ∼10–150 μm from the needle. Table, DIAS computer analysis performed on time-lapse DIC microscopy movies with a 40× objective of cells chemotaxing to cAMP. Speed represents movement of a cell's centroid; change of direction (degrees) is a relative measure of the number and frequency of turns made by the cell. Higher numbers indicate more turns and a less efficient chemotaxis. Directionality is a measure of straightness of cell movement. Persistence is an indirect measure of straightness of cell movement to the chemoattractant source; it is the value divided by direction change values. Lower numbers indicate a less efficient chemotaxis. Roundness is an indication of the cell polarity. Larger numbers indicate the cells are more round and less polarized. (D) KAx-3 and roco2− cells stained with the F-actin marker TRITC-phalloidin. (E) The relative DNA content of DAPI-stained log-phase cells as determined by FACS. Scale bars: 10 μm.
roco2− cells (see Materials and Methods for details) exhibit strong polarity and chemotaxis defects when placed in a cAMP chemoattractant gradient, moving at only ∼50% of the speed of wild-type cells and being less polarized than wild-type cells (Figure 1, B and C; Supplemental Movies M1 and M2). This defect is weaker when cells are very close to the needle where the chemoattractant concentration is highest and the gradient is steepest, as is illustrated in the cell labeled “b” in Figure 1C. The roco2− cell chemotaxis defect is even stronger when cells are examined in a linear gradient in a Dunn chamber (Movies M3 and M4). In roco2− cells, protrusions are smaller and not restricted predominantly to the side closest to the chemoattractant source, occurring over a greater portion of the cortex compared with wild-type cells and resulting in considerably reduced persistence and directionality (Figure 1C and Supplemental Figure S1A). We studied vegetative cells, which undergo random cell motility in the absence of G protein–coupled receptor signaling (Tuxworth et al., 1997; Sasaki et al., 2007), and determined that roco2− cells exhibit general defects in cell motility: they move with < 60% the speed of wild-type cells, do not polarize well, often exhibit F-actin along the majority of the cell's cortex, and lack a prominent pseudopod compared with wild-type cells (Figures 1D and S1B). Expression of wild-type green fluorescent protein (GFP)- (Figure 1C) or T7-tagged Roco2 restores the mobility defects of roco2− cells (Figures 1, C and D, and S1B) and complements all roco2− cell phenotypes described here and below. We also created an inactive (kinase-dead) Roco2 (Roco2KD) by mutating the conserved Asp in the active site of the kinase domain, which resulted in a Roco2 that lacks kinase activity (as shown in Figure 2). As expected, expression of Roco2KD in roco2− cells does not complement the null phenotypes (Figures 1C and S1).
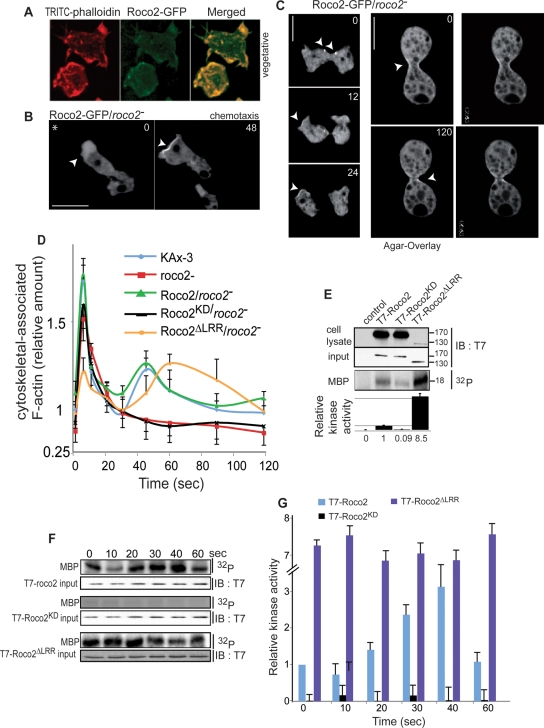
FIGURE 2:
Analysis of roco2− cells. (A) Confocal microscopy images of vegetative GFP-Roco2/roco2− cells fixed and stained with TRITC-phalloidin. (B) Confocal fluorescent images of chemotaxing Roco2-GFP/roco2− cells. Arrows indicate an enrichment of roco2-GFP at the cell's leading edge. (C) Left, confocal fluorescent microscopy images of dividing Roco2-GFP/roco2− cells. Arrows indicate Roco2-GFP appearing on cell poles in late cytokinesis. Right, confocal fluorescent microscopy images of Roco2-GFP/roco2− cells using the “agar-overlay” method. Arrows indicate the cleavage furrow enriched with Roco2-GFP. (D) Kinetics of F-actin polymerization in response to chemoattractant stimulation. (E) Basal Roco2 kinase activity in 6 h pulsed cells. Top, immunoblot and autoradiogram of MBP phosphorylation by Roco2, as resolved by SDS–PAGE. Roco2 protein levels were determined by Western blot analysis, using anti-T7 antibody. Bottom, normalization of incorporated 32P compared with Roco2 protein content. Data are representative of at least three independent experiments, in arbitrary units, where wild-type-Roco2 kinase activity is defined as 1.0. (F) Kinase activity of Roco2, Roco2KD, and Roco2ΔLRR expressed in roco2− cells upon cAMP stimulation of 6 h pulsed cells as described in Materials and Methods. Because Roco2ΔLRR is expressed at a much lower level than wild-type Roco2, the immunoblot to quantify Roco2 protein was exposed for a longer time than the other Western blots. Kinase activity was quantitated using a phosphorimager. (G) Quantitation of kinase activity.
Consistent with these chemotaxis and motility defects, roco2− cells exhibit aggregation defects when plated on nonnutrient agar, with many cells not entering aggregates, particularly when cells are plated at lower densities (Supplemental Figure S2A). When cells are pulsed with cAMP for 6 h prior to plating on nonnutrient agar, the defect is less severe, and at moderate densities the cells start to aggregate but remain as streams even after 24 h (Figure S2B). Previous studies in strain Ax2 indicated that roco2− cells have delayed aggregation but form larger aggregates when plated on nutrient agar (Abe et al., 2003). We did not observe larger aggregates, but this difference could be the result of plating conditions. These aggregation defects are not due to an inability to express genes required for chemotaxis to cAMP, as the level of mRNA for the cAMP G protein–coupled receptor cAR1 in response to cAMP pulsing is the same as in wild-type cells, although the accumulation of cAR1 mRNA is slightly delayed (unpublished data). We found that roco2− cells have the same chemotaxis defects whether they are pulsed for either 6 or 8 h (Figure 1C), indicating that any delay in aggregation-stage gene expression in roco2− cells is not responsible for the roco2− chemotaxis phenotypes.
roco2− cells also exhibit a cytokinesis defect, as suggested previously (Phillips and Gomer, 2010), which, like that of myoII-null (mhcA−) cells, is considerably stronger in cells grown in suspension than in those attached to a substratum (Figures 1E and S3A; see Materials and Methods; De Lozanne and Spudich, 1987). When cell shape is examined during cytokinesis, wild-type cells exhibit “classical” cytoskeletal contraction in the middle of the dividing cell, forming a dumbbell shape with a short (normally <6 μm) cytoplasmic bridge that forms before daughter cell separation (Figure 1F; Movie M5). In contrast, we found that roco2− prospective daughter cells become polarized, often forming lamellipodia-like protrusions on the opposing poles, whereupon they begin to move apart and a thin cytoplasmic bridge forms that is often multiple cell diameters in length (Figures 1F and S3B). In 50% of the cells, this bridge does not cleave; instead, the two presumptive daughter cells retract toward each other and resolve into a single, binucleate cell (retrocytokinesis; Figure S3C; Movie M6). The cells that do complete cytokinesis may do so by traction-mediated cytofission (Uyeda et al., 2000).
Roco2 kinase is a regulator of the actin cytoskeleton and is activated in response to chemoattractant stimulation
Spinning disk confocal images of phalloidin-stained roco2− cells expressing a functional Roco2-GFP reveal that Roco2-GFP is enriched in local F-actin pseudopodial projections in vegetative cells (Figure 2A). In chemotaxing cells, Roco2-GFP is found preferentially in the F-actin-enriched, lamellipodial region and periodically as a tight band at the leading-edge cortex (Figure 2B, left and right panels, respectively). To examine this in more detail, we coexpressed Roco2-GFP with a red fluorescent protein (RFP) fusion of the F-actin binding protein coronin, which helps terminate F-actin polymerization. As can be seen in Movie M7, both Roco2 and coronin are enriched in the anterior cortex, while coronin, but not Roco2, is also found to a lesser extent at the F-actin enriched in the cortex at the posterior of the cell. Although many leading-edge markers rapidly and transiently localize to the cell cortex upon rapid, global chemoattractant stimulation (van Haastert and Devreotes, 2004), we observed no such rapid translocation of Roco2-GFP to the cell cortex (unpublished data). We also found Roco2-GFP at the cleavage furrow during cytokinesis and at the poles of daughter cells immediately before and after cleavage of the cytoplasmic bridge (Figure 2C, left; Movie M8), which is more easily observed using “agar-overlay” to flatten cells (Figure 2C, right). In some cells, fluorescent dots (possibly small vesicles) are present near the cytoplasmic bridge before daughter cell separation (Figure 2C, left, 0 s frame). This localization and the extended cytoplasmic bridge connecting daughter cells in roco2− cells suggest that Roco2 is required for fission of the cytoplasmic bridge and is necessary for efficient daughter cell separation.
The subcellular localization of Roco2 and the phenotypes of roco2− cells suggest that Roco2 regulates the actomyosin cytoskeleton. In response to chemoattractant stimulation, cortical F-actin (F-actin found in the Triton-insoluble fraction) exhibits a biphasic curve with a brief peak at ∼5 s, and a decrease in F-actin to near-basal levels, followed by a lower, broader second peak with a maximum at 20–40 s, which is linked to pseudopod extension (Condeelis et al., 1990). In roco2− cells, the first peak is reduced ∼20% and no detectable second peak is observed (Figure 2D). Expression of full-length Roco2, but not (inactive) Roco2KD, restores these responses (Figure 2D). As the second F-actin peak in Dictyostelium is thought to require phosphatidylinositol-3 kinase (PI3K) function (Chen et al., 2003), which is regulated by Ras (Funamoto et al., 2002), we examined PI3K activity and Ras activation and found that there was little difference between roco2− and wild-type cells, suggesting this is not the basis for the F-actin defect and roco2− cells are competent to activate Ras and PI3K (unpublished data).
To understand a possible linkage between cortical F-actin and Roco2 kinase activity, we developed a kinase activity assay in which T7-Roco2 expressed in roco2− cells is isolated from cells using anti-T7 resin, and the immunocomplex is assayed for kinase activity using [32P]γATP and myelin basic protein (MBP) as a substrate (Materials and Methods). We used pulled-down Roco2KD expressed in roco2− cells to determine background kinase activity in the assay, as this did not exhibit any activity beyond that of a control pull-down (Figure 2, E–G; unpublished data). On chemoattractant stimulation, wild-type Roco2 exhibits a small decrease in kinase activity at ∼10 s, which is followed by a more than threefold increase, peaking at ∼40 s. This broad peak roughly overlaps the second peak of cortical F-actin. In human LRRK2, deletion of the N-terminal domain, which contains the LRRs but retains the Roc and COR domains, results in a kinase with elevated in vitro kinase activity (Jaleel et al., 2007; Greggio et al., 2008). We created a similar N-terminally truncated Roco2 mutant (designated Roco2ΔLRR) by deleting residues 2–424. Figure 2E shows that T7-Roco2ΔLRR expressed in and immunoprecipitated from roco2− cells has a basal (unstimulated cells) activity more than sevenfold higher than that of wild-type Roco2 expressed in and isolated from unstimulated roco2− cells. Unlike wild-type T7-Roco2, the kinase activity of T7-Roco2ΔLRR remains unchanged after cAMP stimulation, indicating that Roco2ΔLRR is a strong gain-of-function and unregulated Roco2 kinase mutant with high constitutive activity (Figure 2, F and G). (The level of Roco2ΔLRR expression is much lower than that of wild-type Roco2. This is observed in all clonal isolates, leading us to assume cells must select against a high level of expression of this activated form of Roco2.) When Roco2ΔLRR is expressed in roco2− cells, we observed that the first F-actin peak is severely reduced and that the second peak is both elevated and extended (Figure 2D). The increase in the second peak in roco2− cells expressing Roco2ΔLRR and the loss of the second F-actin peak in roco2− cells suggest that Roco2 kinase activity correlates with, and is required for, the second F-actin peak. We also examined whether inhibition of PI3K with LY294004 blocked the extended second F-actin peak in roco2− cells expressing Roco2ΔLRR, and found that LY294004 did not have a significant effect on the kinetics of F-actin polymerization in Roco2ΔLRR/roco2− cells (unpublished data).
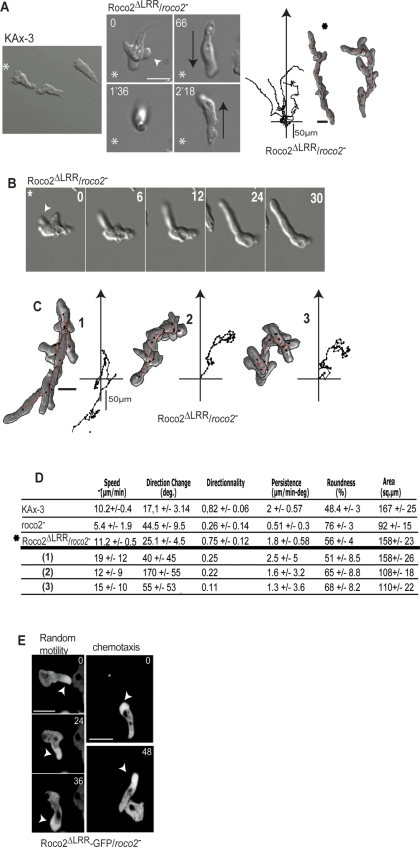
Expression of Roco2ΔLRR in roco2− cells complements the random motility speed-of-movement defect, but the cells exhibit a strong, dominant chemotaxis phenotype when expressed in roco2− or wild-type cells (Figures 3, A–D, and S4, A–D; Movie M9; unpublished wild-type data). During chemotaxis, these cells extend a long, Roco2ΔLRR- and F-actin-containing pseudopod and sporadically extend multiple, dominant pseudopodia simultaneously in different directions (Figure 3E; Movie M9). The extended pseudopod does not effectively adhere to the substratum and, while off of the substratum, the pseudopod shifts its position randomly relative to the gradient (“floating” leading edge), resulting in an inability to effectively move up the chemotaxis gradient (Figure 3, A–C; Movie M9). Even for cells in which the anterior appears to adhere normally, the directionality is considerably reduced. As depicted in Movie M9, the posterior of the cell often remains attached for an extended time and then abruptly releases, resulting in the cell scooting forward. A similar long, GFP-Roco2ΔLRR -containing pseudopod is also observed during random cell movement (Figure 3E).
FIGURE 3:
Analysis of Roco2ΔLRR chemotaxis. (A) DIC images of chemotaxing wild-type and Roco2ΔLRR cells and Roco2ΔLRR cells trajectories. Asterisk, position of the micropipette; black arrows, cell direction; white arrow, nascent pseudopod. (B) Time-lapse images of the formation of an extended pseudopod in Roco2ΔLRR/roco2− cells. (C) Trajectories of adhesive Roco2ΔLRR cells, individual cells in which the pseudopod is not associated with the substrate and is in within the plane of focus (designated as “floating” cells). These cells are labeled 1, 2, and 3, and DIAS analysis of the cells is provided in part (D). (D) DIAS computer analysis performed on time-lapse DIC microscopy as described previously (Wessels et al., 1998). (E) Fluorescent images of confocal microscopy of Roco2ΔLRR-GFP/roco2− cells randomly moving (left panels) or migrating toward a chemoattractant source (right panels). The arrow points to a cell exhibiting a smooth pseudopod.
Filamin (abp10) lies downstream from Roco2
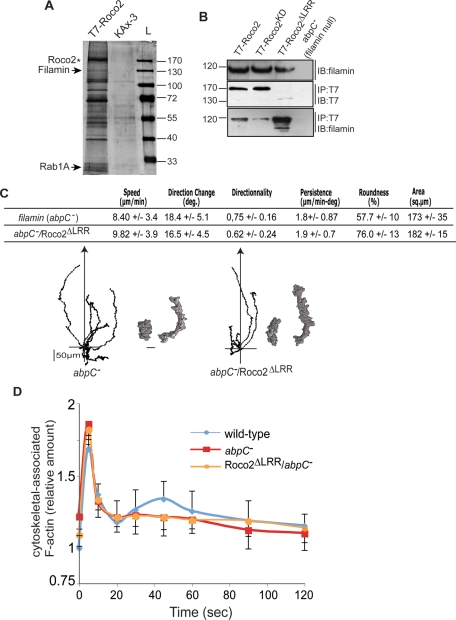
To better understand the regulation and function of Roco2, we performed Roco2 pull-downs followed by mass spectrometry (MS) analyses to identify putative Roco2-interacting proteins. Figure 4A depicts a silver-stained gel of proteins from pull-down experiments with T7-Roco2 and from control cells; Figure S5A lists some of the proteins identified in the T7-Roco2 (not control) pull-downs. One of these is the redox-dependent molecular chaperone DJ-1, mutations of which are linked to PD and which also binds LRRK2 in mammalian and Drosophila cells (Venderova et al., 2009). Two of the other more abundant proteins are Rab1A and filamin (ABP120/ABPc/gelation factor). Because of the potential importance of Rab1A and filamin in directly controlling cell motility, we focused our analysis on these proteins.
FIGURE 4:
Roco2-associated proteins. (A) Roco2-associated proteins were coimmunoprecipitated from T7-Roco2 aggregation-competent cells, separated on an acrylamide gel, and detected by silver nitrate staining. Wild-type KAx-3 cells were treated similarly and used as a control. T7-Roco2 band (asterisk) is indicated as well as filamin/ABP120 and Rab1A (arrows). (B) Coimmunoprecipitation assay results from cells transformed with plasmids expressing T7-tagged Roco2 (wild-type, kinase-dead, or Roco2ΔLRR). Protein extracts were precipitated with anti-T7 antibody and were subjected to a Western blotting analysis using either anti-T7 or an anti-filamin antibody. (C) Chemotaxis of filamin (abpC)-null cells and abpC− cells expressing Roco2ΔLRR. The two tracings show a cell closer to (left) and further away (right) from the needle. (D) Kinetics of chemoattractant-induced F-actin polymerization in abpC− cells and abpC− cells expressing Roco2ΔLRR.
Filamin is a conserved, orthogonal F-actin cross-linking protein that cross-links F-actin filaments within a meshwork and is involved in phagocytosis, cell motility, and morphogenesis required for the second peak of cortical F-actin peak and for subsequent pseudopod extension (Cox et al., 1992, 1995; Cox, 1995; Stossel et al., 2001; Knuth et al., 2004; Khaire et al., 2007; Washington and Knecht, 2008; Baldassarre et al., 2009). Figure 4B demonstrates that endogenous filamin coimmunoprecipitates with T7-Roco2. The amount of filamin that coimmunoprecipitates with the inactive T7-Roco2KD is reduced, compared with wild-type T7-Roco2, while the relative amount of filamin coimmunoprecipitating with the highly activated T7-Roco2ΔLRR is greatly increased. To examine possible functional interactions between Roco2 and filamin, we expressed the dominant, activated Roco2ΔLRR mutant in filamin-null (abpC−) cells and studied chemotaxis of these cells. As previously reported, abpC− cells are considerably less polarized and move more randomly than wild-type cells (Figures 4C and S5B; Cox, 1995; Cox et al., 1992, 1995; Washington and Knecht, 2008). abpC− cells expressing Roco2ΔLRR exhibit the same phenotype as abpC− cells but do not produce the extended pseudopodial projections observed in Roco2ΔLRR-expressing roco2− or wild-type cells (Figure S5B). Previous studies indicated that abpC− cells do not have a defined second peak of cortical F-actin, consistent with their defect in pseudopod extension (Cox et al., 1995; Figure 4D; see Discussion). Although roco2− and wild-type cells expressing Roco2ΔLRR have an elevated second peak of cortical F-actin, Roco2ΔLRR/abpC− cells show a cortical F-actin profile similar to that of abpC− cells, a slightly elevated basal level of F-actin, normal first F-actin peak, and no discernible second peak (Figure 4D). It is worth emphasizing that whereas roco2− cells expressing Roco2ΔLRR exhibit a severely reduced first F-actin peak, this is not observed in an abpC− cell background (Roco2ΔLRR/abpC− cells), suggesting that the inhibitory effect of Roco2ΔLRR on the first peak may be filamin-dependent. Together with findings that Roco2 and filamin coimmunoprecipitate, these observations suggest that filamin lies downstream from and is a possible effector of Roco2 or that the Roco2 and filamin pathways are independent and the abpC− phenotype is epistatic. We favor the possibility that filamin lies downstream from Roco2 and is probably a Roco2 substrate, because of our finding that filamin and Roco2 coimmunoprecipitate. Roco2-GFP localizes normally to pseudopodia in filamin-null (abpC−) cells, indicating filamin is not required for Roco2 localization (unpublished data).
Rab1A binds to and regulates Roco2 activity
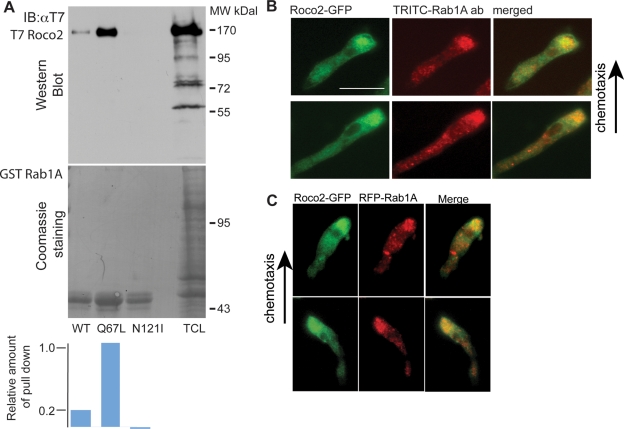
Rab1A is known to be associated with endoplasmic reticulum (ER)/Golgi membrane trafficking (Davidson and Balch, 1993) and is involved in linking the cell periphery to the Golgi in neurites (Sannerud et al., 2006), but little information is available on Dictyostelium Rab1A. Human and Dictyostelium Rab1A are highly conserved (76% identity; Figure S6A). To confirm the potential interaction between Roco2 and Rab1A, we examined the ability of recombinant glutathione S-transferase (GST)-tagged, wild-type Rab1A (GST-Rab1A), constitutively active Rab1A (GST-Rab1AQ67L), and a mutant Rab1A with reduced affinity for both GDP and GTP (GST-Rab1AN121I; Alvarez et al., 2003; Pind et al., 1994) to pull down T7-Roco2 from T7-Roco2/roco2− cell extracts (Figure 5A). We found that Roco2 is pulled down using Rab1A and Rab1AQ67L but not Rab1AN12I. The amount of Roco2 pulled down is more than fivefold higher using Rab1AQ67L than with Rab1A, suggesting that Roco2 directly interacts with Rab1A, and preferentially with its active form.
FIGURE 5:
Roco2 associates with Rab1A. (A) Pull-down assay results from roco2− cells expressing T7-tagged Roco2 using recombinant, GST-fused wild-type Rab1A, Rab1AN121I, and Rab1AQ67L, as described in Materials and Methods. Quantification of the data is shown at the bottom. (B) Confocal microscopy images of fixed chemotaxing roco2− cells expressing Roco2-GFP costained for endogenous Rab1A with α-Rab1A. (C) In vivo images of chemotaxing roco2− cells expressing Roco2-GFP and RFP-Rab1A. Images were taken 12 s apart. Scale bars: 10 μm.
Using a commercial anti-Rab1A antibody that we demonstrated recognizes Dictyostelium Rab1A (Figure S6B), we found that Rab1A is partly associated with small vesicles that are enriched in the pseudopodial region of chemotaxing cells, which overlaps with the localization of GFP-Roco2 (Figure 5B). We also observe background staining, suggesting that a portion of Rab1A is associated with submicroscopic vesicles or membranous structures and/or is cytosolic, presumably associated with RabGDI (http://dictybase.org/gene/DDB_G0268034). In addition, we examined Rab1A localization using RFP-tagged Dictyostelium Rab1A coexpressed with GFP-Roco2 in roco2− cells and found that RFP-Rab1A exhibits a localization similar to that of endogenous Rab1A (Figure 5C).
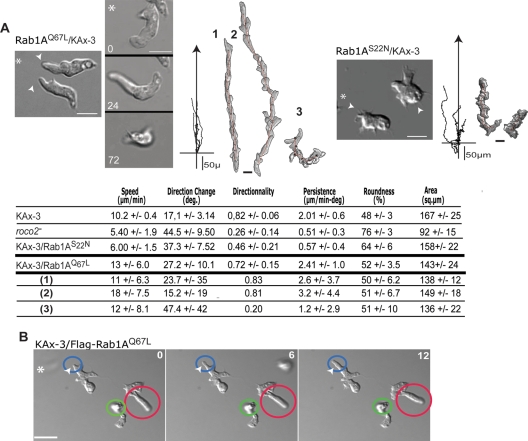
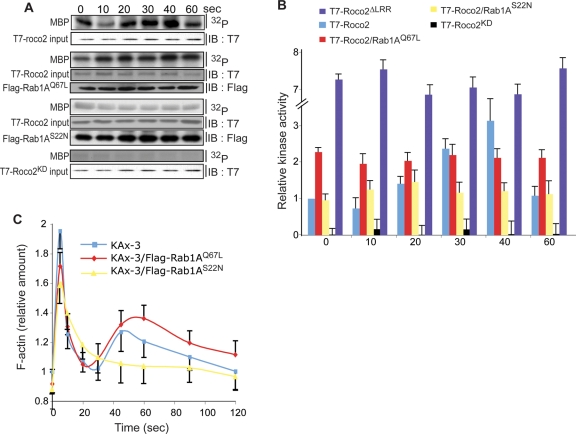
Because we were unable to create a rab1A knockout strain, possibly because rab1A is an essential gene, we used (constitutively active) Rab1AQ67L and (dominant negative) Rab1AS22N to dissect the potential roles of Rab1A in regulating chemotaxis and Roco2 function. Expression of Rab1AQ67L in wild-type cells results in cells producing extended pseudopodia and making an increased number of turns, with many cells displaying a “floating” leading edge, a substrate-association defect that is similar to, but not as severe as, that of Roco2ΔLRR/roco2− cells (Figures 6, A, B, and D, and S7, A and B). Furthermore, we found that coexpression of FLAG-Rab1AQ67L with T7-Roco2 in roco2− cells results in an ∼2.4-fold elevated basal activity of Roco2 that does not change upon chemoattractant stimulation (Figure 7, A and B). As with Roco2ΔLRR/roco2− cells, Rab1AQ67L cells exhibit an elevated second peak of cortical F-actin (Figure 7C), although the first F-actin peak is not reduced as much as it is in Roco2ΔLRR/roco2− cells. roco2− cells expressing Rab1AQ67L cells have the roco2− cell chemotaxis phenotype and aggregation defect and do not show the defects that are associated with overexpression of Rab1AQ67L in wild-type cells (Figures 8A and S7C), suggesting that Rab1A lies upstream from Roco2. Furthermore, coexpressing Rab1AQ67L with Roco2ΔLRR does not increase the already elevated Roco2ΔLRR kinase activity (Figure 8B). These data are consistent with Roco2ΔLRR being constitutively active and with Rab1A lying upstream from Roco2. Our findings suggest that Rab1A-GTP positively regulates Roco2 activity.
FIGURE 6:
DIC images, cell trajectories, and DIAS computer analysis of chemotaxing KAx-3 cells expressing either FLAG-Rab1AQ67L (A) or FLAG-Rab1AS22N (B). In (A), the trajectories of three cells are shown: two of the cells (1, 2) exhibited good anterior adherence during the time of the plots, while cell 3 exhibited poor anterior adherence. (C) DIAS computer analysis. (D) Occurrence of a greatly extended leading edge in wild-type cells expressing FLAG-Rab1AQ67L. The pseudopodial protrusions are highlighted by a colored circle. The red circle shows an already extended pseudopod. The blue circle indicates a newly forming pseudopod. The green circle identifies a pseudopod that is extended upward, out of the plane of focus.
FIGURE 7:
Effect of Rab1A on Roco2 function. (A) Top, immunoblot and autoradiogram of MBP phosphorylation by Roco2 upon cAMP stimulation. Western blot analysis was used to determine Roco2 protein levels (anti-T7 antibody) and Rab1A protein levels (anti-FLAG antibody). (B) Normalization of incorporated 32P compared with Roco2 protein content. Data are representative of at least three independent experiments, in arbitrary units, where wild-type-Roco2 kinase activity prior to cAMP stimulation is defined as 1 (t = 0). Data for Roco2ΔLRR and Roco2KD are shown for comparison. (C) Kinetics of F-actin polymerization in response to chemoattractant stimulation in KAx-3 cells expressing either FLAG-Rab1AS22N or FLAG-Rab1AQ67L.
FIGURE 8:
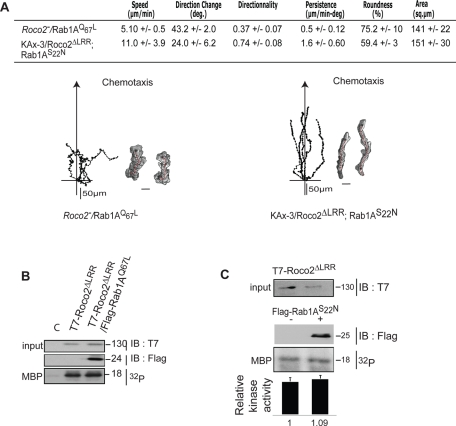
Effect of coexpressing Rab1A mutants on the chemotaxis phenotype of Roco2 mutant strains. (A) Chemotaxis of roco2− cells expressing Rab1AQ67L and roco2− cells coexpressing GFP-Roco2ΔLRR and FLAG- Rab1AS22N. Scale bar: 10 μm for cell images. Coexpression of Rab1AQ67L (B) or Rab1AS22N (C) with Roco2ΔLRR in roco2− cells does not affect Roco2ΔLRR kinase activity. The average of the kinase activity from three sets of cells is shown with the average for wild-type cells set at 1.0. Error bars give the SD.
In agreement with our model, we determined that expression of Rab1AS22N in wild-type cells results in cells that are unable to properly polarize in chemoattractant gradients and that move much more slowly than wild-type cells and with a decrease in directionality (Figure 6A). In addition, Rab1AS22N cells exhibit a slightly reduced first F-actin peak and a dramatically reduced or absent second peak (Figure 7C). These phenotypes are similar to, but not as severe as, those of roco2− cells. Furthermore, basal and chemoattractant-stimulated kinase activity of T7-Roco2 isolated from roco2− cells expressing Rab1AS22N is reduced compared with that of T7-Roco2 isolated from roco2− cells that do not express exogenous Rab1AS22N, and is not modulated by chemoattractant stimulation (Figure 7, A and B). Cells coexpressing Rab1AS22N and Roco2ΔLRR do not exhibit a decrease in Roco2ΔLRR kinase activity and have a chemotaxis phenotype similar to that of Roco2ΔLRR cells (Figures 8, A and C, and S7D). Together, our observations suggest that Rab1A is an upstream activator of Roco2, identifying a previously undescribed pathway regulating pseudopod formation.
DISCUSSION
We demonstrate that Roco2 is a key regulator of the actin cytoskeleton during chemotaxis and provide evidence that Rab1A functions upstream from Roco2 to regulate its activity and that Roco2 acts through filamin to control pseudopod extension (Figure 9). Consistent with this model, roco2− cells lack the second peak of cortical F-actin, which has been linked to pseudopod formation (Hall et al., 1988), and move slowly and exhibit impaired directionality during chemotaxis, consistent with an inability of these cells to effectively form a dominant pseudopod in the direction of the gradient. Similarly, roco2− cells exhibit reduced persistence during random cell motility. In contrast, cells expressing Roco2ΔLRR, which has constitutively high kinase activity, have an elevated second cortical F-actin peak and an extended pseudopod. Furthermore, Roco2ΔLRR/roco2− cells produce extended pseudopodia, a phenotype that is consistent with an elevated second cortical F-actin peak. Because Roco2 kinase activity is induced upon chemoattractant stimulation, with a broad peak that is cotemporaneous with the second F-actin peak linked to pseudopod formation, we suggest that chemoattractant-mediated Roco2 activity is essential for the formation of a stable, functional pseudopod. Our finding that roco2− cells have a normal first F-actin peak indicates that Roco2 is not required for the cringe response, in which there is a uniform polymerization of actin along the cell cortex. Although Roco2 is not required for this response, high Roco2 activity impairs this response, as is observed in cells expressing Roco2ΔLRR. The second F-actin peak is depressed in cells lacking PI3K in Dictyostelium (Chen et al., 2003). However, we found that inhibition of PI3K activity by LY294002 does not affect Roco2 kinase activity (unpublished data), suggesting that the PI3K and Roco2 pathways are independent. Roco2 also plays a role during cytokinesis, which helps explain the growth defects of roco2− cells reported previously (Abe et al., 2003).
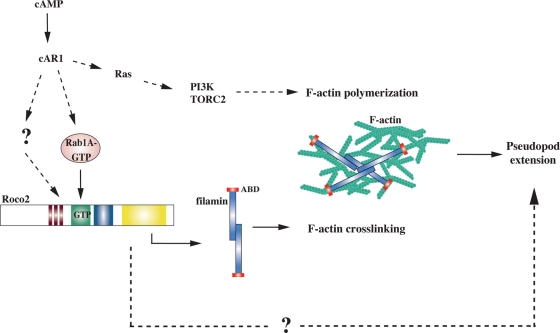
FIGURE 9:
Model for Roco2 regulation of pseudopod extension. The chemoattractant cAMP, through the GPCR cAR1, activates multiple signaling pathways that lead to F-actin polymerization, including Ras-mediated activation of PI3K and TORC2. We provide evidence that chemoattractant-mediated Roco2 activation requires Rab1A-GTP and this functions to control pseudopod extension by regulating filamin. We expect Roco2 activation requires GTP binding to the ROC domain as with other Roco family members. Roco2 may have inputs in addition to Rab1A that directly regulate its activation downstream from the receptor and heterotrimeric G proteins. Our results also do not exclude that Roco2 functions to control pseudopod extension via other effectors in addition to filamin. We suggest that the control of pseudopod extension is independent of pathways that directly control localized F-actin polymerization. We expect that, but do not know whether, chemoattractant stimulation leads to Rab1A activation. ABD, F-actin binding domain of filamin.
Filamin is an F-actin cross-linking protein that is required for the pseudopod extension, functioning to stabilize the F-actin network at the cortex (Cox et al., 1992, 1995; Cox, 1995; Stossel et al., 2001; Khaire et al., 2007; Washington and Knecht, 2008; Baldassarre et al., 2009). We found that Roco2 interacts with the F-actin cross-linking protein filamin (abpC) and that activated Roco2 (Roco2ΔLRR) pulls down more filamin than wild-type Roco2 in a coimmunoprecipitation assay, while the kinase-dead form pulls down less protein, consistent with filamin binding to active Roco2. Results from our studies presented here on roco2− and filamin-null cells, and previous studies on Dictyostelium filamin, support a model in which filamin lies downstream from Roco2 to control pseudopod extension. Disruption of filamin (abpC− cells) renders cells unable to polarize effectively or extend a pseudopod (this paper; Cox, 1995; Cox et al., 1992, 1995). abpC− cells can polymerize F-actin, but they lack a second peak of cortically localized F-actin, possibly because the F-actin is unstable or cannot be stably localized to the cortex in the absence of filamin, resulting in the lack of a stable pseudopod (Cox et al., 1992, 1995; Cox, 1995) and chemotaxis phenotypes similar to those of roco2− cells. Furthermore, the kinetics of filamin incorporation into the cytoskeleton coincide with the second F-actin peak and Roco2 activity (Cox et al., 1992, 1995; Cox, 1995). Consistent with a model in which filamin is a downstream Roco2 effector, abpC− cells expressing Roco2ΔLRR have defects similar to those of abpC− cells, show reduced polarity, lack the long pseudopodial protrusion observed in Roco2ΔLRR-expressing wild-type cells, and have a reduced cortical second F-actin peak. We suggest that a major function of Roco2 is to stabilize the nascent pseudopod and enhance F-actin polymerization, in part through filamin. Although Roco2 appears to directly interact with filamin, we do not know if filamin is a direct substrate of Roco2. In any case, we expect that Roco2 has substrates and/or downstream effectors in addition to filamin that are required for pseudopod extension and stabilization. The Dictyostelium Roco kinase GbpC, and possibly Pats1, in addition to Roco2, control the cytoskeleton (Bosgraaf et al., 2002; van Egmond and van Haastert, 2010). Interestingly, LRRK2 phosphorylates ezrin–radixin–moesin (ERM) cytoskeletal proteins and interacts with F-actin, and gain- and loss-of-function LRRK2 proteins alter the interactions between ERM proteins and F-actin and neuronal morphogenesis, suggesting Roco kinases in other systems may have similar functions in controlling the cytoskeleton (Jaleel et al., 2007; Parisiadou et al., 2009; Zechel et al., 2010; Meixner et al., 2011). We also found that Roco2, like LRRK2 in both mammalian and Drosophila cells, binds to the redox-dependent molecular chaperone DJ-1 (Venderova et al., 2009), which may be a regulator of some Roco kinases.
We provide evidence that, unexpectedly, Rab1A binds to and regulates Roco2. Both Rab1A and Roco2 localize to the pseudopodium, and constitutively active Rab1AQ67L, but not the Rab1A with reduced affinity for guanine nucleotide (GST-Rab1AN121I), pulls down Roco2 from cell extracts. Overexpression of Rab1AS22N results in reduced Roco2 kinase activity and phenotypes similar to those of roco2− cells, including the decrease or loss of the second cortical actin peak and decreased cell polarization, whereas expression of Rab1AQ67L causes elevated Roco2 activity and phenotypes similar to those of Roco2ΔLRR cells, including a high second actin peak, increased cell speed, and the formation of extended pseudopodia that have reduced adhesion to the substratum. We demonstrate that coexpression of Roco2ΔLRR with Rab1AS22N in roco2− cells produces a Roco2ΔLRR phenotype, whereas expression of Rab1AQ67L in roco2− cells yields a roco2− phenotype, consistent with Rab1A-GTP being upstream from and activating Roco2. As Rab1AQ67L pulls down Roco2, Rab1A may function as a direct activator of Roco2. We cannot exclude the possibility of multiple signaling inputs into Roco2 activation, one of which is Rab1A-GTP. At present, we do not know if Rab1A is activated in response to chemoattractant stimulation, although we expect that it is. Furthermore, we presume that Rab1A also has functions in membrane trafficking, some of which may be involved in plasma membrane homeostasis during chemotaxis. Rab1A may help integrate F-actin-mediated pseudopod protrusion with the leading-edge membrane trafficking that has been suggested as a prerequisite for chemotaxis (Traynor and Kay, 2007; Zanchi et al., 2010). We do not observe Roco2 on vesicles and do not know if the Rab1A that activates Roco2 is membrane-associated or cytosolic.
Our findings identify a novel pathway by which pseudopod extension is regulated and represent the first demonstration of the role of a Rab in regulating a kinase, identifying a new function for Rab1A. We suggest that other Roco proteins (including possibly LRRK2) may be regulated by a similar mechanism. Our finding that Roco2 is regulated by Rab1A indicates that the Rab family of GTPases are, unexpectedly, regulators of protein kinase activity and thus may function similarly to Rho and Ras superfamily GTPases in controlling cytoskeletal function through the regulation of effector kinases.
MATERIALS AND METHODS
Strains and constructs
In our wild-type strain KAx-3, the gene encoding Roco2 is duplicated. We created a roco2−/− (designated roco2−) strain by sequentially disrupting the two copies using knockout constructs in which the BsrR or the HygR cassette was inserted into the unique BamHI site within the Roco2 gene. Potential knockouts were selected in HL5 medium containing blasticidin or hygromycin, screened by PCR, and confirmed by Southern blot hybridization (unpublished data). The filamin-null strain was obtained from the Dictyostelium Stock Center (Northwestern University, Evanston, IL).
The full genomic sequence of roco2 was generated by PCR to remove the termination codon, cloned into the SpeI-PstI site of pBluescript KS(−), sequenced, and subcloned in frame into the expression vector EXP-4(+) containing the open reading frame (ORF) of GFP at its C-terminus. For expression of various versions of the Roco2 protein, Roco2 sequences were amplified by PCR with the appropriate oligonucleotides, sequenced, and cloned into the SpeI-PstI site of an EXP-4(+) vector containing a C-term GFP fragment generated previously. The T7 construct was made using a Dictyostelium T7-Exp4 (N-term) plasmid in which the coding sequence of the T7 epitope is preceded by an ATG start codon and 5 A residues. The roco2 sequence was reamplified to modify the start codon and add the appropriate restriction sites (EcoRI and XhoI) for in-frame cloning into the expression vector.
Chemotaxis and motility assays
Chemotaxis and motility assays have been described previously (Sasaki et al., 2007; Charest et al., 2010). For Digital Image Analysis System (DIAS; David Soll, University of Iowa, Iowa City, IA) analysis, we calculated the parameters of at least five (usually 7) cells from each of at least three separate movies recorded on separate days. Because DIAS' autotrace does not work as well as we needed with differential image contrast (DIC) images, the cells were all hand-traced and then analyzed. The SD for the statistics was calculated by the program on the total number of cells used as described previously.
In vivo pull-down and purification of Roco2-interacting proteins
Vegetative and aggregation-competent cells were washed with Na/K phosphate buffer and resuspended at a density of 1 × 108 cells/ml in Na/K phosphate buffer. Cells were lysed (1% NP-40, 300 mM NaCl, 40 mM MOPS [pH 7.0], 20% glycerol, 2 mM Na3VO4, 2 μg/ml leupeptin, and 5 μg/ml aprotinin) and incubated in 2.5 mg/ml dithio-bis-succinimidylpropionate (DSP; EMD Chemicals, La Jolla, CA) for 10 min. The cross-linking reactions were quenched with 200 mM Tris-HCl (pH 7.4). Proteins in total cell extracts were subjected to immunoprecipitation with 25 μl resin of anti-T7 antibody agarose conjugate (Sigma-Aldrich, St. Louis, MO). The coimmunoprecipitated proteins were eluted with 0.1 M glycine (pH 2.5), analyzed by silver staining, and subsequently precipitated with methanol/chloroform.
MS was performed on the total protein sample eluted from the beads as described previously (Du et al., 2008; Charest et al., 2010).
Immunofluorescence microscopy
Pulsed or vegetative cells were allowed to settle onto glass coverslips for 30 min. Cells were quickly fixed and permeabilized with 100% cold methanol at −20°C for 10 min. Cells were washed and incubated with phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) and 0.05% Tween 20 for 1 h. Cells were first incubated with anti-Rab1A (1:500; cat# H00005861-M07A; Novus Biologicals, Littleton, CO) for 1 h. Cells were stained with fluorescein isothiocyanate 1 (FITC)-conjugated anti–mouse immunoglobulin G (1:200) or red fluorescent label rhodamine (TRITC)-labeled phalloidin for 1 h, washed, and mounted for observation with a 60× oil immersion lens. For cells expressing GFP fusion Roco2 proteins, cells were directly stained with TRITC-labeled phalloidin for 1 h, washed, and mounted for observation with a 60× oil immersion lens. For nuclei staining, after fixation and permeabilization, cells were mounted with mounting medium containing 0.0001% Hoechst dye. Images were taken on a spinning-disk confocal microscope.
Biochemical measurements of F-actin, Ras, and Akt/PKB
Chemoattractant-induced Ras activity, PKB kinase activity, and F-actin measurements were performed as described previously in detail (Insall et al., 1996; Meili et al., 1999; Sasaki et al., 2004, 2007; van Haastert, 2006; Zhang et al., 2008). Briefly, for the measurement of F-actin levels in the cortical fraction, vegetative or pulsed cells before and after stimulation in the Triton-insoluble fraction were grown on Petri plates, harvested, washed, and suspended. For examination of F-actin levels upon chemoattractant stimulation, cells were starved for 2 h, pulsed with cAMP for 5 h, washed, and then treated with 1 mM caffeine for 30 min before stimulation with 100 mM cAMP. The cytoskeletal fraction was isolated as proteins insoluble in the detergent Triton X-100 as described previously (Steimle et al., 2001). The protein pellets were separated by SDS–PAGE on 8% acrylamide gels, and stained with Coomassie Blue. The actin band was scanned, and the actin content was quantified using Image Gauge software (Fuji, Stamford, CT). All experiments were repeated at least three times.
Data analysis
For the analysis of kinase or small GTPase activation assays, at least 3 independent experiments were performed for all strains (on different days) using protocols that have been previously described (Sasaki et al., 2007; Charest et al., 2010). A wild-type control was always run within each experiment along with the mutant strains being examined to act as an internal control and standard. Mutants were compared with the internal wild-type control done on the same day. All samples were run in parallel, and we used the same cocktail mix for the assay for all samples, so the data are internally comparable.
For such assays, the activity was normalized to the activity of wild-type cells at the peak for kinases such as PKB, as the activity in unstimulated cells is low and variable. In the case of Roco2, which has a moderate and accurately measurable basic activity at 0 s (unstimulated cells), we set the activity in unstimulated wild-type cells as 1.0 within each experiment. Intensity of the bands was quantified using a phosphorimager or densitometry tracings of autoradiographs within the linear range, with all blots/gels being autoradiographed on the same file, background subtracted, and normalized at each time point for protein levels. For each experiment, the relative activity was then calculated using the wild-type as the internal comparison in each experiment. The values at each time point (averaged if duplicates are done) were then averaged for the 3 (or more) different, independent experiments to obtain the value for the plot. The SD was calculated on the repeats.
For F-actin polymerization analysis, the level of F-actin in wild-type unstimulated cells was set at 1.0. The level of F-actin in the Triton-X 100 insoluble fraction was analyzed using SDS–PAGE followed by staining, as described above in this paper. F-actin levels were quantified within each experiment and averaged at each time point over the three repeats.
Protein isolation and Western blot analysis
Cells were harvested from growing cultures via centrifugation at 1500 × g for 2 min. The pelleted cells were washed in Na/K buffer, resuspended in 500 μl Na/K buffer, and lysed by adding same amount of lysis buffer (1% NP-40, 300 mM NaCl, 40 mM MOPS [pH 7.0], 20% glycerol, 2 mM Na3VO4, 2 μg/ml leupeptin, and 5 μg/ml aprotinin) and incubated on ice for 15 min. The lysate was then centrifuged at 10,000 × g for 10 min at 4°C to pellet the cell debris and the supernatant was transferred to a new tube and stored at −70°C. The concentration of the protein samples was determined using the Bradford method (Bio-Rad Protein Assay, Bio-Rad, Irvine, CA) with BSA as a standard. Laemmli buffer was added to the protein samples that were then boiled for 5 min and subjected to SDS–PAGE on 8 or 12% gels. The separated proteins were transferred to PVDF membranes (Amersham, Arlington Heights, IL) and anti-T7, anti-FLAG, or anti-filamin (Bandala-Sanchez et al., 2006) antibodies were used to detect Roco2, Rab1A, and filamin, respectively. Membranes were blocked with Tris-buffered saline (TBS; 10 mM Tris-HCl, 150 mM NaCl, pH 7.5) containing 5% skim milk powder for at least 1 h at room temperature. The membranes were briefly washed in TBS-Tween (TBST; 20 mM Tris-HCl [pH 7.5], 500 mM NaCl, 0.05% Tween20) and incubated with primary antibody (1:1000) in TBS containing 3% skim milk powder overnight at 4°C. The membrane was washed three times with TBST and incubated with anti-rabbit or anti–mouse horseradish peroxidase (HRP)-conjugated secondary antibody (Promega, Madison, WI) 1:10,000 for 1 h at room temperature followed by three washes in TBST. The signals were detected using the enhanced chemiluminescence system (ECL, Amersham).
Lysis, immunoprecipitation, and pull-down
The samples were lysed by mixing with an equal volume of 2× lysis buffer (1% NP-40, 300 mM NaCl, 40 mM MOPS [pH 7.0], 20% glycerol, 2 mM Na3VO4, 2 μg/ml leupeptin, and 5 μg/ml aprotinin) and incubated on ice for 15 min. The lysate was centrifuged at 10,000 × g for 10 min at 4°C to pellet the cell debris, and the supernatant was transferred to a new tube. Antibody-conjugated agarose beads (20 μl, FLAG or T7; Sigma) were added to 1 ml supernatant and rotated overnight at 4°C. The beads were washed five times with lysis buffer and Laemmli buffer was added to the protein samples. Samples were boiled for 5 min and subjected to SDS–PAGE on 8 or 12% gels. The coimmunoprecipitated proteins were eluted with 0.1 M glycine (pH 2.5), analyzed by silver staining, and subsequently precipitated with methanol/chloroform.
An N-terminal GST fusion of Dictyostelium wild-type Rab1A was constructed using Rab1A cloned from cDNA and then used to create a Q67L and N121I substitution mutants. GST fusions of the wild-type, constitutively active (Q67L), and non-guanine nucleotide-binding (N121I) Rab1A were expressed in Escherichia coli grown at 37°C that had been induced with 0.2 mM IPTG at 37°C for 3 h. Cells were harvested, washed once with cold PBS, and resuspended in PBS containing 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT), aprotinin, and leupeptin, and the suspension was sonicated. NP40 was added to 1% and the cell lysates were cleared by centrifugation. The GST-Rap1A fusions were purified by incubating with glutathione Sepharose with gentle rocking for 1 h at 4°C. The beads were washed four times with PBS and eluted with glutathione. The beads were stored in 50% glycerol at −20°C.
For the pull-down assays, cells expressing T7-Roco2 were lysed in 100 mM Tris (pH 7.5), 300 mM NaCl, 50 mM MgCl2, 20% glycerol, 1% NP40, 2 mM DTT, and 2 mM Vanadate, aprotinin, and leupeptin. The lysates were spun for 10 min and the supernatants were transferred to microtube to which 10 μg GST-Rab1A Sepharose was added and incubated with rocking for 1 h at 4°C. The beads were washed four times with lysis buffer. The samples were subjected to SDS-PAGE and the T7-Roco2 was visualized by Western blotting using mAb against the T7 epitope. The membrane was stained with Coomassie Blue to observe the loaded protein.
Roco2 kinase activity assay
T7-tagged Roco2 was immunoprecipitated from cell lysates using anti-T7 resin. To measure the Roco2 kinase activity, the beads were incubated with 75 μl of kinase buffer containing 5 μCi [32P]ATP, 5 μM ATP, and 5 μg MBP as substrate. After 90 min, the reaction was stopped by the addition of 25 μl 4× Laemmli buffer and boiling for 5 min. The samples were separated by SDS–PAGE (15%), blotted onto a PVDF membrane (Millipore), and exposed to film. All assays were repeated independently. Autoradiograms were obtained by exposure of phosphorimager plates to the membranes, which were then quantified by a Typhoon Trio Reader (GE Healthcare, Waukesha, WI). The signals were normalized to Roco2 levels determined by Western blotting. Data analysis was performed using Image Quant software (GE Healthcare). For the comparison of different Roco2 mutants, at least four independent data points were used for each mutant.
Supplementary Material
Acknowledgments
We thank Angelika Noegel for the GFP-filamin construct and the Firtel lab members and Colin Jamora for constructive discussions and critical reading of the manuscript. This work was supported by U.S. Public Health Service grant GM037830 to R.A.F.
Abbreviations used:
- BSA
bovine serum albumin
- COR
C-terminal of Roc
- DIC
differential image contrast
- DSP
dithio-bis-succinimidylpropionate
- DTT
dithiothreitol
- ERM
ezrin–radixin–moesin
- FITC
fluorescein isothiocyanate 1
- GFP
green fluorescent protein
- LRR
leucine-rich repeat
- MBP
myelin basic protein
- MEKK
mitogen-activated protein kinase kinase kinase
- MLK
mixed-lineage kinase
- MS
mass spectrometry
- MyoII
myosin II
- ORF
open reading frame
- PBS
phosphate-buffered saline
- PD
Parkinson's disease
- PI3K
phosphatidylinositol-3 kinase
- PMSF
phenylmethylsulfonyl fluoride
- RFP
red fluorescent protein
- Roc
Ras of complex proteins
- TBS
Tris-buffered saline
- TBST
TBS-Tween
- TRITC
red fluorescent label rhodamine
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-12-0937) on May 5, 2011.
REFERENCES
- Abe T, Langenick J, Williams JG. Rapid generation of gene disruption constructs by in vitro transposition and identification of a Dictyostelium protein kinase that regulates its rate of growth and development. Nucleic Acids Res. 2003;31:e107. doi: 10.1093/nar/gng095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abysalh JC, Kuchnicki LL, Larochelle DA. The identification of pats1, a novel gene locus required for cytokinesis in Dictyostelium discoideum. Mol Biol Cell. 2003;14:14–25. doi: 10.1091/mbc.E02-06-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez C, Garcia-Mata R, Brandon E, Sztul E. COPI recruitment is modulated by a Rab1b-dependent mechanism. Mol Biol Cell. 2003;14:2116–2127. doi: 10.1091/mbc.E02-09-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandala-Sanchez E, Annesley SJ, Fisher PR. A phototaxis signalling complex in Dictyostelium discoideum. Eur J Cell Biol. 2006;85:1099–1106. doi: 10.1016/j.ejcb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Baldassarre M, Razinia Z, Burande CF, Lamsoul I, Lutz PG, Calderwood DA. Filamins regulate cell spreading and initiation of cell migration. PLoS One. 2009;4:e7830. doi: 10.1371/journal.pone.0007830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosgraaf L, Russcher H, Smith JL, Wessels D, Soll DR, van Haastert PJ. A novel cGMP signalling pathway mediating myosin phosphorylation and chemotaxis in Dictyostelium. EMBO J. 2002;21:4560–4570. doi: 10.1093/emboj/cdf438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosgraaf L, van Haastert PJ. Roc, a Ras/GTPase domain in complex proteins. Biochim Biophys Acta. 2003;1643:5–10. doi: 10.1016/j.bbamcr.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Bosgraaf L, Waijer A, Engel R, Visser AJ, Wessels D, Soll D, van Haastert PJ. RasGEF-containing proteins GbpC and GbpD have differential effects on cell polarity and chemotaxis in Dictyostelium. J Cell Sci. 2005;118:1899–1910. doi: 10.1242/jcs.02317. [DOI] [PubMed] [Google Scholar]
- Charest PG, Shen Z, Lakoduk A, Sasaki AT, Briggs SP, Firtel RA. A Ras signaling complex controls the RasC-TORC2 pathway and directed cell migration. Dev Cell. 2010;18:737–749. doi: 10.1016/j.devcel.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Janetopoulos C, Huang YE, Iijima M, Borleis J, Devreotes PN. Two phases of actin polymerization display different dependencies on PI(3,4,5)P3 accumulation and have unique roles during chemotaxis. Mol Biol Cell. 2003;14:5028–5037. doi: 10.1091/mbc.E03-05-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis J, Bresnick A, Demma M, Dharmawardhane S, Eddy R, Hall AL, Sauterer R, Warren V. Mechanisms of amoeboid chemotaxis: an evaluation of the cortical expansion model. Dev Genet. 1990;11:333–340. doi: 10.1002/dvg.1020110504. [DOI] [PubMed] [Google Scholar]
- Cox D. An In Vivo Analysis of the Function of abp-120 in Dictyostelium Amoebae Using Molecular Genetics (Actin Binding Protein) New York:: Yeshiva University; 1995. p. 257. [Google Scholar]
- Cox D, Condeelis J, Wessels D, Soll D, Kern H, Knecht DA. Targeted disruption of the ABP-120 gene leads to cells with altered motility. J Cell Biol. 1992;116:943–955. doi: 10.1083/jcb.116.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D, Ridsdale JA, Condeelis J, Hartwig J. Genetic deletion of ABP-120 alters the three-dimensional organization of actin filaments in Dictyostelium pseudopods. J Cell Biol. 1995;128:819–835. doi: 10.1083/jcb.128.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson HW, Balch WE. Differential inhibition of multiple vesicular transport steps between the endoplasmic reticulum and trans Golgi network. J Biol Chem. 1993;268:4216–4226. [PubMed] [Google Scholar]
- De Lozanne A, Spudich JA. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- Du F, Edwards K, Shen Z, Sun B, De Lozanne A, Briggs S, Firtel RA. Regulation of contractile vacuole formation and activity in Dictyostelium. EMBO J. 2008;27:2064–2076. doi: 10.1038/emboj.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;109:611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- Goldberg JM, Bosgraaf L, van Haastert PJ, Smith JL. Identification of four candidate cGMP targets in Dictyostelium. Proc Natl Acad Sci USA. 2002;99:6749–6754. doi: 10.1073/pnas.102167299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio E, et al. The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J Biol Chem. 2008;283:16906–16914. doi: 10.1074/jbc.M708718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AL, Schlein A, Condeelis J. Relationship of pseudopod extension to chemotactic hormone-induced actin polymerization in amoeboid cells. J Cell Biochem. 1988;37:285–299. doi: 10.1002/jcb.240370304. [DOI] [PubMed] [Google Scholar]
- Insall RH. Understanding eukaryotic chemotaxis: a pseudopod-centred view. Nat Rev Mol Cell Biol. 2010;11:453–458. doi: 10.1038/nrm2905. [DOI] [PubMed] [Google Scholar]
- Insall RH, Borleis J, Devreotes PN. The aimless RasGEF is required for processing of chemotactic signals through G-protein-coupled receptors in Dictyostelium. Curr Biol. 1996;6:719–729. doi: 10.1016/s0960-9822(09)00453-9. [DOI] [PubMed] [Google Scholar]
- Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev Cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A, Alessi DR. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson's disease mutants affect kinase activity. Biochem J. 2007;405:307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetopoulos C, Firtel RA. Directional sensing during chemotaxis. FEBS Lett. 2008;582:2075–2085. doi: 10.1016/j.febslet.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedrin D, van Rheenen J, Hernandez L, Condeelis J, Segall JE. Cell motility and cytoskeletal regulation in invasion and metastasis. J Mammary Gland Biol Neoplasia. 2007;12:143–152. doi: 10.1007/s10911-007-9046-4. [DOI] [PubMed] [Google Scholar]
- Khaire N, Muller R, Blau-Wasser R, Eichinger L, Schleicher M, Rief M, Holak TA, Noegel AA. Filamin-regulated F-actin assembly is essential for morphogenesis and controls phototaxis in Dictyostelium. J Biol Chem. 2007;282:1948–1955. doi: 10.1074/jbc.M610262200. [DOI] [PubMed] [Google Scholar]
- Knuth M, Khaire N, Kuspa A, Lu SJ, Schleicher M, Noegel AA. A novel partner for Dictyostelium filamin is an alpha-helical developmentally regulated protein. J Cell Sci. 2004;117:5013–5022. doi: 10.1242/jcs.01366. [DOI] [PubMed] [Google Scholar]
- Marin I. The Parkinson disease gene LRRK2: evolutionary and structural insights. Mol Biol Evol. 2006;23:2423–2433. doi: 10.1093/molbev/msl114. [DOI] [PubMed] [Google Scholar]
- Marin I. Ancient origin of the Parkinson disease gene LRRK2. J Mol Evol. 2008;67:41–50. doi: 10.1007/s00239-008-9122-4. [DOI] [PubMed] [Google Scholar]
- Meili R, Ellsworth C, Lee S, Reddy TBK, Ma H, Firtel RA. Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J. 1999;18:2092–2105. doi: 10.1093/emboj/18.8.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meixner A, Boldt K, van Troys M, Askenazi M, Gloeckner CJ, Bauer M, Marto JA, Ampe C, Kinkl N, Ueffing M. A QUICK Screen for Lrrk2 interaction partners–leucine-rich repeat kinase 2 is involved in actin cytoskeleton dynamics. Mol Cell Proteomics. 2011;10, M110001172 doi: 10.1074/mcp.M110.001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paisan-Ruiz C, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Parisiadou L, et al. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J Neurosci. 2009;29:13971–13980. doi: 10.1523/JNEUROSCI.3799-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Gomer RH. The ROCO kinase QkgA is necessary for proliferation inhibition by autocrine signals in Dictyostelium discoideum. Eukaryot Cell. 2010;9:1557–1565. doi: 10.1128/EC.00121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pind SN, Nuoffer C, McCaffery JM, Plutner H, Davidson HW, Farquhar MG, Balch WE. Rab1 and Ca2+ are required for the fusion of carrier vesicles mediating endoplasmic reticulum to Golgi transport. J Cell Biol. 1994;125:239–252. doi: 10.1083/jcb.125.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannerud R, Marie M, Nizak C, Dale HA, Pernet-Gallay K, Perez F, Goud B, Saraste J. Rab1 defines a novel pathway connecting the pre-Golgi intermediate compartment with the cell periphery. Mol Biol Cell. 2006;17:1514–1526. doi: 10.1091/mbc.E05-08-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki AT, Chun C, Takeda K, Firtel RA. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J Cell Biol. 2004;167:505–518. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki AT, Janetopoulos C, Lee S, Charest PG, Takeda K, Sundheimer LW, Meili R, Devreotes PN, Firtel RA. G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J Cell Biol. 2007;178:185–191. doi: 10.1083/jcb.200611138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin N, et al. LRRK2 regulates synaptic vesicle endocytosis. Exp Cell Res. 2008;314:2055–2065. doi: 10.1016/j.yexcr.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Steimle PA, Yumura S, Cote GP, Medley QG, Polyakov MV, Leppert B, Egelhoff TT. Recruitment of a myosin heavy chain kinase to actin-rich protrusions in Dictyostelium. Curr Biol. 2001;11:708–713. doi: 10.1016/s0960-9822(01)00182-8. [DOI] [PubMed] [Google Scholar]
- Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- Swaney KF, Huang CH, Devreotes PN. Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys. 2010;39:265–289. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toft M, Mata IF, Kachergus JM, Ross OA, Farrer MJ. LRRK2 mutations and Parkinsonism. Lancet. 2005;365:1229–1230. doi: 10.1016/S0140-6736(05)74809-1. [DOI] [PubMed] [Google Scholar]
- Traynor D, Kay RR. Possible roles of the endocytic cycle in cell motility. J Cell Sci. 2007;120:2318–2327. doi: 10.1242/jcs.007732. [DOI] [PubMed] [Google Scholar]
- Tuxworth RI, Cheetham JL, Machesky LM, Spiegelmann GB, Weeks G, Insall RH. Dictyostelium RasG is required for normal motility and cytokinesis, but not growth. J Cell Biol. 1997;138:605–614. doi: 10.1083/jcb.138.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda TQP, Kitayama C, Yumura S. Myosin II-independent cytokinesis in Dictyostelium: its mechanism and implications. Cell Struct Func. 2000;25:1–10. doi: 10.1247/csf.25.1. [DOI] [PubMed] [Google Scholar]
- van Egmond WN, Kortholt A, Plak K, Bosgraaf L, Bosgraaf S, Keizer-Gunnink I, van Haastert PJ. Intramolecular activation mechanism of the Dictyostelium LRRK2 homolog Roco protein GbpC. J Biol Chem. 2008;283:30412–30420. doi: 10.1074/jbc.M804265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Egmond WN, van Haastert PJ. Characterization of the Roco protein family in Dictyostelium discoideum. Eukaryot Cell. 2010;9:751–761. doi: 10.1128/EC.00366-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haastert PJ, Devreotes PN. Chemotaxis: signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- van Haastert PJ, Veltman DM. Chemotaxis: navigating by multiple signaling pathways. Sci STKE. 2007;396:pe40. doi: 10.1126/stke.3962007pe40. [DOI] [PubMed] [Google Scholar]
- van Haastert PJM. Analysis of signal transduction: formation of cAMP, cGMP, and Ins(1,4,5)P3 in vivo and in vitro. Meth Mol Biol. 2006;346:369–392. doi: 10.1385/1-59745-144-4:369. [DOI] [PubMed] [Google Scholar]
- Venderova K, et al. Leucine-rich repeat kinase 2 interacts with Parkin, DJ-1 and PINK-1 in a Drosophila melanogaster model of Parkinson's disease. Hum Mol Genet. 2009;18:4390–4404. doi: 10.1093/hmg/ddp394. [DOI] [PubMed] [Google Scholar]
- Washington RW, Knecht DA. Actin binding domains direct actin-binding proteins to different cytoskeletal locations. BMC Cell Biol. 2008;9:10. doi: 10.1186/1471-2121-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels D, Voss E, von Bergen N, Burns R, Stites J, Soll DR. A computer-assisted system for reconstructing and interpreting the dynamic three-dimensional relationships of the outer surface, nucleus and pseudopods of crawling cells. Cell Motil Cytoskeleton. 1998;41:225–246. doi: 10.1002/(SICI)1097-0169(1998)41:3<225::AID-CM4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Coombes CE, Kilaru A, Li X, Gitler AD, Bowers WJ, Dawson VL, Dawson TM, Moore DJ. GTPase activity plays a key role in the pathobiology of LRRK2. PLoS Genet. 2010;6:e1000902. doi: 10.1371/journal.pgen.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanchi R, Howard G, Bretscher MS, Kay RR. The exocytic gene secA is required for Dictyostelium cell motility and osmoregulation. J Cell Sci. 2010;123:3226–3234. doi: 10.1242/jcs.072876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechel S, Meinhardt A, Unsicker K, von Bohlen Und Halbach O. Expression of leucine-rich-repeat-kinase 2 (LRRK2) during embryonic development. Int J Dev Neurosci. 2010;28:391–399. doi: 10.1016/j.ijdevneu.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Zhang S, Charest PG, Firtel RA. Spatiotemporal regulation of Ras activity provides directional sensing. Curr Biol. 2008;18:1587–1593. doi: 10.1016/j.cub.2008.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.