The basal body is a microtubule-organizing center responsible for organizing the cilium. A widely conserved basal body component is the Ca2+-binding protein centrin. A mutagenic analysis of the Tetrahymena centrin shows that its two domains have distinct basal body functions and that Ca2+ is necessary for both functions.
Abstract
The basal body is a microtubule-organizing center responsible for organizing the cilium, a structure important for cell locomotion and sensing of the surrounding environment. A widely conserved basal body component is the Ca2+-binding protein centrin. Analyses of centrin function suggest a role in basal body assembly and stability; however, its molecular mechanisms remain unclear. Here we describe a mutagenic strategy to study the function and essential nature of the various structural features of Cen1 in the ciliate Tetrahymena. We find that the two domains of Cen1 are both essential, and examination of strains containing mutant CEN1 alleles indicates that there are two predominant basal body phenotypes: misorientation of newly assembled basal bodies and stability defects. The results also show that the two domains of Cen1 are able to bind Ca2+ and that perturbation of Ca2+ binding affects Cen1 function. In all, the data suggest that the two domains of Cen1 have distinct functions.
INTRODUCTION
The basal body is a microtubule-organizing center (MTOC) responsible for organizing and anchoring the cilium at the surface of the cell. It is widely found in eukaryotes, except for yeast and higher plants. The cilium is a microtubule-based structure that projects outward from the surface of the cell (Basto et al., 2006; Marshall and Nonaka, 2006). There are two classes of cilia: 1) motile cilia and 2) primary cilia. Motile cilia generate a sweeping motion, which is important for cell locomotion or fluid flow (Marshall and Nonaka, 2006). The second class of cilia, primary cilia, is immotile and acts as a sensory receptor for the cell. Primary cilia play key roles in mechanosensory, chemical sensory, and photosensory functions (Marshall and Nonaka, 2006). Dysfunction at the basal body or the cilium is associated with several human diseases such as polycystic kidney disease and Bardet–Biedl syndrome (Badano et al., 2006). The importance of the basal body is further highlighted by its close relationship to the centrioles found in the centrosome, a MTOC involved in mitotic spindle formation (Bornens and Azimzadeh, 2007). Given the requirement of basal bodies in the structure and function of cilia, detailed molecular analyses of basal bodies are needed.
The basal body is a cylindrical structure consisting of an array of nine microtubule triplets, giving it ninefold symmetry (Allen, 1969; Wolfe, 1970; Dawe et al., 2007; Pearson and Winey, 2009). The structure can be separated into three domains (Kilburn et al., 2007). At the proximal end of the basal body is a structure called the cartwheel, which is comprised of nine spokes originating from a central hub extending to the microtubule triplets (Gavin, 1984; Strnad and Gönczy, 2008). At the distal end is a region called the transition zone, which marks the transition from the microtubule triplets of the basal body to the microtubule doublets of the axoneme (Marshall and Nonaka, 2006). Between the cartwheel and the transition zone is the cylindrical core of the basal body. Despite our understanding of basal body morphology, the roles of its components in assembling and maintaining the structure remain poorly understood.
Proteomic analyses of isolated centrioles or basal bodies from various organisms show that the small Ca2+-binding protein centrin is a widely conserved component (Andersen et al., 2003; Keller et al., 2005; Kilburn et al., 2007; Liu et al., 2007). Ultrastructural examination by immunoelectron microscopy in different organisms show that centrins can be found at two key regions in the basal body: 1) the distal region that is at or near the transition zone, and 2) sites of new basal body assembly (Sanders and Salisbury, 1994; Laoukili et al., 2000; Geimer and Melkonian, 2005; Stemm-Wolf et al., 2005; Kilburn et al., 2007). Structurally, centrins consist of two independent domains connected by a short linker, with each domain containing a pair of EF hands, a Ca2+-binding motif (Figure 1A) (Veeraraghavan et al., 2002; Hu and Chazin, 2003; Matei et al., 2003; Yang et al., 2005; Li et al., 2006; Thompson et al., 2006). An EF hand consists of an α-helix–loop–α-helix structural unit, with Ca2+ binding occurring within the loop region (Gifford et al., 2007). The EF hands have been numbered, with one and two in the N‑terminal domain and three and four in the C‑terminal domain (Figure 1A). At the beginning of the N-terminal domain is a long tail (containing up to 20 residues) of unknown function (Li et al., 2006). Analyses of centrin function in humans, budding yeast, Chlamydomonas, Paramecium, and Tetrahymena suggest that it is involved in MTOC assembly and/or stability (Spang et al., 1993; Salisbury et al., 2002; Koblenz et al., 2003; Ruiz et al., 2005; Stemm-Wolf et al., 2005; Yang et al., 2010); however, its function and mode of action have not been well characterized. The role of Ca2+ is not completely understood, but in vitro experiments suggest that it regulates interactions with binding partners (Geier et al., 1996; Hu et al., 2004; Martinez-Sanz et al., 2006, 2010).
FIGURE 1:
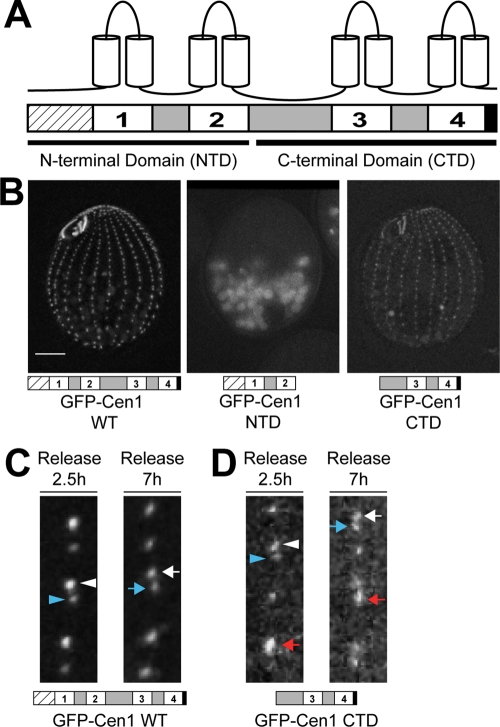
The C-terminal domain of Cen1 localizes to basal bodies. (A) Schematic showing the general structural features in centrins. Hatched box, N-terminal tail; white boxes, EF hands; gray boxes, regions between EF hands; black box, C-terminal tail. (B) Localization of GFP-tagged full-length WT Cen1, the NTD of Cen1, and the CTD of Cen1. Bar, 10 μm. (C) Incorporation of full-length WT GFP-Cen1. At 2.5 h after release from cell cycle arrest, new basal bodies (white arrowhead) are brighter than old basal bodies (blue arrowhead). At 7 h after release from cell cycle arrest, a more uniform labeling is observed for new (white arrow) and old (blue arrow) basal bodies. (D) The incorporation of Cen1's C-terminal domain labeled with GFP is similar to that of full-length WT Cen1. However, some basal bodies were much brighter than others at both time points (red arrow). White arrowhead, new basal body; blue arrowhead, old basal body. In C and D, the width of the insets is 3.5 μm.
The ciliate protist Tetrahymena thermophila is an ideal system in which to study basal bodies, their composition, and the molecular mechanisms of their components. Tetrahymena contains two major basal body populations: 1) the cortical rows, which run along the anterior–posterior axis of the cell, and 2) the oral apparatus, a feeding structure at the anterior end of the cell (Frankel, 2000). We and others previously identified Tetrahymena centrin 1 (Cen1), a human centrin 2 homologue, as a component of basal bodies (Guerra et al., 2003; Stemm-Wolf et al., 2005). A cen1 deletion allele is lethal due to defects in basal body assembly and maintenance (Stemm-Wolf et al., 2005). Immunoelectron microscopy shows that Cen1 localizes to the site of new assembly, the transition zone, and the midzone of the basal body (Stemm-Wolf et al., 2005; Kilburn et al., 2007).
We wanted to gain a better understanding of centrin function and its mode of action by studying Tetrahymena Cen1 in more detail. A mutagenic strategy was used to analyze the essential nature of the various centrin structural features. Through this approach, we found that the two domains of Cen1 have different functions, as well as differing Ca2+ affinities.
RESULTS
Only the C-terminal domain of Cen1 localizes to basal bodies
We asked whether the Cen1 N- or C-terminal domain alone is sufficient to rescue the lethal phenotype for the Tetrahymena cen1 null allele (cen1Δ) (see Supplemental Figure S1 for mutations generated in this study). Although the wild-type (WT) CEN1 allele would rescue the cen1Δ, N- or C-terminal domain deletions of Cen1 were unable to rescue the cen1Δ, indicating that neither domain alone is sufficient to perform Cen1 function. To ascertain whether either domain alone can localize to basal bodies, the domains of Cen1 were tagged with green fluorescent protein (GFP) using an expression vector under the control of the inducible metallothionein promoter. We found that only the C-terminal domain (CTD) of Cen1 localizes to basal bodies (Figure 1B).
The localization of Cen1's CTD was further analyzed by examining its incorporation into basal bodies (Pearson et al., 2009). In this assay, we used the inducible promoter to control the expression of the GFP-tagged proteins. Cells were arrested in G1 by media starvation to inhibit the assembly of new basal bodies. On transfer into growth media, new basal body assembly was initiated, and concurrent with release from arrest, expression of the GFP fusion protein was induced. At 2.5 h, just before the cells had completed one full cell cycle (Pearson et al., 2009; Pearson and Winey, 2009), bright and dim GFP-Cen1 signals were observed at basal bodies (Figure 1C, left, white and blue arrowheads, respectively). New basal body assembly occurs anterior to the old basal body (Perlman, 1973; Kaczanowski, 1978; Frankel, 2000), and, in this case, the bright and dim basal bodies are the new and old basal bodies, respectively, as new basal bodies stably incorporate GFP-Cen1 once it is expressed. The old basal body has some GFP-Cen1 signal because there is an exchangeable population at the basal body transition zone (Pearson et al., 2009). Cells were also examined 7 h after release from starvation. At this time point, many basal bodies have been assembled in the presence of GFP-Cen1, resulting in uniform labeling of new and old basal bodies by GFP (Figure 1C, right, white and blue arrows, respectively) (Pearson et al., 2009).
This assay was performed to analyze the incorporation of Cen1's CTD into basal bodies. After 2.5 h from cell cycle release, the GFP-CTD was present at new and old basal bodies (Figure 1D, left, white and blue arrowheads, respectively). Because the old basal body contained a GFP signal, it is likely that the CTD of Cen1 can exchange at basal bodies like the full-length protein. We observed that some basal bodies were much brighter than the others (Figure 1D, left, red arrow). Seven hours after cell cycle release, the GFP signal from the CTD at new and old basal bodies was more uniform (Figure 1D, right, white and blue arrows, respectively), although there were some brighter basal bodies (Figure 1D, right, red arrow). The overall labeling at basal bodies by the CTD appears dimmer than those labeled by the full‑length protein, suggesting that the CTD has decreased localization efficiency. This assay shows that Cen1's CTD incorporates into basal bodies in a similar manner as full-length Cen1 but with reduced efficiency.
Cen1 EF hand mutant alleles rescue the cen1Δ but exhibit temperature-sensitive phenotypes
Because the domain deletions of Cen1 gave little information about its function at basal bodies, we turned our efforts to mutating its EF hands since they are defining motifs in centrins and evidence indicates that they may have an important role in centrin function (Geier et al., 1996; Middendorp et al., 2000). A point mutation was made by site-directed mutagenesis within the loop regions of Cen1's four EF hands. The loop region contains the residues needed for Ca2+ binding, and the first residue, aspartate, is found in almost all EF hands (Gifford et al., 2007). Mutating this residue into alanine has been shown to perturb Ca2+ binding at an EF hand (Figure 2A) (Geiser et al., 1991). Seven CEN1 EF hand mutant alleles were made: four consisted of only one of the EF hands mutated, two consisted of both EF hands mutated within one domain (i.e., both the first and second EF hands of the N-terminal domain), and one consisted of all four EF hands mutated. All but two of the mutant alleles rescued the cen1Δ. The apparent null alleles were the CTD double EF hand mutant and the all-four EF hand mutant.
FIGURE 2:
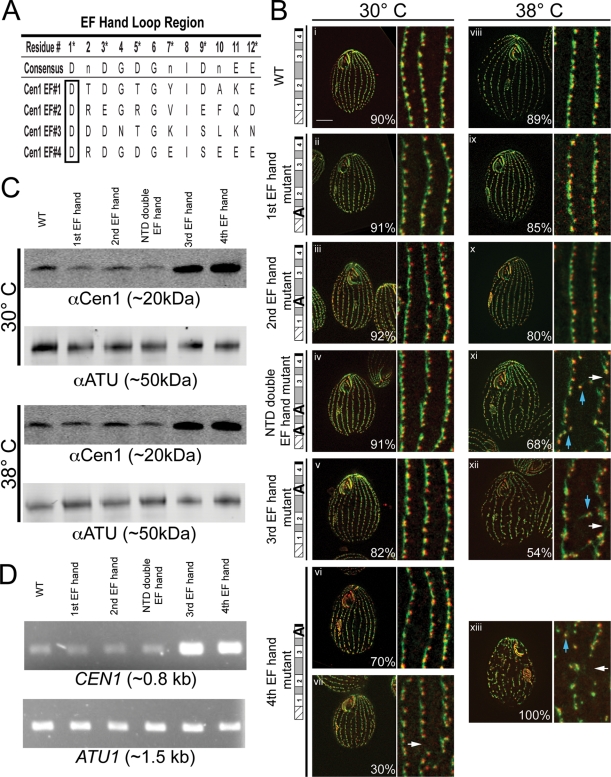
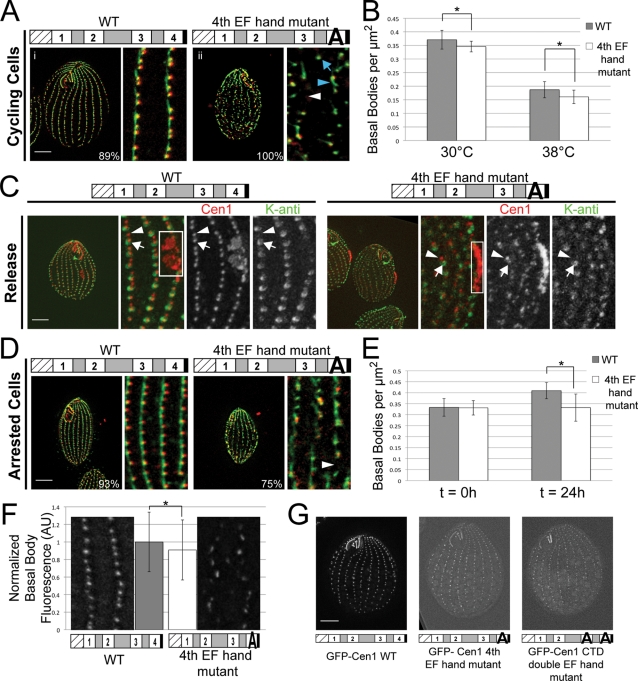
CEN1 EF hand mutant alleles cause temperature-sensitive basal body phenotypes. (A) Residues that make up the loop regions of Cen1's four EF hands. Asterisks denote the residues that form contacts with a Ca2+ ion. The consensus sequence shows the residues typically found in the loop region (Gifford et al., 2007). For the Cen1 EF hand mutations, the first residue of the loop region (boxed) was mutated to alanine. (B) Immunofluorescence images of strains containing CEN1 EF hand mutant alleles. i–vii, Cells grown at 30°C; viii–xiii, cells grown at 38°C; green, Sas6a and kinetodesmal fibers; red, centrin; blue arrows, basal bodies not in cortical rows; white arrows, gaps within cortical rows. Percentages indicate the frequency of observed phenotype for 100 cells. Bar, 10 μm; width of insets, 4.8 μm. (C) Western blots comparing the protein levels of Cen1 (αCen1) in the EF hand mutant strains at 30 and 38°C. Detection of α-tubulin (αATU) served as a loading control. (D) PCR analysis comparing CEN1 gene copy number in the EF hand mutant strains. α-Tubulin (ATU1) served as a PCR control.
Cells rescued with EF hand mutant alleles were fixed and stained with anti-Sas6a and anti-centrin (20H5) antibodies for immunofluorescence microscopy. The anti-Sas6a antibody recognizes the basal body protein Sas6a, which is found at the cartwheel (Kilburn et al., 2007; Culver et al., 2009). The antibody also labels the kinetodesmal (KD) fibers (Culver et al., 2009), which are basal body accessory structures composed of striated rootlets that extend from the basal body, pointing toward the anterior of the cell (Allen, 1969). It should be noted that it is now known that Sas6a is not within the KD fibers (B. P. Culver and M. Winey, personal communication). Thus, the anti-Sas6a antibody serves as a basal body and KD fiber marker. The 20H5 antibody recognizes centrins in various organisms, including Tetrahymena Cen1 (Stemm-Wolf et al., 2005).
We examined the mutants grown at 30°C and observed that the first EF hand, second EF hand, N-terminal domain (NTD) double EF hand, and third EF hand mutants were morphologically similar to the cen1Δ rescued with WT CEN1. Their cortical rows did not have defects in basal body orientation, assembly, or stability (Figure 2B, i–v). The majority of cells with the fourth EF hand mutant allele (70%) were similar to WT; however, 30% had gaps within their cortical rows (Figure 2B, vii, white arrow).
Cells were grown at 38°C to screen for temperature-sensitive EF hand mutant alleles. Both the first EF hand and second EF hand mutant strains remained morphologically similar to WT (Figure 2B, viii–x). However, the NTD double EF hand, third EF hand, and fourth EF hand mutant alleles gave basal body–associated phenotypes. These mutant alleles led to gaps within the cortical rows and basal bodies orientated off from the cortical rows (Figure 2B, xi–xiii, white and blue arrows, respectively). The fourth EF hand mutant allele caused the most severe basal body phenotype. Growth at 38°C was significantly slower for this mutant when compared with WT. The other mutants had growth rates similar to WT (unpublished data).
We examined the levels of the Cen1 protein in cells with the EF hand mutant alleles by Western blot with an anti-Cen1 antibody (Figure 2C). All the mutants have similar levels of Cen1 as WT at 30 and 38°C, except for the third EF hand and fourth EF hand mutants, which had higher levels. Through PCR analysis, we determined that these strains had more copies of CEN1 than WT (Figure 2D). This suggests that the higher levels of the Cen1 protein in the third and fourth EF hand mutants are a result of higher gene copy numbers.
EF hand mutations perturb Ca2+ binding in Cen1
We performed further analyses on the NTD double EF hand, third EF hand, and fourth EF hand mutant alleles because they gave temperature-sensitive basal body phenotypes. We included the CTD double EF hand and all four EF hand mutant alleles with our analysis because they were unable to rescue the cen1Δ.
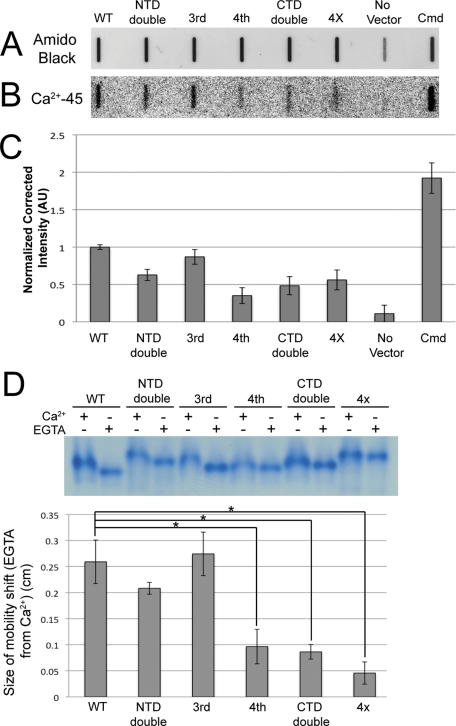
We wanted to determine how Ca2+ affinity in Cen1 was affected by EF hand mutations. Recombinant forms of the mutant proteins were purified from Escherichia coli along with the Tetrahymena Ca2+-binding protein calmodulin to serve as a positive control (Suzuki et al., 1981). The proteins were blotted onto nitrocellulose membranes and stained with amido black to verify equal loading (Figure 3A). Next the ability of the proteins to bind Ca2+ was tested using the radioisotope 45Ca2+ (Figure 3B). The band intensities of the autoradiograph, corrected for the amount of protein added to the membrane (Figure 3C), suggest that Ca2+ affinities are significantly reduced in all of the Cen1 EF hand mutant proteins except for the third EF hand mutant protein. Of interest, the reduction in Ca2+ affinity was greatest when Cen1 contained a mutated fourth EF hand, suggesting that this EF hand has the highest affinity for Ca2+. Supporting this are electrophoretic mobility shifts of the Cen1 mutant proteins in the presence or absence of Ca2+ in native polyacrylamide gels (Geiser et al., 1991). The mobility shifts for Cen1 proteins containing a mutated fourth EF hand were significantly reduced when compared with the WT protein (Figure 3D). In all, our results confirm that Cen1 is a Ca2+-binding protein and that mutations to some of its EF hands perturb its Ca2+-binding affinity. Because mutations to EF hands in the N- and C- terminal domains demonstrate a decreased affinity for Ca2+, it is likely that both domains in WT Cen1 are able to bind Ca2+ but with differing affinities.
FIGURE 3:
Mutations to Cen1's EF hands decrease Ca2+ affinity. (A) Recombinant proteins expressed in E. coli were purified, blotted onto a nitrocellulose membrane, and stained with amido black to show total protein. (B) Autoradiography shows 45Ca2+ bound to proteins on the membrane. (C) Quantification of band intensity in the autoradiograph corrected for the amount of protein. All values except for the third EF hand mutant protein were significantly different from WT (P < 1%). N = 3. (D) Top, electrophoretic mobility shift assays for mutant and WT Cen1 proteins in the presence of Ca2+ or EGTA. Bottom, plot showing the mobility shift for Ca2+-treated vs. EGTA-treated samples. Asterisk, P < 0.01%. N = 5. 4X, all four EF hands mutated; AU, arbitrary units; Cmd, Tetrahymena calmodulin; No Vector, protein purified from E. coli lacking a Cen1 expression plasmid.
The NTD double EF hand mutant allele causes misorientation of newly assembled basal bodies and causes basal body separation defects
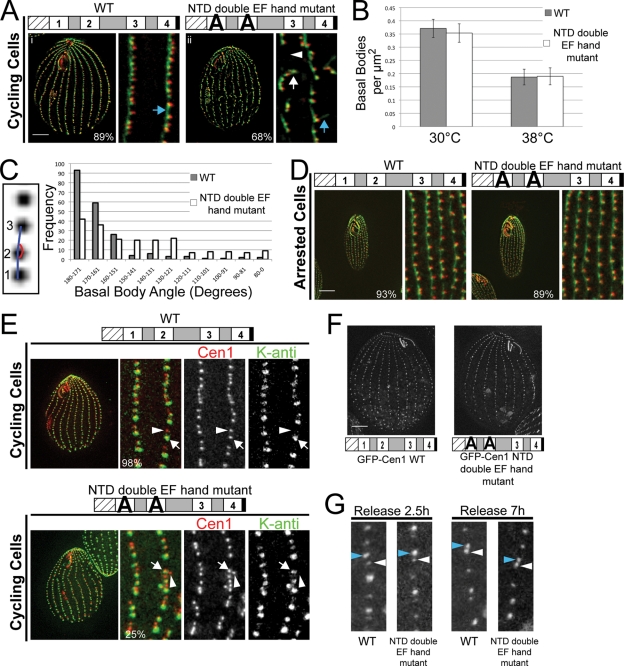
To better understand the function of Cen1 at basal bodies, we performed detailed analyses on the NTD double EF hand mutant allele. Cells progressing through the cell cycle and assembling new basal bodies at 38°C were examined using the anti-Sas6a and 20H5 antibodies (Figure 4A). As expected, cen1Δ rescued with the WT allele had straight cortical rows of basal bodies (Figure 4A, i). Cells with the NTD double EF hand mutant allele had basal bodies branching off from the cortical rows, resulting in gaps within the cortical rows (Figure 4A, ii, white arrow and arrowhead, respectively). We also saw that the KD fibers of some basal bodies within straight cortical rows were orientated improperly in that they no longer point toward the anterior end of the cell (Figure 4A, compare blue arrows in i and ii), suggesting that these basal bodies have been aberrantly rotated.
FIGURE 4:
NTD double EF hand mutant alleles cause defects in the orientation of newly assembled basal bodies. (A) Immunofluorescence images of the NTD double EF hand mutant progressing through the cell cycle at 38°C. The mutant cell has misorientated kinetodesmal fibers (compare blue arrows) and basal bodies branching away from cortical rows (white arrow), causing gaps in the cortical rows (white arrowhead). Green, Sas6a, and kinetodesmal fibers; red, centrin. (B) Plot showing the number of basal bodies per micrometer squared for cells grown at 30 and 38°C. N = 25 cells. (C) The inset shows how basal body angle is measured. Three consecutive basal bodies served as the angle points, with basal bodies 1 and 2 considered to be in the cortical row and basal body 3 in or out of the cortical row. The plot shows the basal body angle frequency for cells grown at 38°C. N = 200 basal bodies. (D) Immunofluorescence images showing the NTD double EF hand mutant whose assembly of basal bodies has been inhibited by cell cycle arrest at 38°C. Green, Sas6a, and kinetodesmal fibers; red, centrin. (E) At 38°C, new basal bodies (arrowhead) are assembled off axis from mature basal bodies (arrow) in the cortical rows in the NTD double EF hand mutant. Red, anti-Cen1, which labels all basal bodies; green, K-antigen, which labels mature basal bodies. (F) Localization of GFP-Cen1 with mutated NTD double EF hands is similar to GFP-Cen1 WT. (G) Incorporation assays show that GFP-Cen1 with mutated NTD double EF hands behaves similarly to GFP-Cen1 WT. Blue arrowheads indicate new basal bodies, and white arrowheads indicate old basal bodies. Bar, 10 μm. Width of insets, 5 μm (A, D, and E) or 3.9 μm (G). Percentages indicate the frequency of observed phenotype for 100 cells (A and D) or 100 basal bodies (E).
The number of basal bodies in the NTD double EF hand mutant was compared with that in WT to ascertain whether the mutant allele affected basal body assembly and/or stability. The number of basal bodies per μm2 for a cell was calculated by counting the foci resulting from the anti-Cen1 antibody. We are confident that the anti-Cen1 antibody is labeling all basal bodies in the mutant because we observed that the 20H5 centrin antibody colocalized with the anti-Sas6a antibody at all basal bodies within the cortical rows (Figure 4A, ii), indicating that all basal bodies in the mutant have Cen1. The number of basal bodies per μm2 in the NTD double EF hand mutant is similar to that in WT at 30 and 38°C (Figure 4B), indicating that there is no loss of basal bodies. Basal body angle was measured to determine whether there is a basal body orientation defect for the NTD double EF hand mutant allele. We found that the mutant had more variation in basal body angle, with an average angle of 143 ± 31°. This was significantly different from WT, which had an average angle of 164 ± 21° (Figure 4C) (P < 0.01%, N = 200 basal bodies). Our data indicate that the main phenotype for the NTD double EF hand mutant allele is a basal body orientation defect.
Next we asked whether the orientation defects would arise in mutant cells not undergoing new basal body assembly. Cells were grown at 30°C, arrested at 30°C by starvation to inhibit new basal body assembly, and shifted to 38°C. After 24 h at 38°C, the cells were examined by immunofluorescence microscopy. The NTD double EF hand mutant had straight cortical rows of basal bodies, which was similar to cells with the WT allele (Figure 4D). This suggests that the orientation defects arise from the assembly of new basal bodies off axis from the cortical rows. To test this, cells were grown at 30°C, arrested at 30°C to inhibit basal body assembly, shifted to 38°C for 24 h, and released into growth media at 38°C to initiate new basal body assembly at the restrictive temperature. Four hours after release at 38°C, cells were fixed and stained for K-antigen, which labels only mature basal bodies (Williams et al., 1990; Shang et al., 2005), and Cen1, which labels all basal bodies (Stemm-Wolf et al., 2005; Pearson et al., 2009). Cells with the WT allele had new basal bodies unlabeled by K-antigen that assemble anterior to mature basal bodies labeled by K-antigen (Figure 4E; arrow, mature basal body; arrowhead, newly assembled basal body). In contrast, the NTD double EF hand mutant had new basal bodies that assembled off axis from mature basal bodies in the cortical rows (Figure 4E; arrow, mature basal body; arrowhead, newly assembled basal body). Thus, the NTD double EF hand mutant allele causes misorientation of newly assembled basal bodies.
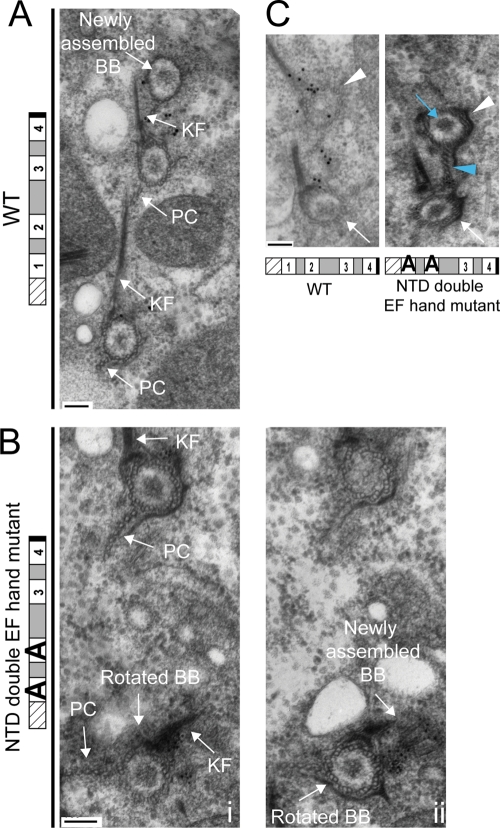
EM was used to examine basal bodies in the NTD double EF hand mutant grown at 38°C. Serial sections from mutant and WT were stained with the anti-Cen1 antibody, revealing that the localization of Cen1 to basal body domains for the NTD double EF hand mutant allele was similar to the WT allele (Supplemental Figure S2A). We also observed a similar incorporation pattern for GFP-Cen1 with the NTD double EF hand mutations as WT, suggesting that the mutant allele does not affect the localization and incorporation of the mutant protein into basal bodies (Figure 4, F and G). EM analysis of the mutant found cases of a new basal body being assembled off axis from an aberrantly rotated mother basal body in the cortical rows (Figure 5, A and B), confirming the phenotype we observed by immunofluorescence microscopy. It suggests that the assembly orientation defect might be a result of aberrantly rotated basal bodies. We also found many examples in the mutant showing a mature daughter basal body closer to its mother basal body than what is observed for WT (Figure 5C), indicating that the NTD double EF hand mutant allele causes basal body separation defects. In several of these examples, some accessory structures of the daughter basal body, such as the postciliary microtubules, were impinging on the mother basal body (Figure 5C, blue arrowhead). In all, the EM data suggest that Cen1 has a role in the separation of a daughter basal body from its mother basal body.
FIGURE 5:
Electron microscopy analysis reveals that the NTD double EF hand mutant allele causes basal body rotation and separation defects at 38°C. (A) Three basal bodies in a cortical row from a WT cell are shown, with the topmost basal body being a newly assembled basal body. (B, i, ii) Serial sections from a NTD double EF hand mutant cell are shown, with (ii) being more proximal. The basal body toward the bottom has been aberrantly rotated, as indicated by its kinetodesmal fiber (KF) and postciliary microtubule (PC) structures. (B, ii) The aberrantly rotated basal body has a new basal body being assembled away from the cortical row. (C) Immunoelectron micrographs showing the basal body separation defect. The white arrows and arrowheads indicate the mother and daughter basal bodies, respectively. Note that the daughter basal body in the mutant is close to its mother basal body, yet the density within the mutant daughter basal body (blue arrow) indicates that it is more mature than the WT daughter basal body (Allen, 1969). The postciliary microtubules of the mutant's daughter basal body (blue arrowhead) appear to be impinging on the mother basal body. Particle labeling is Cen1. Bar, 100 nm. BB, basal body; KF, kinetodesmal fiber; PC, postciliary microtubules.
Because of the domain nature of Cen1's structure, we wondered whether other mutations to the NTD of Cen1 would give similar phenotypes as the NTD double EF hand mutant allele. Nine constructs containing NTD CEN1 mutant alleles were made (Supplemental Figure S1). One of these consisted of an N-terminal tail deletion, and the rest were charged residues to alanine mutations. We avoided making any mutations within the EF hand loop regions. Six of the nine mutant alleles were able to rescue the cen1Δ. One of the apparent null alleles was the N-terminal tail deletion, suggesting that it has an important role in Cen1 function. Five of the six rescued alleles displayed temperature-sensitive basal body orientation defects that were similar to the NTD double EF hand mutant allele (Supplemental Figure S3; CEN1–29 is a representative NTD mutant allele). In all, our data suggest that the NTD of Cen1 has a role in the orientation of newly assembled basal bodies.
The fourth EF hand mutant allele causes basal body stability defects due to decreased localization efficiency of the mutant protein
We performed similar analyses on the fourth EF hand mutant allele as we did on the NTD double EF hand mutant allele. Cycling cells with the fourth EF hand mutant allele at 38°C had many basal bodies outside of the cortical rows, enlarged KD fibers, and large gaps within the cortical rows (Figure 6A, ii, blue arrow, blue arrowhead, and white arrowhead, respectively). Large gaps within cortical rows are usually indicative of a loss of basal bodies due to assembly and/or stability defects (Shang et al., 2002). The mutant strain had ∼7 ± 5% fewer basal bodies per μm2 than WT at 30°C, and that difference increased to 14 ± 13% at 38°C (Figure 6B), confirming that the fourth EF hand mutant has a loss of basal bodies. Along with a loss of basal bodies, cells with the fourth EF hand mutant allele were significantly shorter than WT at 38°C (44 ± 3 vs. 53 ± 3 μm, respectively, P < 0.001%, N = 25 cells).
FIGURE 6:
Fourth EF hand mutant allele causes basal body stability defects due to poor localization of the mutant protein. (A) Immunofluorescence images showing the fourth EF hand mutant progressing through the cell cycle at 38°C. The mutant has gaps in the cortical rows (white arrowhead), misorientated basal bodies (blue arrow), and enlarged kinetodesmal fibers (blue arrowhead). Green, Sas6a and kinetodesmal fibers; red, centrin. (B) Plot showing the number of basal bodies per micrometer squared for cells grown at 30 and 38°C. Asterisk, P < 1%; N = 25 cells. (C) Fourth EF hand mutant can assemble new basal bodies (arrowhead) from mature basal bodies (arrow) at 38°C. Red, anti-Cen1, labels all basal bodies; green, K-antigen, which labels mature basal bodies; white box, oral primordium, which is indicative of new basal body assembly. (D) Immunofluorescence images showing gaps in the cortical rows (arrowhead) of the fourth EF hand mutant whose assembly of basal bodies has been inhibited by cell cycle arrest at 38°C. Green, Sas6a and kinetodesmal fibers; red, centrin. (E) Plot showing the number of basal bodies per micrometer squared in arrested cells before (t = 0 h) and after 24 h (t = 24 h) at 38°C. Asterisk, P < 0.01%; N = 25 cells. (F) Quantification of anti-Cen1 staining shows that the fourth EF hand mutant has less Cen1 at basal bodies than WT. Asterisk, P < 1%; N = 200 basal bodies. AU, arbitrary units. (G) Localization efficiency of GFP-tagged Cen1 with a mutated fourth EF hand or mutated CTD double EF hands is less than that of GFP-Cen1 WT. Bar, 10 μm. Width of insets, 5 μm. Percentages indicate the frequency of observed phenotype for 100 cells.
Because the fourth EF hand mutant has a loss of basal bodies, we wanted to determine whether it is due to an inability to assemble new basal bodies. Cells were grown at 30°C, arrested at 30°C to inhibit new basal body assembly, shifted to 38°C for 24 h, and released into growth media to initiate new basal body assembly at 38°C. Four hours after release at 38°C, cells were fixed and stained for K-antigen, which labels only mature basal bodies (Williams et al., 1990; Shang et al., 2005), and Cen1, which labels all basal bodies (Stemm-Wolf et al., 2005, Pearson et al., 2009). This assay reveals that at 38°C, the fourth EF hand mutant strain is able to assemble new basal bodies from mature basal bodies (Figure 6C; arrow, mature basal body; arrowhead, newly assembled basal body). Further confirmation that the fourth EF hand mutant can assemble new basal bodies is the observation of oral primordia (Figure 6C, box). The appearance of the oral primordium is indicative of new basal body assembly, for it is a cellular structure that results from new basal body assembly, which then develops into an oral apparatus as a cell undergoes cell division (Wolfe, 1970; Kaczanowski, 1978; Frankel, 2000).
Because the fourth EF hand mutant strain can assemble new basal bodies at 38°C, we hypothesize that the loss of basal bodies in this mutant at 38°C is due to basal body stability defects. To test this, cells were grown at 30°C, arrested at 30°C to inhibit new basal body assembly, and shifted to 38°C. After 24 h at 38°C in an arrested state, cells were examined by immunofluorescence microscopy. The majority of cells with the fourth EF hand mutant allele (75%) had gaps within the cortical rows (Figure 6D, white arrowhead), suggesting a loss of basal bodies. The mutant had about the same number of basal bodies per μm2 as WT before the shift to 38°C (Figure 6E, t = 0 h). However, after 24 h at 38°C, the mutant had a 19 ±15% decrease in basal bodies per μm2 (Figure 6E, t = 24 h). This demonstrates that the fourth EF hand mutant loses basal bodies due to stability defects and confirms the role for Cen1 in basal body maintenance (Stemm-Wolf et al., 2005).
Similar analyses on the third EF hand mutant allele showed that cycling cells at 38°C had the same phenotypes as the fourth EF hand mutant strain, including gaps that are indicative of a loss of basal bodies (Supplemental Figure S4A). However, the number of cells observed with basal body–related phenotypes was much smaller for the third EF hand mutant strain (54%) compared with the fourth EF hand mutant strain (100%). Because new oral primordia were observed in the third EF hand mutant (Supplemental Figure S4A), we expect that they are able to assemble new basal bodies at 38°C. When the mutant strain was arrested to inhibit new basal body assembly, a large fraction (68%) was morphologically similar to WT; however, 32% displayed gaps within the cortical rows of basal bodies (Supplemental Figure S4B). Our data suggest that the third EF hand mutant allele results in a loss of basal bodies due to a slight defect in basal body stability, although it is not nearly as severe as the fourth EF hand mutant allele.
We noticed that basal bodies in the fourth EF hand mutant strain had varying intensity when labeled with anti-Cen1 antibodies compared with WT (see Figure 6C). Quantification of fluorescence intensity showed that the Cen1 signal at basal bodies was significantly reduced by almost 10% for the fourth EF hand mutant strain when compared with WT (Figure 6F). The percentage of reduction underreports the localization efficiency of the mutant protein since WT and mutant cells have unequal levels of Cen1 (Figure 2C). Furthermore, the localization efficiency of GFP-Cen1 with a mutated fourth EF hand was reduced when compared with WT GFP-Cen1 (Figure 6G). Even with a decrease in localization efficiency, GFP-Cen1 with a mutated fourth EF hand shows an incorporation pattern into basal bodies that is similar to the WT protein (Supplemental Figure S5). Of interest, GFP-Cen1 with a mutated third EF hand shows a slight decrease in localization efficiency, which is not nearly as severe as the fourth EF hand mutant protein (Supplemental Figure S4, C-F). Immunoelectron microscopic analysis on basal bodies in the mutant strain determined that the mutant protein localized to the same basal body domains as the WT protein but with a significant reduction in the labeling (Supplemental Figure S2A). In addition, the mutant protein mislocalized to regions other than the site of new assembly at the proximal end of the basal body (Supplemental Figure S2, A and B). In all, our data suggest that the basal body stability defects in the fourth EF hand mutant are due to poor and defective localization of the mutant Cen1 protein. Consistent with our conclusion that CTD EF hand mutations decrease Cen1 localization was our finding that GFP-Cen1 with CTD double EF hand mutations has decreased localization compared with WT GFP-Cen1 (Figure 6G). Examination of the mutant protein's incorporation into basal bodies shows that it does not incorporate bimodally into basal bodies like the WT protein (Supplemental Figure S5), suggesting that the ability to incorporate bimodally is necessary for correct Cen1 function.
To further test the domain nature of Cen1, we generated 10 alleles containing mutations to Cen1's CTD or the linker region between the two domains (Supplemental Figure S1). Three of the alleles contained deletions within CEN1: the first four residues of the linker region, the last four residues of the linker region, and the three-residue C-terminal tail. The rest of the mutant alleles contained charged residues to alanine mutations. Of the deletions, only the C-terminal tail deletion allele rescued the cen1Δ. This mutant allele caused a basal body stability defect, similar to the fourth EF hand mutant allele (Supplemental Figure S3). Four charged residues to alanine mutant alleles rescued the cen1Δ, and they all caused temperature-sensitive basal body stability defects similar to the fourth EF hand mutant allele (Supplemental Figure S3; CEN1-126 is a representative CTD mutant allele). In all, the data suggest that the CTD of Cen1 has a role in basal body stability.
DISCUSSION
We have used a mutagenic strategy to study the function of centrin at Tetrahymena basal bodies. Two predominant basal body–associated phenotypes were observed. One encompasses defects in basal body stability, further supporting the hypothesis that Cen1 is important for basal body maintenance (Stemm-Wolf et al., 2005). The other observed phenotype is misorientation of newly assembled basal bodies. Centrins have been suggested to be involved in the assembly of MTOCs in various species (Spang et al., 1993; Salisbury et al., 2002; Koblenz et al., 2003; Ruiz et al., 2005; Yang et al., 2010), including Tetrahymena (Stemm-Wolf et al., 2005), and our data further suggest that one role centrins have in the assembly of MTOCs is the orientation of the newly assembled MTOC. It may appear that none of the mutant alleles give basal body assembly defects; however, we observed that the frequency of new basal body assembly is reduced for the fourth EF hand mutant allele compared with WT (unpublished data). Therefore, it is possible that this mutant allele causes basal body assembly defects in addition to stability defects.
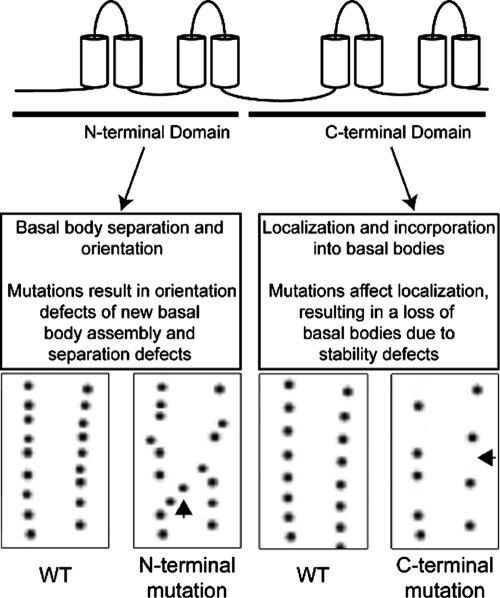
The results from our mutagenic approach give key information about the structure–function relationship of centrins and suggest distinct functions for the two domains (Figure 7). Neither domain alone is sufficient to rescue the cen1Δ, indicating that both domains are required to carry out Cen1 function. CTD mutant alleles cause basal body stability defects, and further examination of CTD EF hand mutant alleles shows that the localization of the mutant proteins to basal bodies is reduced. We also found that only the CTD of Cen1 localizes to basal bodies, suggesting that the CTD is responsible for basal body localization. This hypothesis is further supported by the crystal structure showing the interaction between the budding yeast centrin (Cdc31p) and Sfi1p, which contains multiple centrin-binding sites (Kilmartin, 2003; Li et al., 2006). The structure shows that the CTD of Cdc31p makes the bulk of contacts with Sfi1p. Furthermore, a screen for CDC31 temperature-sensitive mutant alleles showed that the majority of CTD mutant alleles resulted in protein mislocalization (Ivanovska and Rose, 2001). It has also been shown that only the CTD of human centrin 2 localizes to centrioles (Gavet et al., 2003) and interacts with the human Sfi1 (Martinez-Sanz et al., 2010). Taken together, it appears that the primary function of centrin's CTD is to localize and incorporate the protein into MTOCs and that the basal body stability defects in the Cen1 CTD mutants are likely due to reduced levels of Cen1 at basal bodies.
FIGURE 7:
Working model showing the function of Cen1's two domains. Mutations to the NTD of Cen1 cause basal body orientation and separation defects, suggesting that the NTD has a role in those processes. Mutations to the CTD of Cen1 lead to decreased localization efficiency of the mutant Cen1 protein, which then causes defects in basal body stability.
CEN1 NTD mutant alleles alter basal body orientation, suggesting that the NTD has a role in this process. None of the NTD mutant alleles affect the localization of the mutant protein, indicating that the observed basal body defect is a direct result of Cen1 dysfunction. How the NTD of centrin accomplishes its function remains to be determined. However, the NTD of Cdc31p, although largely unbound, does make some contacts with Sfi1p and the CTD of a neighboring centrin protein (Li et al., 2006). Furthermore, it has been observed that the NTD for the human centrin 2 forms oligomers (Tourbez et al., 2004; Yang et al., 2005). This may have a role in the NTD's function in regulating the orientation of new basal body assembly.
We also examined the Ca2+-binding properties of Cen1. The decreased Ca2+-binding affinities observed in the mutant proteins might be the cause of the phenotypes observed in the mutant strains, suggesting that Ca2+ is necessary for proper function. The CTD—more specifically, the fourth EF hand—has the higher Ca2+-binding affinity. The CTDs of the human centrin 2 and Chlamydomonas centrin also have higher affinity for Ca2+ than their NTDs (Veeraraghavan et al., 2002; Matei et al., 2003; Yang et al., 2005), suggesting that this specificity is evolutionarily conserved. On the basis of the Ca2+ affinities in the Chlamydomonas centrin, it has been postulated that the two domains of centrin have different functions (Veeraraghavan et al., 2002). The ability of the CTD to bind Ca2+ at low concentrations suggests that Ca2+ acts as a structural component that shifts the CTD into a conformation that favors interactions with a binding partner. The lower Ca2+ affinity in the NTD indicates that it behaves as a Ca2+ sensor. Our data further support this hypothesis and suggest that the NTD regulates MTOC orientation of assembly by acting as a Ca2+ sensor. How Ca2+ binding in the NTD of centrins regulates its function remains to be determined.
The majority of basal bodies in cells containing the NTD double EF hand mutant allele have separation defects, suggesting that Cen1 also has a role in the separation of a daughter basal body from its mother. Cen1 localization by immunoelectron microscopy has been observed between a mother and a daughter basal body, making it plausible that it regulates the separation of the two basal bodies. It is possible that a pair of basal bodies too close to one another due to a separation defect can lead to an orientation defect. The close proximity may cause an accessory structure of one basal body, such as postciliary microtubules, to impinge on the other basal body, subsequently leading a basal body to aberrantly rotate. When a new basal body begins to assemble from an aberrantly rotated basal body, the new basal body will be off axis from the cortical rows. Thus, the primary defect observed for the NTD mutant alleles may be the failure of basal bodies to separate correctly. There are reports of centrin involved in basal body or centriole separation (Lutz et al., 2001; Koblenz et al., 2003; Ruiz et al., 2005). Our data suggest that the ability to separate correctly may be necessary to ensure proper basal body orientation.
Correct basal body orientation in vertebrate multiciliated cells is critical for driving the flow of fluid in the proper direction across the surface of epithelial tissue (Marshall and Kintner, 2008). For example, basal bodies in the multiciliated cells of the oviduct are specifically orientated to produce directional fluid flow that transports the egg to the uterus (Marshall and Kintner, 2008). The planar cell polarity pathway is a key regulator of basal body positioning within the cell (Mitchell et al., 2009), but there are likely more components involved in this process. The fact that we observed basal body orientation defects for the CEN1 mutant alleles suggests that centrin may also be a key regulator of basal body positioning in vertebrate multiciliated cells. Our data also suggest that centrins are involved in the stability of basal bodies. The ability to maintain stable basal bodies is likely critical for vertebrate multiciliated cells to perform their essential functions. However, the role for centrins at basal bodies in vertebrate cells is not well understood.
We have shown distinct functions for the two domains of Cen1. How the domains are involved in their respective functions remains to be discovered. Several Sfi1 homologues have been identified in Tetrahymena, and their functions are unknown (A. J. Stemm-Wolf and M. Winey, personal communication). It will be key to elucidate how the mutant Cen1 proteins interact with these Sfi1 homologues in order to fully understand centrin's mode of action at basal bodies.
MATERIALS AND METHODS
Strains and culture conditions
The CEN1 knockout heterokaryon T. thermophila strains UCB8 and UCB9 were used to generate the cen1 null allele (cen1Δ) (see later discussion). The wild-type B2086 strain (Tetrahymena Stock Center, Cornell University, Ithaca, NY) was also used in this study. Cultures were grown in SPP media (2% proteose peptone, 0.1% yeast extract, 0.2% glucose, 0.003% FeEDTA) at 24, 30, or 38°C, depending on the experiment. For starvation experiments, cells were grown in SPP to mid-log phase, washed, and resuspended in 10 mM Tris, pH 7.5, at 24, 30, or 38°C. Macronuclear biolistic transformation was performed to introduce constructs into Tetrahymena cells by homologous recombination (Cassidy-Hanley et al., 1997).
Strains containing GFP-tagged Cen1 were generated by cloning CEN1 into the pENTR-D Gateway Entry Vector (Invitrogen, Carlsbad, CA). The coding sequence was then cloned into pBSmttGFPgtw (Doug Chalker, Washington University, St. Louis, MO) using the Gateway cloning system (Invitrogen). The resulting construct adds the GFP tag to the N-terminus of Cen1, is under control of a metallothionein-inducible promoter, targets for integration into the rpl29 locus, and provides cycloheximide resistance. Site-directed mutagenesis was performed to pENTR-D–Cen1 to generate the mutant forms of GFP-Cen1. All mutations were confirmed by sequencing (Macrogen USA, Rockville, MD), and were cloned into pBSmttGFPgtw. Tetrahymena transformants were selected by growth in SPP containing cycloheximide (15 μg/ml).
The expression of GFP-Cen1 and its mutants was induced by adding CdCl2 (0.5 μg/ml) to cells grown to mid-log phase at 30°C. To determine the localization of the protein, cells were examined 7 h after induction. The GFP-Cen1 incorporation assay was performed as described previously (Pearson et al., 2009). Briefly, cells were grown to mid-log phase at 30°C and arrested by starvation. After overnight arrest at 30°C, cells were washed into fresh SPP containing CdCl2 (0.5 μg/ml) to induce expression of GFP-Cen1. Cells were examined 2.5 and 7 h after cell cycle release at 30°C.
cen1Δ rescue
The cen1Δ rescue constructs consisted of the CEN1 locus (1038 and 981 base pairs upstream and downstream from the start and stop codons of CEN1, respectively) cloned into pUC18. The plasmid pUC18-CEN1 targets the CEN1 locus, and the expression of CEN1 is under control of its endogenous promoter. Mutations and deletions to pUC18-CEN1 were generated by site-directed mutagenesis and confirmed by sequencing. Supplemental Figure S1 shows all of the CEN1 mutant alleles that were generated in this study.
The cen1Δ was generated as described in Stemm-Wolf et al. (2005). Briefly, UCB 8 and 9 have both copies encoding CEN1 replaced by the NEO2 cassette in their micronuclei. On mating of the two strains, their micronuclei form new macronuclei that have the NEO2 cassette at the CEN1 locus instead of the coding regions of CEN1. The cen1Δ is selected for resistance to paromomycin, which is provided by the NEO2 cassette (Weide et al., 2007). The cen1 deletion allele eventually causes cells to die due to basal body stability and assembly defects (Stemm-Wolf et al., 2005).
The cen1Δ was rescued by introducing pUC18-CEN1 plasmids containing WT CEN1 or mutant alleles into cells by biolistics 10.5 h after mating UCB 8 to UCB 9 (Cassidy-Hanley et al., 1997). Rescue was selected by resistance to paromomycin (100 μg/ml) in SPP and viability after seven days at 30°C. Once rescued strains were identified, genomic DNA was isolated by phenol:chloroform:isoamyl alcohol extraction followed by precipitation with isopropanol as described in Gaertig et al. (1994). The CEN1 locus was sequenced to confirm the presence of CEN1 and any mutant alleles. Alleles that did not rescue the cen1Δ in three transformations were considered to be lethal.
Fluorescence microscopy
Live-cell imaging was performed to examine cells expressing GFP-Cen1. Cells were washed with 10 mM Tris, pH 7.5, concentrated by pelleting, and placed on microscope slides (VWR, Radnor, PA). For immunofluorescence, cells were chemically fixed with paraformaldehyde and ethanol as described in Stuart and Cole (2000). Fixed cells were added to poly-l-lysine coated antibody slides (Bellco Glass, Vineland, NJ). All primary antibodies were diluted in phosphate-buffered saline (PBS) plus 1% bovine serum albumin (BSA). The rabbit polyclonal Tetrahymena Cen1 antibody (Stemm-Wolf et al., 2005) was diluted 1:1000. The mouse monoclonal 20H5 antibody raised against the Chlamydomonas centrin (provided by J. Salisbury, Mayo Clinic, Rochester, MN) was diluted 1:1000. The rabbit polyclonal Tetrahymena Sas6a antibody (Culver et al., 2009) was diluted 1:2500. The mouse monoclonal K-antigen antibody 10D12 (provided by J. Frankel, University of Iowa, Iowa City, IA) was diluted 1:50. The mouse monoclonal KD fiber antibody F1-5D8 (provided by J. Frankel) was diluted 1:250. For experiments using the K-antigen antibody, cells were chemically fixed only with 70% ethanol as described in Pearson et al. (2009). All primary antibody incubations were carried out overnight at 4°C, except for the K-antigen antibody, which was incubated at 4°C for 3 d. After primary antibody incubations, cells were washed five times with PBS plus 0.1% BSA. The secondary antibodies used were anti–rabbit FITC, anti–mouse Texas Red, anti-rabbit Texas Red, anti–mouse FITC (Jackson ImmunoResearch Labs, West Grove, PA), and anti–mouse Alexa 488 (Invitrogen/Molecular Probes, Carlsbad, CA). All secondary antibodies were diluted 1:1000 in PBS plus 1% BSA and incubated with cells at room temperature for 1 h. Cells were washed five times with PBS plus 0.1% BSA and mounted in Citifluor (Citifluor, London, United Kingdom).
All imaging was performed at room temperature using an Eclipse Ti inverted microscope (Nikon, Tokyo, Japan) fitted with a CFI Plan Apo VC 60x H numerical aperture–1.4 objective (Nikon) and a CoolSNAP hq2 charge-coupled device camera (Photometrics, Tucson, AZ). The MetaMorph Imaging software (Molecular Devices, Sunnyvale, CA) was used to collect and analyze images. For live-cell imaging, an exposure time of 800 ms was used. For immunofluorescence, an exposure time of 250 ms was used. Images were subjected to the nearest-neighbors deconvolution algorithm using the MetaMorph Imaging software.
Basal body fluorescence intensities were measured as described in Pearson et al. (2009). Briefly, 5 × 5 (inner fluorescence, Fi) and 9 × 9 (outer fluorescence, Fo) pixel regions were placed around a basal body, and MetaMorph measured the integrated fluorescence intensity for each region. Background fluorescence (Fbkgd) was calculated by subtracting Fi from Fo to get the fluorescence of the surrounding region not contained in the 5 × 5 region. The value was then corrected for area (Fbkgd = [Fo – Fi] × [25/56]). Basal body fluorescence intensity was calculated by subtracting Fbkgd from Fi. For experiments involving the Cen1 antibody, 200 basal bodies were analyzed for each condition. To measure all statistical differences, a Student's t test was performed using the Excel spreadsheet software (Microsoft, Redmond, WA).
Cen1 mutant strain analysis
Number of basal bodies per μm2. Images of cells whose basal bodies were labeled by the Cen1 antibody were collected. For growing cells, cells in oral primordium stages 1–2 (Bakowska et al., 1982) were analyzed to ensure that the quantification was done at a consistent cell cycle stage. ImageJ (National Institutes of Health, Bethesda, MD) was used to measure cell length and cell surface area. The Count Particles feature in ImageJ was used to count the basal bodies within the cell surface area. The number of basal bodies per μm2 was calculated by dividing the basal body count by the cell surface area. A total of 25 cells were analyzed for each condition.
Basal body angle. ImageJ was used to measure basal body angle. Three consecutive basal bodies in a cortical row were used as the points for angle measurements. The first two basal bodies were considered to be within the cortical row, and the third basal body was either in or out of the cortical row (Figure 4C). For each condition, 200 basal bodies were measured.
Western blot. Whole-cell extracts were prepared by lysing ∼30,000 cells in sample buffer (20% glycerol, 2% SDS, 125 mM Tris, pH 6.8, 5% β-mercaptoethanol) and heating to 80°C for 4 min. Approximately 2250 cells were added to each lane in 4–20% Precise Protein precast gels (Pierce, Rockford, IL). Proteins were transferred to Immobilon membranes (Millipore, Billerica, MA) using a transfer apparatus (Bio-Rad Labs, Hercules, CA). Membranes were blotted in Tris-buffered saline (TBS) containing 0.05% Tween and 2% BSA. Primary antibodies (rabbit polyclonal Cen1 antibody and mouse monoclonal B-5-1-2 α-tubulin antibody [Sigma-Aldrich, St. Louis, MO]) were diluted 1:1000 into TBS containing 0.05% Tween and 2% BSA. Primary antibody incubations were carried overnight at 4°C, and then the membrane was washed three times with TBS containing 0.05% Tween. Secondary antibodies (anti–rabbit IR800 and anti–mouse IR680 [LI-COR Biosciences, Lincoln, NE]) were diluted 1:10,000 into TBS containing 0.05% Tween and 2% BSA. Secondary antibody incubations were performed at room temperature for 1 h, and then the membrane was washed three times with TBS containing 0.05% Tween. Blots were visualized on a LI-COR Odyssey scanner.
CEN1 copy number. Genomic DNA was isolated from Tetrahymena cells by phenol:chloroform:isoamyl alcohol extraction, followed by precipitation with isopropanol as described previously. PCR with Phusion polymerase (New England Biolabs, Ipswich, MA) was performed with 0.25 μg of genomic DNA and primers for α-tubulin and Cen1. PCR was carried out according to the manufacturer's instructions, except that 20 cycles were performed. Cen1 forward primer: TGTCAGAATCATGAATTCAG. Cen1 reverse primer: TACGAGTAACGTTAATTAGC. α-Tubulin forward primer: AGAGAAGTTATTTCAATTCACGTC. α-Tubulin reverse primer: CTAAATTTTTAATCGCAGGCACA. PCR samples were run in a 1% Tris-acetate-EDTA agarose gel. The gel was stained with ethidium bromide and viewed through a PHOTO/Analyst Investigator Eclipse workstation (FOTODYNE, Hartland, WI).
Electron microscopy
Tetrahymena cells were prepared for ultrastructural analysis and immunolocalization of Cen1 by high-pressure freezing followed by freeze substitution (Giddings et al., 2010; Meehl et al., 2010). Briefly, cells were centrifuged into a cryoprotectant solution consisting of 15% dextran (9–11 kDa; Sigma-Aldrich) and 5% BSA in 10 mM Tris-HCl (pH 7.4). The resulting loose pellet was high-pressure frozen in a Bal-Tec HPM-010 (Leica Microsystems, Wetzlar, Germany), then freeze-substituted in 0.25% glutaraldehyde and 0.1% uranyl acetate in acetone and embedded in Lowicryl HM20.
Nickel grids containing ribbons of 15–20 serial 60-nm-thick sections were prepared for immuno‑electron microscopy by incubating them in blocking solution (1% nonfat dry milk dissolved in PBS–Tween 20 [0.1%]) and then in blocking solution containing the rabbit polyclonal Cen1 antibody diluted 1:200. The 10- or 15-nm gold-conjugated anti–rabbit secondary antibody was applied to the grids (Ted Pella, Redding, CA). Grids were poststained with 2% uranyl acetate and lead citrate. Samples were imaged using a Philips CM 10 (FEI, Hillsboro, OR) equipped with a Gatan BioScan digital camera (Gatan, Pleasanton, CA) or a Philips CM 100 transmission electron microscope equipped with an AMT V600 digital camera (Advanced Microscopy Techniques, Danvers MA). Six Cen1-containing basal body domains were identified by morphological criteria (Allen, 1969; Kilburn et al., 2007). Gold particles on each serial cross section through the basal bodies were counted and assigned to these domains. To measure statistical differences, a chi-square test was performed using the Excel spreadsheet software.
In vitro Ca2+-binding assays
The E. coli plasmid expressing recombinant 6x-histidine-Cen1, pQE10-Cen1, was generated in a previous study (Stemm-Wolf et al., 2005). EF hand mutations were made to the plasmid by site-directed mutagenesis and confirmed by sequencing. The Tetrahymena calmodulin gene was cloned into pQE10 (Qiagen, Valencia, CA). Recombinant proteins were purified as described in Stemm-Wolf et al. (2005). E. coli strain M15 was grown in 500 ml of Luria broth plus 0.2% glucose for 4 h at 37°C. Cultures were equilibrated to room temperature, and isopropyl β-d-thiogalactoside was added to 0.3 mM to induce expression of recombinant proteins. Induction was carried out for 3 h at room temperature. Cells were pelleted and washed once with PBS. The pellets were resuspended in PBS plus protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, 1 μg/ml aprotinin, and 1 μg/ml pepstatin A). Lysozyme was added to 200 μg/ml, and samples were incubated for 20 min at room temperature. Samples were then sonicated five times for 15 s with 1-min cooldowns on ice. Lysates were pelleted by centrifugation at 10,000 × g at 4°C for 15 min. The supernatant was loaded onto a column containing Talon resin (BD Biosciences Clontech, Palo Alto, CA) washed with PBS plus protease inhibitors. Proteins were eluted from the column in PBS plus 200 mM imidazole. All purified proteins were dialyzed into 10 mM 3-(N-morpholino)propanesulfonic acid (MOPS) , 50 mM KCl, pH 8, treated with Chelex-100 (Bio-Rad) to remove Ca2+ contaminants.
Nitrocellulose membranes (Amersham Biosciences, GE Healthcare, Piscataway, NJ) were prewetted in MOPS buffer (10 mM MOPS, 50 mM KCl, pH 8). Proteins (100 pmol) were blotted onto the nitrocellulose membrane using a Hybri-Slot Manifold (Whatman Biometra, Göttingen, Germany). The membrane was stained with amido black (Sigma-Aldrich) according to standard protocols (Hornbeck et al., 2001). Images of stained membranes were taken using a CanoScan 8400F scanner (Canon, Lake Success, NY). To test for Ca2+ binding, the membranes were incubated in a Ca2+ solution containing 100 mM CaCl2 buffered to ∼100 μM free Ca2+ with 100 mM ethylene glycol tetraacetic acid (EGTA). A total of 10 μCi of 45Ca2+ (PerkinElmer, Waltham, MA) was added to the Ca2+ solution, and incubations were carried out for 20 min at room temperature. Membranes were then washed three times with distilled H2O and allowed to dry. Dried membranes were exposed overnight to Phosphor Screens (Molecular Dynamics, Piscataway, NJ). Screens were visualized on a Storm 860 PhosphorImager scanner (Molecular Dynamics). Band intensities from the amido black staining and the autoradiograph were quantified using the Gels analytical tool in ImageJ. For sample comparisons, the band intensities from the autoradiographs were corrected for protein amount by dividing the autoradiograph intensity by the amido black intensity. The experiment was performed in triplicate at room temperature.
Electrophoretic mobility shift assays were performed by incubating Cen1 proteins (500 μg/ml) in MOPS buffer (10 mM MOPS, 50 mM KCl, pH 8) containing 1 mM CaCl2 or EGTA overnight at 4°C. Protein samples were diluted in 2× native sample buffer (125 mM Tris, pH 6.8, 20% glycerol). Samples were run on 12% Laemmli acrylamide gels (Ausubel et al., 1997) minus SDS in the gels and running buffer. Gels were washed three times in distilled H2O and stained in Gel Code Blue Stain Reagent (ThermoFisher Scientific, Waltham, MA). Gels were scanned on a Canon CanoScan 8400F scanner. The mobility shift for Ca2+ versus EGTA samples was measured using ImageJ software. The experiment was performed five times.
Supplementary Material
Acknowledgments
We thank Chad Pearson for advice and helpful discussions, Joe Frankel and Jeffrey Salisbury for antibodies, and Christina Clarissa for help with immunoelectron microscopy analysis. This work was supported by National Institutes of Health Grants GM074746 (to M.W.) and T32 GM08759 (to T.V.).
Abbreviations used:
- cen1Δ
cen1 null
- Cen1
Tetrahymena centrin 1
- CTD
C-terminal domain
- KD
kinetodesmal
- MTOC
microtubule-organizing center
- NTD
N-terminal domain
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-10-0844) on April 20, 2011.
REFERENCES
- Allen RD. The morphogenesis of basal bodies and accessory structures of the cortex of the ciliated protozoan Tetrahymena pyriformis. J Cell Biol. 1969;40:716–733. doi: 10.1083/jcb.40.3.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JR, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smoth JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1997. [Google Scholar]
- Bakowska J, Frankel J, Nelsen EM. Regulation of the pattern of basal bodies within the oral apparatus of Tetrahymena thermophila. J Embryol Exp Morphol. 1982;69:83–105. [PubMed] [Google Scholar]
- Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Bornens M, Azimzadeh J. Origin and evolution of the centrosome. Adv Exp Med Biol. 2007;607:119–129. doi: 10.1007/978-0-387-74021-8_10. [DOI] [PubMed] [Google Scholar]
- Cassidy-Hanley D, Bowen J, Lee JH, Cole E, VerPlank LA, Gaertig J, Gorovsky MA, Bruns PJ. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics. 1997;146:135–147. doi: 10.1093/genetics/146.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver BP, Meehl JB, Giddings TH, Jr, Winey M. The two SAS-6 homologs in Tetrahymena thermophila have distinct functions in basal body assembly. Mol Biol Cell. 2009;20:1865–1877. doi: 10.1091/mbc.E08-08-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe HR, Farr H, Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J Cell Sci. 2007;120:7–15. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- Frankel J. Cell biology of Tetrahymena thermophila. Methods Cell Biol. 2000;62:27–125. doi: 10.1016/s0091-679x(08)61528-9. [DOI] [PubMed] [Google Scholar]
- Gaertig J, Thatcher TH, Gu L, Gorovsky MA. Electroporation-mediated replacement of a positively and negatively selectable beta-tubulin gene in Tetrahymena thermophila. Proc Natl Acad Sci USA. 1994;91:4549–4553. doi: 10.1073/pnas.91.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O, Alvarez C, Gaspar P, Bornens M. Centrin4p, a novel mammalian centrin specifically expressed in ciliated cells. Mol Biol Cell. 2003;14:1818–1834. doi: 10.1091/mbc.E02-11-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin RH. In vitro reassembly of basal body components. J Cell Sci. 1984;66:147–154. doi: 10.1242/jcs.66.1.147. [DOI] [PubMed] [Google Scholar]
- Geier BM, Wiech H, Schiebel E. Binding of centrins and yeast calmodulin to synthetic peptides corresponding to binding sites in the spindle pole body components Kar1p and Spc110p. J Biol Chem. 1996;271:28366–28374. doi: 10.1074/jbc.271.45.28366. [DOI] [PubMed] [Google Scholar]
- Geimer S, Melkonian M. Centrin scaffold in Chlamydomonas reinhardtii revealed by immunoelectron microscopy. Eukaryot Cell. 2005;4:1253–1263. doi: 10.1128/EC.4.7.1253-1263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR, van Tuinen D, Brockerhoff SE, Neff MM, Davis TN. Can calmodulin function without binding calcium? Cell. 1991;65:949–959. doi: 10.1016/0092-8674(91)90547-c. [DOI] [PubMed] [Google Scholar]
- Giddings TH, Jr, Meehl JB, Pearson CG, Winey M. Electron tomography and immuno-labeling of Tetrahymena thermophila basal bodies. In: Müller-Reichert T., editor. Methods in Cell Biology. Vol. 96. Burlington, MA: Academic Press; 2010. pp. 117–141. [DOI] [PubMed] [Google Scholar]
- Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem J. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- Guerra C, Wada Y, Leick V, Bell A, Satir P. Cloning, localization, and axonemal function of Tetrahymena centrin. Mol Biol Cell. 2003;14:251–261. doi: 10.1091/mbc.E02-05-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck P, Fleisher TA, Papadopoulos NM. In: Current Protocols in Immunology. New York: John Wiley & Sons; 2001. Isotype determination of antibodies. Chapter 2, Unit 2.2. [DOI] [PubMed] [Google Scholar]
- Hu H, Chazin WJ. Unique features in the C-terminal domain provide caltractin with target specificity. J Mol Biol. 2003;330:473–484. doi: 10.1016/s0022-2836(03)00619-3. [DOI] [PubMed] [Google Scholar]
- Hu H, Sheehan JH, Chazin WJ. The mode of action of centrin. J Biol Chem. 2004;279:50895–50903. doi: 10.1074/jbc.M404233200. [DOI] [PubMed] [Google Scholar]
- Ivanovska I, Rose MD. Fine structure analysis of the yeast centrin, Cdc31p, identifies residues specific for cell morphology and spindle pole body duplication. Genetics. 2001;157:503–518. doi: 10.1093/genetics/157.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczanowski A. Gradients of proliferation of ciliary basal bodies and the determination of the position of the oral primordium in Tetrahymena. J Exp Zool. 1978;204:417–430. doi: 10.1002/jez.1402040313. [DOI] [PubMed] [Google Scholar]
- Keller LC, Romijn EP, Zamora I, Yates JR, Marshall WF. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Jr, Culver BP, Yates JR, 3rd, Winey M. New Tetrahymena basal body protein components identify basal body domain structure. J Cell Biol. 2007;178:905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J Cell Biol. 2003;162:1211–1221. doi: 10.1083/jcb.200307064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblenz B, Schoppmeier J, Grunow A, Lechtreck KF. Centrin deficiency in Chlamydomonas causes defects in basal body replication, segregation and maturation. J Cell Sci. 2003;116:2635–2646. doi: 10.1242/jcs.00497. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Perret E, Middendorp S, Houcine O, Guennou C, Marano F, Bornens M, Tournier F. Differential expression and cellular distribution of centrin isoforms during human ciliated cell differentiation in vitro. J Cell Sci. 2000;113:1355–1364. doi: 10.1242/jcs.113.8.1355. [DOI] [PubMed] [Google Scholar]
- Li S, Sandercock AM, Conduit P, Robinson CV, Williams RL, Kilmartin JV. Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J Cell Biol. 2006;173:867–877. doi: 10.1083/jcb.200603153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Tan G, Levenkova N, Li T, Pugh EN, Rux JJ, Speicher DW, Pierce EA. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6:1299–1317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz W, Lingle W, McCormick D, Greenwood T, Salisbury J. Phosphorylation of centrin during the cell cycle and its role in centriole separation preceding centrosome duplication. J Biol Chem. 2001;276:20774–20780. doi: 10.1074/jbc.M101324200. [DOI] [PubMed] [Google Scholar]
- Marshall WF, Kintner C. Cilia orientation and the fluid mechanics of development. Curr Opin Cell Biol. 2008;20:48–52. doi: 10.1016/j.ceb.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Nonaka S. Cilia: tuning in to the cell's antenna. Curr Biol. 2006;16:R604–R614. doi: 10.1016/j.cub.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Martinez-Sanz J, Kateb F, Assairi L, Blouquit Y, Bodenhausen G, Abergel D, Mouawad L, Craescu CT. Structure, dynamics and thermodynamics of the human centrin 2/hSfi1 complex. J Mol Biol. 2010;395:191–204. doi: 10.1016/j.jmb.2009.10.041. [DOI] [PubMed] [Google Scholar]
- Martinez-Sanz J, Yang A, Blouquit Y, Duchambon P, Assairi L, Craescu CT. Binding of human centrin 2 to the centrosomal protein hSfi1. FEBS J. 2006;273:4504–4515. doi: 10.1111/j.1742-4658.2006.05456.x. [DOI] [PubMed] [Google Scholar]
- Matei E, Miron S, Blouquit Y, Duchambon P, Durussel I, Cox JA, Craescu CT. C-Terminal half of human centrin 2 behaves like a regulatory EF-hand domain. Biochemistry. 2003;42:1439–1450. doi: 10.1021/bi0269714. [DOI] [PubMed] [Google Scholar]
- Meehl JB, Giddings TH, Jr, Winey M. High pressure freezing, electron microscopy, and immuno-electron microscopy of Tetrahymena thermophila basal bodies. In: Gavin RH, editor. In: Cytoskeleton Methods and Protocols. Vol. 586. New York: Humana Press; 2010. pp. 227–241. [DOI] [PubMed] [Google Scholar]
- Middendorp S, Kuntziger T, Abraham Y, Holmes S, Bordes N, Paintrand M, Paoletti A, Bornens M. A role for centrin 3 in centrosome reproduction. J Cell Biol. 2000;148:405–416. doi: 10.1083/jcb.148.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell B, Stubbs JL, Huisman F, Taborek P, Yu C, Kintner C. The PCP pathway instructs the planar orientation of ciliated cells in the Xenopus larval skin. Curr Biol. 2009;19:924–929. doi: 10.1016/j.cub.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Giddings TH, Jr, Winey M. Basal body components exhibit differential protein dynamics during nascent basal body assembly. Mol Biol Cell. 2009;20:904–914. doi: 10.1091/mbc.E08-08-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Winey M. Basal body assembly in ciliates: the power of numbers. Traffic. 2009;10:461–471. doi: 10.1111/j.1600-0854.2009.00885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman BS. Basal body addition in ciliary rows of Tetrahymena pyriformis. J Exp Zool. 1973;184:365–367. doi: 10.1002/jez.1401840310. [DOI] [PubMed] [Google Scholar]
- Ruiz F, Garreau de Loubresse N, Klotz C, Beisson J, Koll F. Centrin deficiency in Paramecium affects the geometry of basal-body duplication. Curr Biol. 2005;15:2097–2106. doi: 10.1016/j.cub.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Salisbury J, Suino K, Busby R, Springett M. Centrin-2 is required for centriole duplication in mammalian cells. Curr Biol. 2002;12:1287–1292. doi: 10.1016/s0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- Sanders M, Salisbury J. Centrin plays an essential role in microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J Cell Biol. 1994;124:795–805. doi: 10.1083/jcb.124.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Li B, Gorovsky MA. Tetrahymena thermophila contains a conventional gamma-tubulin that is differentially required for the maintenance of different microtubule-organizing centers. J Cell Biol. 2002;158:1195–1206. doi: 10.1083/jcb.200205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Tsao C-C, Gorovsky MA. Mutational analyses reveal a novel function of the nucleotide-binding domain of gamma-tubulin in the regulation of basal body biogenesis. J Cell Biol. 2005;171:1035–1044. doi: 10.1083/jcb.200508184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Courtney I, Fackler U, Matzner M, Schiebel E. The calcium-binding protein division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J Cell Biol. 1993;123:405–416. doi: 10.1083/jcb.123.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemm-Wolf AJ, Morgan G, Giddings TH, Jr, White EA, Marchione R, McDonald HB, Winey M. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol Biol Cell. 2005;16:3606–3619. doi: 10.1091/mbc.E04-10-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart KR, Cole ES. Nuclear and cytoskeletal fluorescence microscopy techniques. Methods Cell Biol. 2000;62:291–311. doi: 10.1016/s0091-679x(08)61538-1. [DOI] [PubMed] [Google Scholar]
- Strnad P, Gönczy P. Mechanisms of procentriole formation. Trends Cell Biol. 2008;18:389–396. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nagao S, Abe K, Hirabayashi T, Watanabe Y. Tetrahymena calcium-binding protein is indeed a calmodulin. J Biochem. 1981;89:333–336. doi: 10.1093/oxfordjournals.jbchem.a133200. [DOI] [PubMed] [Google Scholar]
- Thompson JR, Ryan ZC, Salisbury JL, Kumar R. The structure of the human centrin 2-xeroderma pigmentosum group C protein complex. J Biol Chem. 2006;281:18746–18752. doi: 10.1074/jbc.M513667200. [DOI] [PubMed] [Google Scholar]
- Tourbez M, Firanescu C, Yang A, Unipan L, Duchambon P, Blouquit Y, Craescu CT. Calcium-dependent self-assembly of human centrin 2. J Biol Chem. 2004;279:47672–47680. doi: 10.1074/jbc.M404996200. [DOI] [PubMed] [Google Scholar]
- Veeraraghavan S, Fagan PA, Hu H, Lee V, Harper JF, Huang B, Chazin WJ. Structural independence of the two EF-hand domains of caltractin. J Biol Chem. 2002;277:28564–28571. doi: 10.1074/jbc.M112232200. [DOI] [PubMed] [Google Scholar]
- Weide T, Bockau U, Rave A, Herrmann L, Hartmann M. A recombinase system facilitates cloning of expression cassettes in the ciliate Tetrahymena thermophila. BMC Microbiol. 2007;7:12. doi: 10.1186/1471-2180-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams NE, Honts JE, Kaczanowska J. The formation of basal body domains in the membrane skeleton of Tetrahymena. Development. 1990;109:935–942. doi: 10.1242/dev.109.4.935. [DOI] [PubMed] [Google Scholar]
- Wolfe J. Structural analysis of basal bodies of the isolated oral apparatus of Tetrahymena pyriformis. J Cell Sci. 1970;6:679–700. doi: 10.1242/jcs.6.3.679. [DOI] [PubMed] [Google Scholar]
- Yang A, Miron S, Duchambon P, Assairi L, Blouquit Y, Craescu CT. The N-terminal domain of human centrin 2 has a closed structure, binds calcium with a very low affinity, and plays a role in the protein self-assembly. Biochem. 2005;45:880–889. doi: 10.1021/bi051397s. [DOI] [PubMed] [Google Scholar]
- Yang C-H, Kasbek C, Majumder S, Yusof AM, Fisk HA. Mps1 phosphorylation sites regulate the function of centrin 2 in centriole assembly. Mol Biol Cell. 2010;21:4361–4372. doi: 10.1091/mbc.E10-04-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.