AIP1 is a conserved enhancer of ADF/cofilin-dependent actin dynamics. Caenorhabditis elegans has two AIP1 genes. They have overlapping functions, and ablation of both AIP1 isoforms causes embryonic lethality with strong defects in the assembly of muscle sarcomeres.
Abstract
Disassembly of actin filaments by actin-depolymerizing factor (ADF)/cofilin and actin-interacting protein 1 (AIP1) is a conserved mechanism to promote reorganization of the actin cytoskeleton. We previously reported that unc-78, an AIP1 gene in the nematode Caenorhabditis elegans, is required for organized assembly of sarcomeric actin filaments in the body wall muscle. unc-78 functions in larval and adult muscle, and an unc-78–null mutant is homozygous viable and shows only weak phenotypes in embryos. Here we report that a second AIP1 gene, aipl-1 (AIP1-like gene-1), has overlapping function with unc-78, and that depletion of the two AIP1 isoforms causes embryonic lethality. A single aipl-1–null mutation did not cause a detectable phenotype. However, depletion of both unc-78 and aipl-1 arrested development at late embryonic stages due to severe disorganization of sarcomeric actin filaments in body wall muscle. In vitro, both AIPL-1 and UNC-78 preferentially cooperated with UNC-60B, a muscle-specific ADF/cofilin isoform, in actin filament disassembly but not with UNC-60A, a nonmuscle ADF/cofilin. AIPL-1 is expressed in embryonic muscle, and forced expression of AIPL-1 in adult muscle compensated for the function of UNC-78. Thus our results suggest that enhancement of actin filament disassembly by ADF/cofilin and AIP1 proteins is critical for embryogenesis.
INTRODUCTION
The actin cytoskeleton plays major roles in a number of dynamic processes, including cell migration, cytokinesis, morphogenesis, and muscle contraction (Pollard and Cooper, 2009). Actin filaments are often dynamic, and their assembly and disassembly are controlled by a number of actin-regulatory proteins. In particular, disassembly of actin filaments is critical for rapid cytoskeletal reorganization, as well as for persistent turnover of the actin cytoskeleton, because disassembly of actin filaments can be a rate-limiting step of actin filament turnover (Ono, 2007). Actin depolymerizing factor (ADF)/cofilin promotes actin filament turnover by severing and depolymerizing actin filaments (Bernstein and Bamburg, 2010). Severing of actin filaments by ADF/cofilin increases the number of uncapped filament ends where polymerization and depolymerization occur (Maciver et al., 1991; Ichetovkin et al., 2000; Andrianantoandro and Pollard, 2006). In addition, ADF/cofilin enhances dissociation of actin monomers from pointed ends of actin filaments (Carlier et al., 1997; Maciver et al., 1998). ADF/cofilin is conserved among eukaryotes, and knockouts of ADF/cofilin genes cause lethality in yeast (Iida et al., 1993; Moon et al., 1993), Drosophila (Gunsalus et al., 1995), Caenorhabditis elegans (McKim et al., 1994), and mice (Gurniak et al., 2005). Therefore ADF/cofilin–dependent regulation of actin filament dynamics is essential for supporting cell viability and embryonic development.
The function of ADF/cofilin is regulated by multiple mechanisms by posttranslational modifications and cooperation or competition with other actin-regulatory proteins (Van Troys et al., 2008). Among them, actin-interacting protein 1 (AIP1) is a conserved WD-repeat protein that cooperates with ADF/cofilin (Ono, 2003). Although AIP1 by itself has low affinity with actin filaments, it strongly enhances severing of actin filaments in the presence of ADF/cofilin (Aizawa et al., 1999; Iida and Yahara, 1999; Okada et al., 1999; Rodal et al., 1999; Ono et al., 2004). A recent study showed that AIP1 increases the actin monomer pool in cooperation with ADF/cofilin (Okreglak and Drubin, 2010). Correlative analyses of biochemical activity and in vivo phenotypes of mutant forms of AIP1 have shown that the activity of AIP1 to promote disassembly of ADF/cofilin–bound actin filaments is required for actin filament turnover in yeast (Clark et al., 2006; Clark and Amberg, 2007; Okada et al., 2006) and muscle actin organization in C. elegans (Mohri et al., 2004, 2006). AIP1 is also involved in cell migration and cytokinesis in Dictyostelium (Konzok et al., 1999) and mammalian cells (Kato et al., 2008). Thus the collaborative relationship between ADF/cofilin and AIP1 in actin regulation is conserved in eukaryotes. However, requirement of AIP1 for cell viability or development is not consistent in different organisms. Gene knockout or RNA interference (RNAi) depletion of AIP1 is lethal in Arabidopsis (Ketelaar et al., 2004), Drosophila (Ren et al., 2007), and mice (Kile et al., 2007). In contrast, a knockout of an AIP1 gene (unc-78) in C. elegans is homozygous viable and only causes phenotypes in striated muscle, with no detectable defects in embryonic development (Ono, 2001). Also, gene knockout of AIP1 is not lethal in yeast (Iida and Yahara, 1999; Rodal et al., 1999) and Dictyostelium (Konzok et al., 1999). Thus the function of AIP1 at levels of whole organisms is not clearly understood.
C. elegans has two functionally distinct ADF/cofilin isoforms, UNC-60A and UNC-60B, which are generated from the unc-60 gene by alternative splicing (McKim et al., 1994). UNC-60A only weakly severs actin filaments and strongly sequesters actin monomers, whereas UNC-60B strongly severs actin filaments and does not sequester actin monomers (Ono and Benian, 1998; Yamashiro et al., 2005). UNC-60A is widely expressed in many tissues and required for embryonic cytokinesis (Ono et al., 2003) and actin organization in the somatic gonad (Ono et al., 2008). UNC-60B is specifically expressed in striated muscle and required for assembly of myofibrils (Ono et al., 1999 , 2003). An AIP1 protein is encoded by the unc-78 gene that cooperates with UNC-60B for myofibril assembly in striated muscle (Ono, 2001). In vitro, UNC-78 cooperates only with UNC-60B but not with UNC-60A to promote actin filament disassembly (Mohri and Ono, 2003). These results indicate that UNC-78 and UNC-60B collaborate in a muscle-specific manner. However, the C. elegans genome has a second, uncharacterized AIP1 gene, which we designated as aipl-1 (AIP1-like gene-1). A previous genome-wide study suggested that aipl-1 and unc-78 are functionally redundant (Tischler et al., 2006), but biochemical and cell biological functions of aipl-1 were unclear. In this study, we attempted to determine the function of aipl-1 and its functional relationship with unc-78 and ADF/cofilin isoforms. We found that AIPL-1 and UNC-78 have overlapping function by cooperating preferentially with the same ADF/cofilin isoform. Of importance, depletion of the two AIP1 proteins results in embryonic lethality, indicating that AIP1 proteins are required for C. elegans development. These results reveal a previously unknown essential function of AIP1 proteins in C. elegans and suggest that AIP1 proteins are important for morphogenesis of multicellular organisms rather than viability of individual cells.
RESULTS
A second AIP-1 isoform is encoded by aipl-1 and expressed in neurons, body wall muscle, and spermatheca
The C. elegans Genome Sequencing Consortium predicted that a putative gene on chromosome V, K08F9.2 [originally designated as temporally assigned gene (tag)-216], encodes a protein that is highly homologous to AIP1 proteins. We sequenced an Expressed Sequence Tag clone, yk1621e12, from Yuji Kohara's laboratory (National Institute of Genetics, Mishima, Japan) and confirmed that the exon–intron structure of this gene was correct as predicted. The sequence of the encoded protein (600 amino acids; GenBank accession number CAB03187.1) is 66% identical to that of UNC-78 (Supplemental Figure S1). Therefore we designated this gene as actin-interacting protein-1–like gene-1 (aipl-1) (Supplemental Figure S1). We previously identified four residues on UNC-78 (E126, D168, F182, and F192) that are important for its actin-filament-disassembly activity (Mohri et al., 2004, 2006), and all of them are conserved in AIPL-1 (Supplemental Figure S1, arrows). The three-dimensional structure of AIPL-1 was homology-modeled based on the crystal structure of UNC-78 (Protein Data Bank code 1PEV) (Mohri et al., 2004) (Supplemental Figure S2). The two structures are nearly identical, except for minor differences in two loops in blades 5 and 10 (Supplemental Figure S2, arrows). However, these loops are away from the residues that are required for actin disassembly, and it is not clear whether the differences in these loops are significant for their functions.
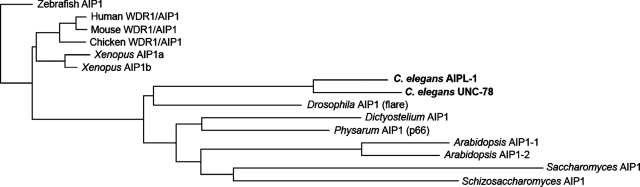
Phylogenetic analysis of AIP1 proteins among eukaryotes suggests that UNC-78 and AIPL-1 relatively recently diverged after nematodes separated from other species (Figure 1). Most species have only one AIP1 gene in the genome. The African clawed frog, Xenopus laevis, has two AIP1 isoforms: AIP1a, the originally cloned AIP1 (XAIP1) (Okada et al., 1999), and AIP1b (GenBank accession number AAH41232.1). These are most likely derived from the pseudotetraploid genome of this species. The green plant Arabidopsis thaliana also has two AIP1 isoforms. AIP1-1 is specifically expressed in reproductive tissues, whereas AIP1-2 is ubiquitously expressed (Allwood et al., 2002). However, whether the two Arabidopsis AIP1 isoforms are functionally different is unknown. The phylogenetic tree suggests that there are no isoform-specific correlations between any of the C. elegans and Arabidopsis or Xenopus AIP1 proteins (Figure 1).
FIGURE 1:
AIPL-1 is a second AIP1 isoform in C. elegans. AIPL-1 and UNC-78 from C. elegans are 66% identical in their amino acid sequences. Sequence alignment is shown in Supplemental Figure S1. A phylogenetic tree of AIP1 sequences from various species was generated by the neighbor-joining method using CLC Sequence Viewer (CLC Bio, Cambridge, MA).
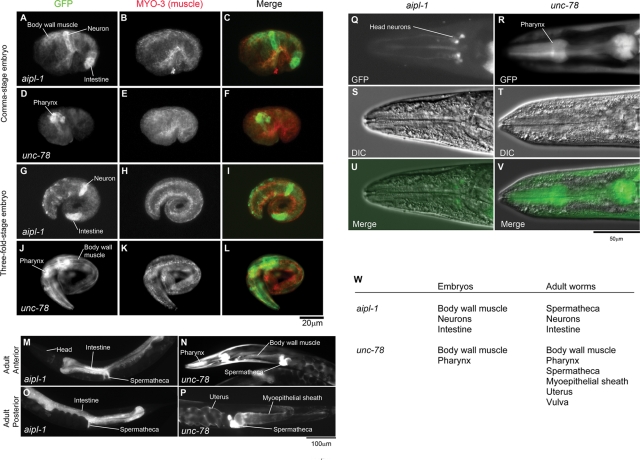
Expression pattern of aipl-1 was examined by a promoter-reporter analysis. The 2-kb upstream sequence of the aipl-1 gene was fused to green fluorescent protein (GFP), and the construct was introduced in wild-type worms. Expression of GFP was initially detected in embryos at the comma-to-1.5-fold stages (310–350 min after first cell division) in the neurons, the intestine, and the body wall muscle (Figure 2, A–C). In older embryos, expression of GFP is gradually diminished in the body wall muscle (Figure 2, G–I), whereas it persisted in the neurons and intestine (Figure 2, G–I). In adult worms, expression of GFP was detected in the intestine (Figure 2M), the spermatheca (Figure 2O), and some of the head neurons (Figure 2, Q, S, and U). This pattern was different from that of unc-78, which is expressed in the pharynx and the body wall muscle from embryos (Figure 2, D–F and J–L) to adults (Figure 2, N, R, T, and V) and the spermatheca, the myoepithelial sheath, the uterus, and the vulva only in adults (Figure 2, N and P) (Mohri and Ono, 2003; Mohri et al., 2006). In the body wall muscle, expression of UNC-78 was first detected at the 1.5-fold stage, but its level was very low (Mohri and Ono, 2003). Thus expression patterns of aipl-1 and unc-78 overlap in the body wall muscle and the spermatheca. We attempted to generate a specific antibody against AIPL-1 using a synthetic peptide corresponding to residues 388–399 that is distinct from UNC-78. However, we were not able to obtain specific antibody that reacts with the AIPL-1 protein in three independent attempts using six rabbits (unpublished data). These results demonstrate that the two AIP1 isoforms are very similar in their sequences but expressed in different tissues with some overlaps.
FIGURE 2:
Expression patterns of aipl-1 and unc-78. The promoter region of aipl-1 or unc-78 was fused with a GFP reporter, and the patterns of GFP expression were examined. (A–L) Promoter activities of aipl-1 (A–C, G–I) and unc-78 (D–F, J–L) in embryos. Embryos were fixed and stained with anti-GFP (A, D, G, J) and anti–MYO-3 (a marker for the body wall muscle) (B, E, H, K). Merged images are shown in C, F, I, and L (GFP in green and MYO-3 in red). Embryos are shown at the comma stage (∼430 min old) (A–F) and the threefold stage (∼550 min old) (G–L). (M–P) Promoter activities of aipl-1 (M, O) and unc-78 (N, P) in adult worms. Adult worms were examined without fixation. The patterns in the anterior (M, N) and posterior halves (O, P) are shown. The unc-78 promoter was also active in the vulva (Mohri et al., 2006), but the data are not shown in this figure. (Q–V) High-magnification images of the head region are shown by GFP fluorescence (Q, R) and Nomarski microscopy (S, T). Merged images are shown in U and V. Scale bars are shown in the figure. (W) Summary of the promoter activities in embryos and adult worms.
The two AIP-1 genes, unc-78 and aipl-1, have overlapping and essential function in embryonic development
To determine the function of aipl-1, we characterized aipl-1 mutant phenotypes. On our request, the C. elegans Gene Knockout Consortium isolated a deletion allele aipl-1(ok1019). aipl-1(ok1019) has a deletion of 2 kb in the aipl-1 gene. The remaining sequence codes for only one-third of the protein from the N-terminus. Because of the tightly packed β-propeller structure of AIP1 (Mohri et al., 2004) (Figure 1A), this deletion allele is unlikely to generate a functional protein. Thus aipl-1(ok1019) is a putative null or strong loss-of-function allele. Nonetheless, aipl-1(ok1019) homozygotes were superficially indistinguishable from wild type. Morphologically, no abnormality was detected under a dissecting microscope in aipl-1(ok1019) homozygotes from embryos to adults (unpublished data). Organization of actin filaments in various tissues also appeared to be normal as examined by staining with phalloidin (unpublished data).
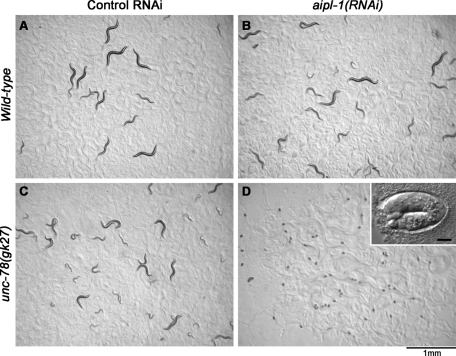
To determine whether the two AIP1 isoforms, AIPL-1 and UNC-78, have overlapping functions, we examined double knockdown or knockout phenotypes. unc-78(gk27) is an unc-78-null allele, and unc-78(gk27) homozygotes are viable but exhibit disorganization of actin filaments in the body wall muscle (Ono, 2001). We crossed unc-78(gk27) with aipl-1(ok1019), but no viable double homozygotes were generated in their F2 progeny (unpublished data), suggesting that an aipl-1 unc-78 double mutant is lethal. To facilitate phenotypic characterization, aipl-1 was depleted by RNA interference in wild-type or unc-78(gk27) worms, and embryonic phenotypes were determined. Embryonic lethality was measured by incubating embryos from wild-type or unc-78(gk27) worms with control RNAi or aipl-1(RNAi) treatment for 24 h on nematode growth medium (NGM) agar plates (Figure 3). Most of embryos from wild-type worms with control RNAi or aipl-1(RNAi) became larvae with very low lethality of 0.77% (3/289) for control RNAi (Figure 3A) and 0.45% (2/447) for aipl-1(RNAi) (Figure 3B). Embryos from unc-78(gk27) worms with control RNAi also hatched with low lethality of 1.2% (5/425) (Figure 3C). However, embryos from unc-78(gk27) with aipl-1(RNAi) were 100% (921/921) lethal (Figure 3D), and most of them were arrested at the twofold or threefold stage (450–520 min old) (Figure 3D, inset; Table 1). These phenotypes were similar to the Pat (paralyzed, arrested elongation at twofold) phenotype in severe muscle-affecting mutations (Williams and Waterston, 1994).
FIGURE 3:
Depletion of the two AIP1 isoforms causes embryonic lethality. Embryos from wild-type (A, B) or unc-78(gk27) (C, D) worms that had been treated with control RNAi (A, C) or aipl-1(RNAi) (B, D) were incubated for 24 h on NGM agar plates. Normally, they hatch and become larvae. However, embryos from unc-78(gk27) with aipl-1(RNAi) were arrested and did not hatch (D). The inset in D is a high-magnification differential interference contrast image of an embryo that was arrested at the twofold stage. Bar in inset, 10 μm. Bar at the bottom of the figure, 1.0 mm.
TABLE 1:
Summary of phenotypes.
| Phenotype | Actin aggregatesa | UNC-60Ba | |
|---|---|---|---|
| Wild type; control RNAi | WT | - | Diffuse |
| Wild type; aipl-1(RNAi) | WT | - | Diffuse |
| unc-78(gk78); control RNAi | Unc | - | Diffuse |
| unc-78(gk78); aipl-1(RNAi) | Pat | +++ | Aggregated |
| Wild type | WT | - | Diffuse |
| unc-60B(r398) | Unc | + | Diffuse |
| aipl-1(ok1019) | WT | - | Partially aggregated |
| unc-60B(r398); aipl-1(ok1019) | Unc | ++ | Diffuse |
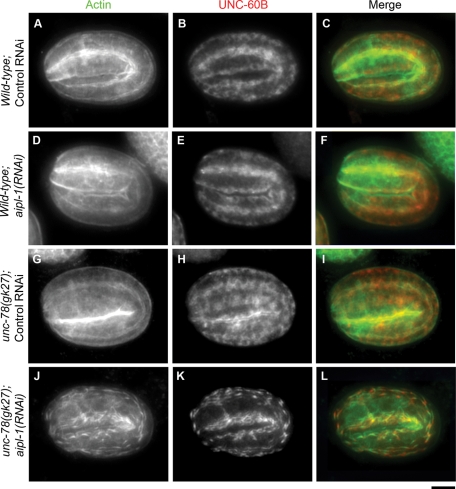
Because the body wall muscle expresses both AIPL-1 and UNC-78 (Figure 2), we further characterized actin organization in embryonic muscle. In wild-type embryos with control RNAi, actin initially accumulated in continuous lines in muscle cells at the 1.5-fold stage (420-min-old embryo) (Supplemental Figure S3B), and the line of actin became wider and more intense as the myofibril assembly continued during subsequent embryonic development (Figure 4A and Supplemental Figure S3, B–D). This pattern of actin assembly was indistinguishable from that for wild-type embryos under standard culture conditions (Epstein et al., 1993). RNAi of aipl-1 in wild-type embryos did not cause detectable changes in actin organization (Figure 4D and Supplemental Figure S3, I–L). In the unc-78–null mutant with control RNAi, actin was assembled into myofibrils in a normal pattern (Figure 4G and Supplemental Figure S3, E–H), as reported previously for the unc-78–null mutant under standard culture conditions (Ono, 2001). However, RNAi of aipl-1 in the unc-78–null mutant caused formation of actin aggregates as early as the twofold stage (450-min-old embryos) (Figure 4J and Supplemental Figure S3O). In arrested embryos, actin filaments were highly disorganized (Supplemental Figure S3P). These phenotypes indicate that aipl-1 and unc-78 have redundant functions in organized assembly of actin filaments in embryonic muscle.
FIGURE 4:
Depletion of the two AIP1 isoforms causes abnormal actin filament assembly in the embryonic body wall muscle. Wild-type (A–F) or unc-78(gk27) (G–L) worms were treated with control RNAi (A–C, G–I) or aipl-1(RNAi) (D–F, J–L), and their F1 embryos were stained for actin (A, D, G, J) and UNC-60B (B, E, H, K). Micrographs of embryos at the twofold stage (450 min old) are shown. Micrographs of embryos at different stages are shown in Supplemental Figure S3. Merged images are shown in C, F, I, and L (actin in green and UNC-60B in red). Bar, 10 μm.
Furthermore, mislocalization of UNC-60B, a muscle-specific ADF/cofilin isoform, was also synergistically enhanced by depletion of the two AIP1 isoforms. In wild-type embryos with control RNAi, UNC-60B was specifically expressed in the body wall muscle and localized to the diffuse cytoplasm (Figure 4B and Supplemental Figure S3, A–D), as previously reported in wild-type embryos under standard culture conditions (Ono et al., 1999). RNAi of aipl-1 in wild-type embryos did not cause detectable changes in UNC-60B localization (Figure 4E and Supplemental Figure S3, I–L) except for the appearance of small aggregates of UNC-60B in late embryos (Figure S3L). In unc-78–null mutant with control RNAi, many aggregates of UNC-60B were formed in late embryos (Supplemental Figure S3H), whereas UNC-60B localization up to the twofold stage was normal (Figure 4H and Supplemental Figure S3, E–G). However, RNAi of aipl-1 in the unc-78–null mutant caused formation of UNC-60B aggregates as early as the comma stage (Supplemental Figure S3M), and the extent of UNC-60B aggregation was enhanced as the embryos became older (Figure 4K and Supplemental Figure S3, N–P). UNC-60B colocalized with actin but unevenly distributed in these aggregates (Figure 4, J–L). These results suggest that the two AIP1 isoforms redundantly regulate the function of UNC-60B in embryonic muscle.
Both UNC-78 and AIPL-1 show preference for the same ADF/cofilin isoform in actin filament disassembly
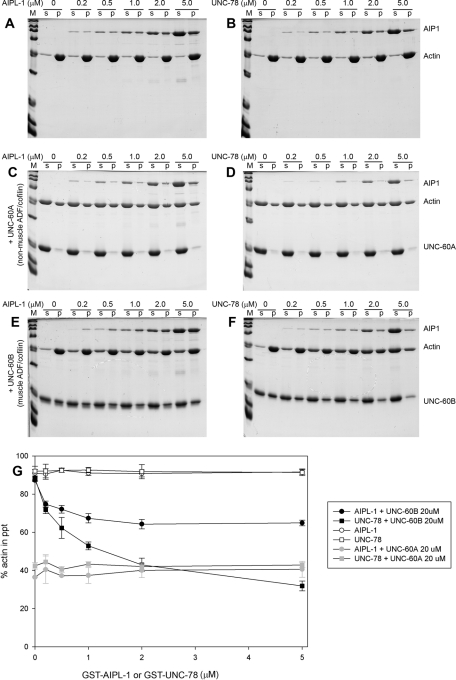
We previously demonstrated that UNC-78 preferentially cooperates with UNC-60B (muscle ADF/cofilin) but not with UNC-60A (nonmuscle ADF/cofilin) in actin filament disassembly in vitro (Mohri and Ono, 2003). Using recombinant AIPL-1 protein, we found that AIPL-1 preferentially cooperated with the same ADF/cofilin isoform as UNC-78 in actin filament disassembly (Figure 5). Both AIPL-1 and UNC-78 were produced as glutathione-S-transferase (GST)–fusion proteins and tested for their effects on actin filament disassembly by sedimentation assays. Cleavage of GST from AIPL-1 was not successful due to poor digestion by thrombin (unpublished data). However, the activity of GST-UNC-78 to disassemble UNC-60B–bound actin filaments is indistinguishable from that of UNC-78 with no tag sequence (Mohri et al., 2004). Therefore we reasoned that GST-AIPL-1 also possessed similar activity to AIPL-1 with no tag.
FIGURE 5:
Both AIPL-1 and UNC-78 cooperate with the same ADF/cofilin isoform to promote actin filament disassembly in vitro. (A–F) Actin filament disassembly activity of GST-AIPL-1 and GST-UNC-78 were examined by F-actin sedimentation assays. F-actin (10 μM) was incubated with various concentrations (0–5 μM) of GST-AIPL-1 (A, C, E) or GST-UNC-78 (B, D, F) in the absence of ADF/cofilin (A, B) or in the presence of 20 μM UNC-60A (C, D) or 20 μM UNC-60B (E, F) for 30 min at room temperature. The mixtures were ultracentrifuged and fractionated into supernatants (s) and pellets (p) and analyzed by SDS–PAGE. Positions of bands of AIP1 (GST-AIPL-1 or GST-UNC-78), actin, UNC-60A, and UNC-60B are indicated on the right. (G) The results were quantitatively analyzed by densitometry. Percentages of actin in the pellets are plotted as a function of the concentrations of GST-AIPL-1 or GST-UNC-78. Data shown are mean ± SD of three independent experiments.
When GST-AIPL-1 or GST-UNC-78 was incubated with F-actin, there was no increase in the amounts of actin in the supernatants (Figure 5, A and B), indicating that either AIPL-1 or UNC-78 did not depolymerize actin filaments in the absence of ADF/cofilin. UNC-60A, a nonmuscle ADF/cofilin isoform, has actin-monomer sequestering activity (Ono and Benian, 1998; Yamashiro et al., 2005), and 20 μM UNC-60A alone depolymerized ∼60% of 10 μM F-actin (Figure 5, C and D). Increasing concentrations of GST-AIPL-1 or GST-UNC-78 did not change the extent of actin depolymerization (Figure 5, C and D). In contrast, UNC-60B, a muscle ADF/cofilin isoform, remains bound to F-actin and does not sequester G-actin (Ono and Benian, 1998; Yamashiro et al., 2005). In the presence of 20 μM UNC-60B, ∼90% of actin remained as filaments and fractionated in the pellets (Figure 5, E and F). However, increasing concentrations of GST-AIPL-1 or GST-UNC-78 increased actin in the supernatants (Figure 5, E and F), indicating that actin filament disassembly was promoted. Quantitative analysis indicates that UNC-78 had stronger activity to enhance actin disassembly than AIPL-1 (Figure 5G). Thus these biochemical studies demonstrate that the two AIP1 isoforms preferentially cooperate with only UNC-60B, a muscle-specific ADF/cofilin, for actin filament disassembly.
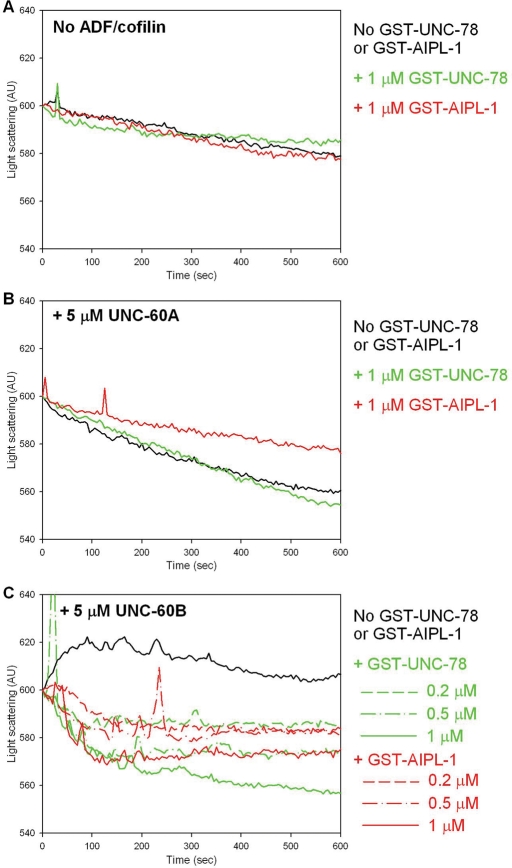
Effects of GST-AIPL-1 and GST-UNC-78 on actin filament disassembly were also analyzed kinetically by light scattering measurements. When F-actin (25 μM) was diluted to 4 μM actin, light scattering of the F-actin solution was gradually decreased due to spontaneous depolymerization to restore an equilibrium state (Figure 6A, black line). Addition of 1 μM GST-UNC-78 (Figure 6A, green line) or 1 μM GST-AIPL-1 (Figure 6A, red line) did not alter the rate of depolymerization. UNC-60A enhanced the rate of depolymerization (Figure 6B, black line), and 1 μM GST-UNC-78 (Figure 6B, green line) did not alter the effect of UNC-60A. However, 1 μM GST-AIPL-1 (Figure 6B, red line) slightly suppressed the rate of UNC-60A–induced depolymerization. Nonetheless, in the sedimentation assays, GST-AIPL-1 did not alter the extent of UNC-60A–induced actin depolymerization (Figure 5G), suggesting that GST-AIPL-1 slowed actin depolymerization in the presence of UNC-60A without affecting the equilibrium levels of depolymerization. The sedimentation assays also indicated that GST-AIPL-1 cosedimented with F-actin in the presence of UNC-60A (Figure 5C), suggesting that apparent suppression of UNC-60A–induced depolymerization by GST-AIPL-1 might be artificial due to increased light scattering by side binding of GST-AIPL-1. UNC-60B increased light scattering of F-actin (Figure 6C, black line) because it binds to the side of actin filaments and increases the thickness of the filaments (Ono and Benian, 1998; Yamashiro et al., 2005). GST-UNC-78 rapidly decreased light scattering of F-actin in the presence of UNC-60B (Figure 6C, green lines) in a dose-dependent manner. GST-AIPL-1 similarly decreased light scattering of F-actin in the presence of UNC-60B (Figure 6C, red lines) in a dose-dependent manner. The activity of GST-AIPL-1 was weaker than that of GST-UNC-78, as the depolymerization curve of 1.0 μM GST-AIPL-1 (Figure 6C, red solid line) was nearly identical to that of 0.5 μM GST-UNC-78 (Figure 6C, green dash-dot line). This difference in their activities was consistent with slightly weaker activity of GST-AIPL-1 than that of GST-UNC-78 in the sedimentation assays (Figure 5G). However, again, the sedimentation assays indicated that GST-AIPL-1 cosedimented with F-actin in the presence of UNC-60B (Figure 5D). Therefore there is possibility that the apparent difference in depolymerization kinetics between UNC-78 and AIPL-1 is artificial due to counteracting enhancement of light scattering by filament binding. Nonetheless, this kinetic analysis provided additional biochemical evidence that both AIPL-1 and UNC-78 preferentially enhance UNC-60B–bound actin filaments.
FIGURE 6:
Both AIPL-1 and UNC-78 cooperate with the same ADF/cofilin isoform to promote the rate of actin filament disassembly in vitro. Kinetics of actin filament disassembly was analyzed by light scattering assays. F-actin was diluted to 4 μM in the absence of ADF/cofilin (A) or in the presence of 5 μM UNC-60A (B) or 5 μM UNC-60B (C). Experiments were performed in the absence of GST-AIPL-1 or GST-UNC-78 (black lines) or in the presence of GST-UNC-78 (green lines) or GST-AIPL-1 (red lines). Light scattering (arbitrary units) is plotted as a function of time (seconds).
AIPL-1 cooperates with UNC-60B in actin filament organization in vivo
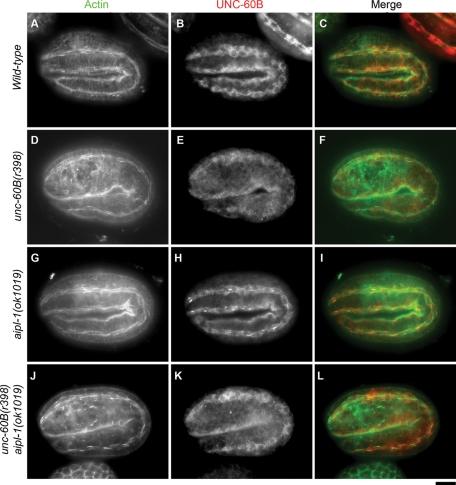
To determine the in vivo functional relationship between AIPL-1 and UNC-60B, we tested whether an aipl-1 mutation enhances an unc-60B mutant phenotype. In wild-type embryos at the twofold stage (450 min old), actin became aligned in continuous myofibrils in the body wall muscle (Figure 7A). unc-60B(r398) is a weak loss-of-function allele, and the mutant embryos at an equivalent stage showed relatively minor disorganization of actin filaments with a few discontinuations of the actin bands (Figure 7D). aipl-1(ok1019) (aipl-1–null) embryos had normally assembled actin filaments in the body wall muscle (Figure 7G). However, an unc-60B(r398) aipl-1(ok1019) double mutant showed an enhanced phenotype with a number of actin aggregates in the body wall muscle (Figure 7J).
FIGURE 7:
AIPL-1 cooperates with UNC-60B in organizing sarcomeric actin filaments in embryonic body wall muscle. Twofold-stage embryos of wild-type (A–C), unc-60B(r398) (D–F), aipl-1(ok1019) (G–I), and unc-60B(r398) aipl-1(ok1019) (J–L) worms were stained for actin (A, D, G, J) and UNC-60B (B, E, H, K). Merged images are shown in C, F, I, and L (actin in green and UNC-60B in red). Bar, 10 μm.
The aipl-1 mutation also affected the localization of UNC-60B (Table 1). In wild-type embryos, UNC-60B was specifically expressed in the body wall muscle and localized to the diffuse cytoplasm (Figure 7B). In the aipl-1–null mutant, although the majority of UNC-60B remained in the cytoplasm, aggregates of UNC-60B were frequently formed in the muscle cells (Figure 7H). This phenotype is consistent with the biochemical activity of AIPL-1 to disassemble UNC-60B–bound actin filaments (Figures 5 and 6). Aggregates of UNC-60B were not found in the unc-60B(r398) aipl-1(ok1019) double mutants (Figure 7K). Previously, we showed that the unc-60B(r398) mutation reduces affinity of UNC-60B with F-actin (Ono et al., 1999, 2001). Therefore our interpretation is that the mutant UNC-60B protein remained localized in the diffuse cytoplasm with low affinity with actin in the myofibrils even in the absence of AIPL-1. These results suggest that localization of only F-actin–bound UNC-60B is affected by AIPL-1.
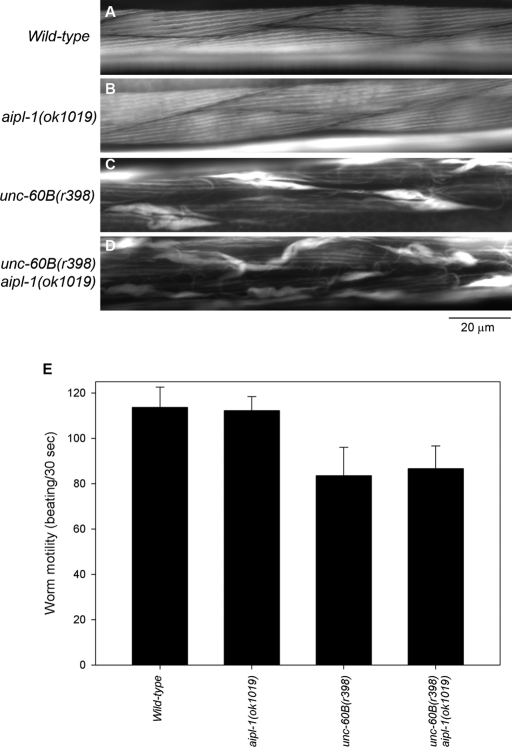
In adult worms, the aipl-1 mutation did not enhance the unc-60B mutant phenotype. Actin organization in the body wall muscle of the aipl-1-null mutant was indistinguishable from that of wild type (Figure 8, A and B). The unc-60B(r398) single mutant had disorganization of actin filaments with formation of actin aggregates (Figure 8C). The unc-60B(r398) aipl-1(ok1019) double mutant showed actin disorganization to a similar extent to the unc-60B(r398) single mutant (Figure 8D). Quantitative analysis of worm motility as a measurement of muscle activity also showed that the aipl-1–null mutation did not alter motility in either wild-type or unc-60B(r398) worms (Figure 8E). These results strongly suggest that AIPL-1 is the major AIP1 isoform in early embryonic muscle and cooperates with UNC-60B for actin organization and that AIPL-1 is not required in postembryonic muscle, most likely due to expression of UNC-78 as the major AIP1 isoform in larval and adult muscle.
FIGURE 8:
AIPL-1 is not required for organized sarcomeric actin assembly in adult body wall muscle. Adult wild-type (A), aipl-1(ok1019) (B), unc-60B(r398) (C), and unc-60B(r398) aipl-1(ok1019) (D) worms were stained for F-actin by tetramethylrhodamine-phalloidin. Bar, 20 μm. (E) Motility of wild-type, aipl-1(ok1019), unc-60B(r398), and unc-60B(r398) aipl-1(ok1019) worm. The aipl-1(ok1019) mutation did not significantly alter motility in wild-type or unc-60B(r398) background. Data are mean ± SD, n = 10.
Overexpression of AIPL-1 in body wall muscle compensates for the function of UNC-78
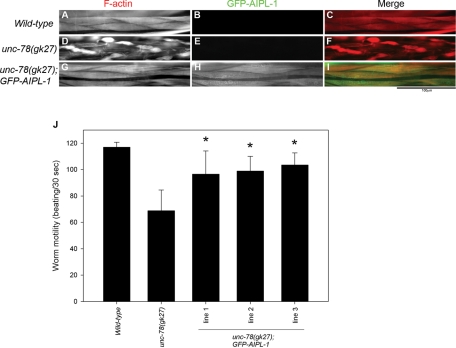
Mutational analyses of aipl-1 and unc-78 strongly suggest that the two AIP1 isoforms have overlapping functions. To determine their functional redundancy more directly, we tested whether forced expression of AIPL-1 compensates for the function of UNC-78. An unc-78–null mutant showed severe disorganization of actin filaments in the body wall muscle (Figure 9D) as compared with striated organization of actin filaments in wild-type worms (Figure 9A). Transgenic expression of GFP-tagged AIPL-1 in the adult body wall muscle of the unc-78–null mutant by a muscle-specific promoter (Pmyo-3) rescued the actin disorganization and restored nearly normal sarcomeric actin organization (Figure 9G). GFP-AIPL-1 colocalized with actin in a striated manner (Figure 9, H and I), which is similar to the localization of UNC-78 (Mohri and Ono, 2003). The unc-78–null worms moved more slowly than wild-type worms due to disorganized myofibrils in the body wall muscle (Figure 9J). However, expression of GFP-AIPL-1 in the unc-78–null mutant partially rescued worm motility (Figure 9J). Many of the transgenic worms had a few cells with no expression of GFP-AIPL-1, and these cells had disorganized actin filaments (unpublished data). Therefore this mosaic expression of the transgene is a likely explanation for incomplete restoration of worm motility. Three independent transgenic lines showed similar extents of restoration of worm motility (Figure 9J). These results indicate that AIPL-1 can compensate for the function of UNC-78 in the body wall muscle and support the conclusion that AIPL-1 and UNC-78 have overlapping functions.
FIGURE 9:
AIPL-1 compensates for the function of UNC-78 in the body wall muscle. Adult worms of wild type (A–C), unc-78(gk27) (D–F), or unc-78(gk27) with a transgene for expression of GFP-AIPL-1 (G–I) were stained for F-actin by tetramethylrhodamine-phalloidin (A, D, G). Micrographs of the GFP fluorescence are shown in B, E, and H. Merged images are shown in C, F, and I (F-actin in red and GFP in green). Bar, 100 μm. Expression of GFP-AIPL-1 was forced in adult body wall muscle by the myo-3 promoter (Okkema et al., 1993). (J) Worm motility of wild type, unc-78(gk27), and unc-78(gk27) with a transgene for expression of GFP-AIPL-1 (three independent transgenic lines). Data are mean ± SD, n = 10. Motility of unc-78(gk27) with the transgene was significantly different (p < 0.001, asterisks) from that of unc-27(gk27) without the transgene.
DISCUSSION
In this study, we identified a second AIP1 gene, aipl-1, in C. elegans and demonstrated that the two AIP1 genes, aipl-1 and unc-78, have overlapping and essential function for embryonic development. Gene knockout of either one of the AIP1 isoforms was homozygous viable. However, simultaneous ablation of the two AIP1 genes caused embryonic lethality. AIP1-depleted animals were arrested at a late embryonic stage and failed to elongate their bodies due to severe disorganization of myofibrils in the body wall muscle. In vitro, both AIPL-1 and UNC-78 cooperated with UNC-60B, a muscle-specific ADF/cofilin isoform, in actin filament disassembly, but not with UNC-60A, a nonmuscle ADF/cofilin isoform. These biochemical observations are consistent with the phenotypic observations that AIP1 proteins are essential for actin organization in muscle but not in early embryogenesis, in which UNC-60A plays an essential role. These results demonstrate that AIP1 proteins are essential for viability in C. elegans and suggest that isoform-specific and tissue-specific cooperation of AIP1 and ADF/cofilin is important for morphogenetic events that involve actin filament reorganization.
Our results strongly suggest that AIP1 proteins play a critical role in assembly of myofibrils in embryonic muscle cells. Previously, we demonstrated that unc-78 encodes an AIP1 protein that is required for organized actin assembly into myofibrils (Ono, 2001; Mohri et al., 2006). However, an unc-78–null mutant is homozygous viable and shows only mild phenotypes in embryonic muscle (Ono, 2001). In this study, we demonstrated that depletion of two AIP1 isoforms causes embryonic arrest with severe disorganization of actin filaments in the body wall muscle. This phenotype is similar to the Pat phenotypes that are caused by defects in critical components for the function of the body wall muscle (Williams and Waterston, 1994). Mutations in components of the attachment structures (integrin, vinculin, perlecan) and thin and thick filaments (myosin heavy chain A, tropomyosin, troponin) are known to cause the Pat phenotypes (Moerman and Williams, 2006), but no regulators of actin dynamics had been known as a Pat gene. Even a null mutation of UNC-60B (a muscle-specific ADF/cofilin) is homozygous viable and does not cause a Pat phenotype (Ono et al., 2003). This is probably due to the presence of UNC-60A (a nonmuscle ADF/cofilin), which may be sufficient to support embryonic viability (Ono et al., 2003; Anyanful et al., 2004), although the precise role of UNC-60A in the body wall muscle is unknown. Although aipl-1 is expressed in neurons, the Pat phenotype of the unc-78(gk78) aipl-1(RNAi) animals is unlikely to be caused by a neuronal defect. The aipl-1–null mutant did not show a detectable motility defect, and severe neuronal defects such as presynaptic defects in unc-13 mutants do not cause a Pat phenotype (Richmond et al., 1999). Enhancement of unc-60B mutant phenotypes by depletion of unc-78 or aipl-1 strongly suggests that UNC-60B and the two AIP1 isoforms cooperate for organized assembly of actin filaments, and our biochemical data suggest that promotion of actin filament disassembly by UNC-60B and the two AIP1 isoforms is important for their in vivo function. These are consistent with the observations that UNC-60B and actin accumulate in abnormal aggregates in the absence of AIP1 proteins (Figure 4, J–L), suggesting that AIP1 proteins are required for disassembly of UNC-60B–bound actin filaments in vivo.
Essential function of ADF/cofilin for myofibril assembly and maintenance has also been demonstrated in vertebrates (Ono, 2010), but the function of AIP1 in vertebrate muscle is unknown. Recently, AIP1 has been shown to be important for myofibril formation in Drosophila striated muscle (Schnorrer et al., 2010), but a role for ADF/cofilin in Drosophila muscle is unknown. Mammalian cofilin-2 (CFL2) is predominantly expressed in striated muscle (Ono et al., 1994; Thirion et al., 2001; Vartiainen et al., 2002), and knockdown of cofilin induces disorganization of myofibrils in cultured myocytes (Skwarek-Maruszewska et al., 2009). A mutation in the human CFL2 gene causes nemaline myopathy (Agrawal et al., 2007). Although mammals have three ADF/cofilin isoforms, only a single gene for AIP1 (also known as WDR1) is present in each mammalian genome. Mutations in the mouse AIP1 gene (WDR1) cause blood disorders, and a severe loss-of-function mutation is embryonic lethal (Kile et al., 2007). Therefore it is of great interest to see whether the WDR1-mutant mice exhibit any phenotypes in skeletal and cardiac muscles.
Comparison of the unc-60B–null and AIP1-depletion phenotypes also raises a new question. The unc-60B–null mutant is homozygous viable, whereas depletion of the two AIP1 isoforms is embryonic lethal. Our in vitro studies indicate that the interaction of UNC-78 or AIPL-1 with actin filaments is dependent on UNC-60B (Figure 5) (Mohri and Ono, 2003). UNC-60B alone can enhance actin filament disassembly and promote actin turnover in vitro (Yamashiro et al., 2005), but the strong AIP1-depletion phenotypes indicate that AIP1 proteins are required for mediating rapid actin turnover in vivo. On the other hand, viability of the unc-60B–null mutant should be supported by UNC-60A or AIP1 proteins or cooperation of them. However, our in vitro studies show that neither AIP1 isoform cooperates with UNC-60A in actin filament disassembly. In the unc-60B–null mutant, UNC-60A is not overexpressed in the body wall muscle (Ono et al., 2003), and compensatory expression of UNC-60A is not a likely mechanism of maintaining viability. A possible mechanism is that AIP1 proteins have an ADF/cofilin–independent function and support viability in the absence of UNC-60B. However, an ADF/cofilin–independent function of AIP1 proteins is unknown. In the absence of ADF/cofilin, AIP1 proteins only weakly interact with F-actin in vitro and show no actin-severing activity (Mohri and Ono, 2003; Ono et al., 2004). Another possible mechanism is that UNC-60A and AIP1 proteins cooperate in vivo in the presence of an unknown factor that allows their functional interaction. Coronin is a candidate factor that promotes cooperation of UNC-60A and AIP1 proteins. Coronin is a WD-repeat protein and enhances actin filament turnover synergistically with ADF/cofilin (Brieher et al., 2006; Cai et al., 2007b; Kueh et al., 2008; Gandhi et al., 2009) and AIP1 (Ishikawa-Ankerhold et al., 2010). In vitro, coronin is reported to enhance binding of ADF/cofilin to F-actin (Brieher et al., 2006), although discrepant results have also been reported (Cai et al., 2007a). C. elegans has a coronin gene (cor-1) that encodes multiple splice variants (Yonemura and Mabuchi, 2001), but its function is not understood. Thus functional analysis of coronin in C. elegans may reveal an additional regulatory mechanism for actin turnover during embryonic development.
Our results demonstrated overlapping functions of AIPL-1 and UNC-78 in the body wall muscle. However, expression patterns of the two AIP1 isoforms are different in other tissues, suggesting that they also have distinct functions. AIPL-1 is expressed in the head neurons and the intestine, whereas UNC-78 is in the pharynx and the vulva. Although AIPL-1 specifically cooperates with UNC-60B (ADF/cofilin) in actin filament disassembly in vitro, UNC-60B is not detected in the head neurons and the intestine by immunohistochemistry (Ono et al., 2003). Instead, UNC-60A is expressed in these tissues (Ono et al., 2003). Again, these observations suggest that AIPL-1 has an ADF/cofilin–independent function or that a third factor enhances functional cooperation of AIPL-1 with UNC-60A. Nonetheless, we did not detect any phenotypes in the neurons and the intestine in the aipl-1–null mutant, and the function of aipl-1 in nonmuscle tissues is unknown.
We demonstrated that AIP1 proteins are essential in C. elegans. This is consistent with lethal phenotypes caused by AIP1 mutation or knockdown in Arabidopsis (Ketelaar et al., 2004), Drosophila (Ren et al., 2007), and mice (Kile et al., 2007). However, AIP1 is not essential in budding yeast (Iida and Yahara, 1999; Rodal et al., 1999) and Dictyostelium (Konzok et al., 1999). AIP1 is required for cell proliferation in Drosophila wings (Ren et al., 2007), and AIP1 knockdown moderately increases the rate of cytokinesis failure in Dictyostelium and mammalian cultured cells (Konzok et al., 1999; Kato et al., 2008). However, in AIP1-depleted C. elegans, early embryogenesis was superficially normal. Rather, lethality was caused by severe muscle defects that arrested body elongation during embryogenesis. Thus, although AIP1 proteins are involved in fundamental cell biological events, including cytokinesis and cell migration, its role apparently becomes more critical in multicellular organisms in which tissue-specific reorganization of the actin cytoskeleton is required for complex morphogenetic processes. Therefore analysis of functions of ADF/cofilin and AIP1 during development of other tissues and organs in multicellular organisms may reveal new regulatory mechanisms of morphogenesis.
MATERIALS AND METHODS
Nematode strains
Wild-type strain N2 and VC701 aipl-1(ok1019) were obtained from the Caenorhabditis Genetics Center (Minneapolis, MN). aipl-1(ok1019) was outcrossed with wild-type worms three times before the experiments. unc-78(gk27) was described previously (Ono, 2001). unc-60B(r398) was described previously (McKim et al., 1988), and an additionally outcrossed strain was used (Yu and Ono, 2006). An unc-60B(r398) aipl-1(ok1019) double mutant was generated by standard crosses and isolating recombinants that have both mutations on chromosome V. Nematodes were grown under standard conditions at 20ºC as described previously (Brenner, 1974).
Promoter analysis
Promoter::GFP constructs for analysis of the aipl-1 promoter were made using fusion PCR as previously described (Hobert, 2002). Promoter-containing sequences (1961 or 2961 base pairs from the initiation codon) were amplified by PCR and fused upstream of the GFP-coding region in the pPD95.67 GFP-coding cassette (kindly provided by A. Fire, Stanford University, Stanford, CA). The PCR constructs (30 ng/μl) and a transformation marker pCeh361 [dpy-5(+)] (Thacker et al., 2006) (100 ng/μl) were injected into the syncytial region of the gonad of the target strain dpy-5(e907). Transgenic F1 worms (Dpy-5 rescued) were isolated, and F2 worms that stably inherited transgenes were selected to establish transgenic lines. A promoter::GFP construct for analysis of the unc-78 promoter was made by inserting the 1.4-kb upstream region of the unc-78 gene at the 5′ end of the GFP-coding sequence of a GFP-expression vector pPD117.01 as described previously (Mohri et al., 2006). The plasmid vector (20 ng/μl) and a transformation marker pRF4 (80 ng/μl) were injected into the syncytial region of the gonad of the wild-type strain N2. Transgenic F1 worms with dominant roller phenotype were isolated, and F2 worms that stably inherited transgenes were selected to establish transgenic lines. Patterns of GFP expression were analyzed as described in Fluorescence microscopy.
RNAi experiments
RNAi experiments were performed by feeding with Escherichia coli–expressing double-stranded RNA as described previously (Ono and Ono, 2002). An RNAi clone for aipl-1 (V-9L16) was obtained from MRC Geneservice (Cambridge, United Kingdom) (Kamath et al., 2003). L4 larvae were treated with RNAi, and phenotypes were characterized in the F1 generation.
Fluorescence microscopy
Immunofluorescence staining of embryos was performed as described previously (Ono et al., 2003). Briefly, worm embryos were collected by cutting gravid adults and picking from agar plates on polylysine-coated slides, permeabilized by a freeze-crack method (Epstein et al., 1993), and fixed with methanol at −20ºC for 5 min. They were washed by phosphate-buffered saline (PBS) for 10 min and stained with mouse antiactin (C4; MP Biomedicals, Solon, OH) and rabbit anti–UNC-60B (Ono et al., 1999) monoclonal antibodies (mAbs) diluted in 1% bovine serum albumin in PBS. The primary antibodies were visualized by staining with Alexa 488–conjugated goat anti–mouse immunoglobulin G (IgG) (Invitrogen, Carlsbad, CA) and Cy3-conjugated donkey anti–rabbit IgG (Jackson ImmunoResearch, West Grove, PA).
Staining of whole worms with tetramethylrhodamine-phalloidin (Sigma-Aldrich, St. Louis, MO) was performed as described previously (Ono, 2001).
To localize GFP in transgenic embryos, they were immunostained by anti-GFP rabbit polyclonal (Rockland Immunochemicals, Gilbertsville, PA) and anti–MYO-3 mouse mAb 5–6 (Miller et al., 1983). To observe the GFP fluorescence in live adult worms, they were anesthetized in 0.1% tricaine/0.01% tetramisole in M9 buffer for 30 min and mounted on 2% agarose pads.
Samples were mounted with ProLong Gold (Invitrogen) and observed by epifluorescence using a Nikon Eclipse TE2000 inverted microscope with a CFI Plan Fluor ELWD 40× (dry; numerical aperture [NA], 0.60) or Plan Apo 60× (oil; NA 1.40) objective. Images were captured by a SPOT RT monochrome charge-coupled device camera (Diagnostic Instruments, Sterling Heights, MI) and processed by IPLab imaging software (BD Biosciences, Franklin Lakes, NJ) and Photoshop CS3 (Adobe, San Jose, CA).
Motility assay
Worm motility was quantified as described previously (Epstein and Thomson, 1974). Briefly, adult worms were placed in M9 buffer. Then the total number of beats in 30 s was recorded.
Proteins
Rabbit muscle actin was prepared from rabbit muscle acetone powder (Pel-Freez Biologicals, Rogers, AR) as described (Pardee and Spudich, 1982). Recombinant UNC-60A, UNC-60B (Ono and Benian, 1998), and GST-UNC-78 (Mohri et al., 2004) were purified as described. To produce recombinant GST-AIPL-1, the entire coding region of AIPL-1 was amplified from the cDNA clone yk1612e12 (kindly provided by Yuji Kohara) by PCR and cloned into pGEX-2T between the BamHI and SmaI sites. The insert was verified by DNA sequencing. The E. coli strain BL21(DE3) was transformed with the expression vector and cultured in M9ZB medium containing 50 μg/ml ampicillin at 37ºC until A600 reached 0.6 cm−1. Then, the culture was cooled, and protein expression was induced by adding 0.1 mM isopropyl β-d-thiogalactopyranoside overnight at 15ºC. The cells were harvested by centrifugation at 5000 × g for 10 min and disrupted by sonication in PBS containing 0.2 mM dithiothreitol (DTT) and 1 mM phenylmethanesulfonyl fluoride. The homogenates were centrifuged at 20,000 × g for 20 min, and the supernatants were applied to a Glutathione-Uniflow Column (Clontech, Mountain View, CA). Bound proteins were eluted with 10 mM glutathione, 20 mM Tris-HCl, and 0.2 mM DTT, pH 8.0. Fractions containing pure GST-AIPL-1 were dialyzed against 0.1 M KCl, 2 mM MgCl2, 1 mM DTT, 50% glycerol, and 20 mM HEPES-NaOH, pH 7.5, overnight at 4ºC and stored at −20ºC.
F-actin sedimentation assay
Sedimentation assays were performed as described previously (Mohri et al., 2004). Briefly, 10 μM F-actin was incubated with various concentrations of GST-AIPL-1, GST-UNC-78, UNC-60A, and/or UNC-60B in F-buffer (0.1 M KCl, 2 mM MgCl2, 1 mM DTT, 20 mM HEPES-NaOH, pH 7.5) for 30 min at room temperature and centrifuged in a TLA-100 rotor (Beckman Coulter, Brea, CA) at 80,000 rpm for 20 min. The supernatants and pellets were adjusted to the same volumes and analyzed by SDS–PAGE. Gels were stained with Coomassie Brilliant Blue R-250 (National Diagnostics, Atlanta, GA) and scanned by an Perfection V700 scanner (Epson, Long Beach, CA) at 300 dots per inch, and the band intensity of actin was quantified by ImageJ.
Light scattering assay
F-actin (25 μM stock) was diluted to 4 μM in F-buffer without or with UNC-60A, UNC-60B, GST-UNC-78, and/or GST-AIPL-1, and light scattering at an angle of 90º and a wavelength of 400 nm was measured with an F-4500 fluorescence spectrophotometer (Hitachi High-Technologies, Tokyo, Japan). Slit width was set at 5 and 2.5 nm for excitation and emission, respectively.
Transgenic expression of GFP-AIPL-1
Transgenic expression of GFP-AIPL-1 was driven by the myo-3 promoter (Okkema et al., 1993). The entire coding sequence of AIPL-1 was amplified from the AIPL-1 cDNA (yk1612e12) by PCR and cloned at the EcoRI–NheI sites of pPD118.20, an expression vector with the myo-3 promoter and the GFP coding sequence (provided by Andrew Fire, Stanford University, Stanford CA). The entire coding region was sequenced to confirm the insert and the absence of PCR-induced errors. Transgenic nematodes were generated as described previously (Mello and Fire, 1995). The plasmid vector at 20 μg/ml was mixed with 80 μg/ml pGEM-3Zf (Promega, Madison, WI) as carrier DNA. The DNA mixture was injected into the distal arm of the hermaphroditic gonad of unc-78(gk27) as described (Mohri et al., 2006). Transformants were selected by expression of GFP as observed by epifluorescence, and the transgenes were maintained as extrachromosomal arrays.
Supplementary Material
Acknowledgments
We thank Ronen Zaidel-Bar and Jeff Hardin for communicating their preliminary results on aipl-1 RNAi. Monoclonal antibody 5–6 was developed by Henry Epstein (University of Texas Medical Branch, Galveston, TX) and was obtained from the Developmental Studies Hybridoma Bank developed under auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences, University of Iowa, Iowa City, IA. Some C. elegans strains were provided by the Caenorhabditis Genetics Center, which is supported by the National Institutes of Health National Center for Research Resources. D.L.B. holds a Canada Research Chair. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada to D.L.B. and the National Institutes of Health (R01 AR48615) and University Research Committee of Emory University to S.O.
Abbreviations used:
- ADF
actin-depolymerizing factor
- AIP1
actin-interacting protein 1
- GFP
green fluorescent protein
- GST
glutathione-S-transferase
- NGM
nematode growth medium
- RNAi
RNA interference
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-12-0934) on May 5, 2011.
REFERENCES
- Agrawal PB, et al. Nemaline myopathy with minicores caused by mutation of the CFL2 gene encoding the skeletal muscle actin-binding protein, cofilin-2. Am J Hum Genet. 2007;80:162–167. doi: 10.1086/510402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H, Katadae M, Maruya M, Sameshima M, Murakami-Murofushi K, Yahara I. Hyperosmotic stress-induced reorganization of actin bundles in Dictyostelium cells over-expressing cofilin. Genes Cells. 1999;4:311–324. doi: 10.1046/j.1365-2443.1999.00262.x. [DOI] [PubMed] [Google Scholar]
- Allwood EG, Anthony RG, Smertenko AP, Reichelt S, Drobak BK, Doonan JH, Weeds AG, Hussey PJ. Regulation of the pollen-specific actin-depolymerizing factor LlADF1. Plant Cell. 2002;14:2915–2927. doi: 10.1105/tpc.005363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Anyanful A, Ono K, Johnsen RC, Ly H, Jensen V, Baillie DL, Ono S. The RNA-binding protein SUP-12 controls muscle-specific splicing of the ADF/cofilin pre-mRNA in C. elegans. J Cell Biol. 2004;167:639–647. doi: 10.1083/jcb.200407085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieher WM, Kueh HY, Ballif BA, Mitchison TJ. Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J Cell Biol. 2006;175:315–324. doi: 10.1083/jcb.200603149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Makhov AM, Bear JE. F-actin binding is essential for coronin 1B function in vivo. J Cell Sci. 2007a;120:1779–1790. doi: 10.1242/jcs.007641. [DOI] [PubMed] [Google Scholar]
- Cai L, Marshall TW, Uetrecht AC, Schafer DA, Bear JE. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 2007b;128:915–929. doi: 10.1016/j.cell.2007.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MG, Amberg DC. Biochemical and genetic analyses provide insight into the structural and mechanistic properties of actin filament disassembly by the Aip1p cofilin complex in Saccharomyces cerevisiae. Genetics. 2007;176:1527–1539. doi: 10.1534/genetics.107.072066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MG, Teply J, Haarer BK, Viggiano SC, Sept D, Amberg DC. A genetic dissection of Aip1p's interactions leads to a model for Aip1p-cofilin cooperative activities. Mol Biol Cell. 2006;17:1971–1984. doi: 10.1091/mbc.E05-10-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Casey DL, Ortiz I. Myosin and paramyosin of Caenorhabditis elegans embryos assemble into nascent structures distinct from thick filaments and multi-filament assemblages. J Cell Biol. 1993;122:845–858. doi: 10.1083/jcb.122.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Thomson JN. Temperature-sensitive mutation affecting myofilament assembly in Caenorhabditis elegans. Nature. 1974;250:579–580. doi: 10.1038/250579a0. [DOI] [PubMed] [Google Scholar]
- Gandhi M, Achard V, Blanchoin L, Goode BL. Coronin switches roles in actin disassembly depending on the nucleotide state of actin. Mol Cell. 2009;34:364–374. doi: 10.1016/j.molcel.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus KC, Bonaccorsi S, Williams E, Verni F, Gatti M, Goldberg ML. Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J Cell Biol. 1995;131:1243–1259. doi: 10.1083/jcb.131.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurniak CB, Perlas E, Witke W. The actin depolymerizing factor n-cofilin is essential for neural tube morphogenesis and neural crest cell migration. Dev Biol. 2005;278:231–241. doi: 10.1016/j.ydbio.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Ichetovkin I, Han J, Pang KM, Knecht DA, Condeelis JS. Actin filaments are severed by both native and recombinant Dictyostelium cofilin but to different extents. Cell Motil Cytoskeleton. 2000;45:293–306. doi: 10.1002/(SICI)1097-0169(200004)45:4<293::AID-CM5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Iida K, Moriyama K, Matsumoto S, Kawasaki H, Nishida E, Yahara I. Isolation of a yeast essential gene, COF1, that encodes a homologue of mammalian cofilin, a low-Mr actin-binding and depolymerizing protein. Gene. 1993;124:115–120. doi: 10.1016/0378-1119(93)90770-4. [DOI] [PubMed] [Google Scholar]
- Iida K, Yahara I. Cooperation of two actin-binding proteins, cofilin and Aip1, in Saccharomyces cerevisiae. Genes Cells. 1999;4:21–32. doi: 10.1046/j.1365-2443.1999.00235.x. [DOI] [PubMed] [Google Scholar]
- Ishikawa-Ankerhold HC, Gerisch G, Muller-Taubenberger A. Genetic evidence for concerted control of actin dynamics in cytokinesis, endocytic traffic, and cell motility by coronin and Aip1. Cytoskeleton. 2010;67:442–455. doi: 10.1002/cm.20456. [DOI] [PubMed] [Google Scholar]
- Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kato A, Kurita S, Hayashi A, Kaji N, Ohashi K, Mizuno K. Critical roles of actin-interacting protein 1 in cytokinesis and chemotactic migration of mammalian cells. Biochem J. 2008;414:261–270. doi: 10.1042/BJ20071655. [DOI] [PubMed] [Google Scholar]
- Ketelaar T, Allwood EG, Anthony R, Voigt B, Menzel D, Hussey PJ. The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr Biol. 2004;14:145–149. doi: 10.1016/j.cub.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Kile BT, et al. Mutations in the cofilin partner Aip1/Wdr1 cause autoinflammatory disease and macrothrombocytopenia. Blood. 2007;110:2371–2380. doi: 10.1182/blood-2006-10-055087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konzok A, Weber I, Simmeth E, Hacker U, Maniak M, Muller-Taubenberger A. DAip1, a Dictyostelium homologue of the yeast actin-interacting protein 1, is involved in endocytosis, cytokinesis, and motility. J Cell Biol. 1999;146:453–464. doi: 10.1083/jcb.146.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueh HY, Charras GT, Mitchison TJ, Brieher WM. Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J Cell Biol. 2008;182:341–353. doi: 10.1083/jcb.200801027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver SK, Pope BJ, Whytock S, Weeds AG. The effect of two actin depolymerizing factors (ADF/cofilins) on actin filament turnover: pH sensitivity of F-actin binding by human ADF, but not of Acanthamoeba actophorin. Eur J Biochem. 1998;256:388–397. doi: 10.1046/j.1432-1327.1998.2560388.x. [DOI] [PubMed] [Google Scholar]
- Maciver SK, Zot HG, Pollard TD. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Heschl MF, Rosenbluth RE, Baillie DL. Genetic organization of the unc-60 region in Caenorhabditis elegans. Genetics. 1988;118:49–59. doi: 10.1093/genetics/118.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Matheson C, Marra MA, Wakarchuk MF, Baillie DL. The Caenorhabditis elegans unc-60 gene encodes proteins homologous to a family of actin-binding proteins. Mol Gen Genet. 1994;242:346–357. doi: 10.1007/BF00280425. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Miller DM, Ortiz I, Berliner GC, Epstein HF. Differential localization of two myosins within nematode thick filaments. Cell. 1983;34:477–490. doi: 10.1016/0092-8674(83)90381-1. [DOI] [PubMed] [Google Scholar]
- Moerman DG, Williams BD. Sarcomere assembly in C. elegans muscle. WormBook. 2006:1–16. doi: 10.1895/wormbook.1.81.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri K, Ono K, Yu R, Yamashiro S, Ono S. Enhancement of actin-depolymerizing factor/cofilin-dependent actin disassembly by actin-interacting protein 1 is required for organized actin filament assembly in the Caenorhabditis elegans body wall muscle. Mol Biol Cell. 2006;17:2190–2199. doi: 10.1091/mbc.E05-11-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri K, Ono S. Actin filament disassembling activity of Caenorhabditis elegans actin-interacting protein 1 (UNC-78) is dependent on filament binding by a specific ADF/cofilin isoform. J Cell Sci. 2003;116:4107–4118. doi: 10.1242/jcs.00717. [DOI] [PubMed] [Google Scholar]
- Mohri K, Vorobiev S, Fedorov AA, Almo SC, Ono S. Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. J Biol Chem. 2004;279:31697–31707. doi: 10.1074/jbc.M403351200. [DOI] [PubMed] [Google Scholar]
- Moon AL, Janmey PA, Louie KA, Drubin DG. Cofilin is an essential component of the yeast cortical cytoskeleton. J Cell Biol. 1993;120:421–435. doi: 10.1083/jcb.120.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Obinata T, Abe H. XAIP1: a Xenopus homologue of yeast actin interacting protein 1 (AIP1), which induces disassembly of actin filaments cooperatively with ADF/cofilin family proteins. J Cell Sci. 1999;112:1553–1565. doi: 10.1242/jcs.112.10.1553. [DOI] [PubMed] [Google Scholar]
- Okada K, Ravi H, Smith EM, Goode BL. Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: A coordinated mechanism for severing and capping filaments. Mol Biol Cell. 2006;17:2855–2868. doi: 10.1091/mbc.E06-02-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics. 1993;135:385–404. doi: 10.1093/genetics/135.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V, Drubin DG. Loss of Aip1 reveals a role in maintaining the actin monomer pool and an in vivo oligomer assembly pathway. J Cell Biol. 2010;188:769–777. doi: 10.1083/jcb.200909176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Parast M, Alberico C, Benian GM, Ono S. Specific requirement for two ADF/cofilin isoforms in distinct actin-dependent processes in Caenorhabditis elegans. J Cell Sci. 2003;116:2073–2085. doi: 10.1242/jcs.00421. [DOI] [PubMed] [Google Scholar]
- Ono K, Yamashiro S, Ono S. Essential role of ADF/cofilin for assembly of contractile actin networks in the C. elegans somatic gonad. J Cell Sci. 2008;121:2662–2670. doi: 10.1242/jcs.034215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J Cell Biol. 2001;152:1313–1319. doi: 10.1083/jcb.152.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1: new blades for twisted filaments. Biochemistry. 2003;42:13363–13370. doi: 10.1021/bi034600x. [DOI] [PubMed] [Google Scholar]
- Ono S. Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int Rev Cytol. 2007;258:1–82. doi: 10.1016/S0074-7696(07)58001-0. [DOI] [PubMed] [Google Scholar]
- Ono S. Dynamic regulation of sarcomeric actin filaments in striated muscle. Cytoskeleton. 2010;67:677–692. doi: 10.1002/cm.20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Baillie DL, Benian GM. UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J Cell Biol. 1999;145:491–502. doi: 10.1083/jcb.145.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Benian GM. Two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins, encoded by the unc-60 gene, differentially regulate actin filament dynamics. J Biol Chem. 1998;273:3778–3783. doi: 10.1074/jbc.273.6.3778. [DOI] [PubMed] [Google Scholar]
- Ono S, McGough A, Pope BJ, Tolbert VT, Bui A, Pohl J, Benian GM, Gernert KM, Weeds AG. The C-terminal tail of UNC-60B (ADF/cofilin) is critical for maintaining its stable association with F-actin and is implicated in the second actin-binding site. J Biol Chem. 2001;276:5952–5958. doi: 10.1074/jbc.M007563200. [DOI] [PubMed] [Google Scholar]
- Ono S, Minami N, Abe H, Obinata T. Characterization of a novel cofilin isoform that is predominantly expressed in mammalian skeletal muscle. J Biol Chem. 1994;269:15280–15286. [PubMed] [Google Scholar]
- Ono S, Mohri K, Ono K. Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/cofilin-bound actin filaments. J Biol Chem. 2004;279:14207–14212. doi: 10.1074/jbc.M313418200. [DOI] [PubMed] [Google Scholar]
- Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156:1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren N, Charlton J, Adler PN. The flare gene, which encodes the AIP1 protein of Drosophila, functions to regulate F-actin disassembly in pupal epidermal cells. Genetics. 2007;176:2223–2234. doi: 10.1534/genetics.107.072959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond JE, Davis WS, Jorgensen EM. UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat Neurosci. 1999;2:959–964. doi: 10.1038/14755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal AA, Tetreault JW, Lappalainen P, Drubin DG, Amberg DC. Aip1p interacts with cofilin to disassemble actin filaments. J Cell Biol. 1999;145:1251–1264. doi: 10.1083/jcb.145.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorrer F, et al. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature. 2010;464:287–291. doi: 10.1038/nature08799. [DOI] [PubMed] [Google Scholar]
- Skwarek-Maruszewska A, Hotulainen P, Mattila PK, Lappalainen P. Contractility-dependent actin dynamics in cardiomyocyte sarcomeres. J Cell Sci. 2009;122:2119–2126. doi: 10.1242/jcs.046805. [DOI] [PubMed] [Google Scholar]
- Thacker C, Sheps JA, Rose AM. Caenorhabditis elegans dpy-5 is a cuticle procollagen processed by a proprotein convertase. Cell Mol Life Sci. 2006;63:1193–1204. doi: 10.1007/s00018-006-6012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirion C, Stucka R, Mendel B, Gruhler A, Jaksch M, Nowak KJ, Binz N, Laing NG, Lochmuller H. Characterization of human muscle type cofilin (CFL2) in normal and regenerating muscle. Eur J Biochem. 2001;268:3473–3482. doi: 10.1046/j.1432-1327.2001.02247.x. [DOI] [PubMed] [Google Scholar]
- Tischler J, Lehner B, Chen N, Fraser AG. Combinatorial RNA interference in Caenorhabditis elegans reveals that redundancy between gene duplicates can be maintained for more than 80 million years of evolution. Genome Biol. 2006;7:R69. doi: 10.1186/gb-2006-7-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Troys M, Huyck L, Leyman S, Dhaese S, Vandekerkhove J, Ampe C. Ins and outs of ADF/cofilin activity and regulation. Eur J Cell Biol. 2008;87:649–667. doi: 10.1016/j.ejcb.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Mustonen T, Mattila PK, Ojala PJ, Thesleff I, Partanen J, Lappalainen P. The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Mol Biol Cell. 2002;13:183–194. doi: 10.1091/mbc.01-07-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BD, Waterston RH. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S, Mohri K, Ono S. The two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins differently enhance actin filament severing and depolymerization. Biochemistry. 2005;44:14238–14247. doi: 10.1021/bi050933d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura I, Mabuchi I. Heterogeneity of mRNA coding for Caenorhabditis elegans coronin-like protein. Gene. 2001;271:255–259. doi: 10.1016/s0378-1119(01)00509-1. [DOI] [PubMed] [Google Scholar]
- Yu R, Ono S. Dual roles of tropomyosin as an F-actin stabilizer and a regulator of muscle contraction in Caenorhabditis elegans body wall muscle. Cell Motil Cytoskeleton. 2006;63:659–672. doi: 10.1002/cm.20152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.