The mechanism of collagen secretion is not completely understood. It is found that cTAGE5 binds to TANGO1, and it is suggested that collagen VII export from the ER is driven by a cTAGE5/TANGO1 complex.
Abstract
Cutaneous T-cell lymphoma-–associated antigen 5 (cTAGE5), an originally identified tumor antigen, is overexpressed in various cancer cell lines. The cDNA encodes an integral membrane protein containing two coiled-coil motifs and a proline-rich domain. We show that cTAGE5 specifically localizes to the endoplasmic reticulum (ER) exit sites. In addition, cTAGE5 forms a complex with TANGO1 (MIA3), a previously characterized cargo receptor for collagen VII, by the interaction of their coiled-coil motifs. Of interest, cTAGE5, as well as TANGO1, is capable of interacting with the inner-layer coatomer of COPII Sec23/24 complex through their C-terminal proline-rich domains and required for collagen VII secretion. We propose that cTAGE5 acts as a coreceptor of TANGO1 for collagen VII export from the ER.
INTRODUCTION
The newly synthesized secretory proteins exit the endoplasmic reticulum (ER) in coat protein II (COPII)–coated vesicles. The process of COPII-coated carrier formation is relatively well characterized (Lee et al., 2004). The activation of small GTPase Sar1 (Nakano and Muramatsu, 1989; Barlowe et al., 1993) by a guanine-nucleotide exchange factor, Sec12 (Nakano et al., 1988; Barlowe and Schekman, 1993), leads to the recruitment of inner-layer coatomers of COPII, Sec23/24 complex, to the membranes (Hicke et al., 1992). Subsequently, the outer-layer coatomer Sec13/31 complex binds and finishes the coat assembly (Salama et al., 1993). Recently, the importance of Sec16 in the organization of ER exit sites has been reported in several organisms, including Pichia partoris (Connerly et al., 2005; Bhattacharyya and Glick, 2007; Iinuma et al., 2007; Ivan et al., 2008).
In mammals, there are four isoforms of Sec24, and each seems to have a different capacity to interact with transmembrane cargoes or cargo receptors (Wendeler et al., 2007). Although several receptors have been identified, molecules recognized by these receptors are only the portions of diverse secretory proteins (Dancourt and Barlowe, 2010). It is unclear whether all secretory proteins need to be captured by receptors or bulk fluid flow can contribute to the selective transport (Thor et al., 2009; Dancourt and Barlowe, 2010). In this context, it is potentially important to identify new cargo receptors that are involved in the trafficking of different spectra of cargo molecules.
Special interest has been directed at the trafficking of cargoes such as procollagens and chylomicrons since these are too large to fit into conventional COPII-coated carriers (Fromme and Schekman, 2005). Recently, several molecules have been found to mediate macromolecule trafficking, such as Sec23A, Sec24D and Sec13, and structural models have also been proposed (Bi et al., 2007; Stagg et al., 2008; Jones et al., 2003; Boyadjiev et al., 2006; Townley et al., 2008; Lang et al., 2006; Fromme et al., 2007; Ohisa et al., 2010; Sarmah et al., 2010). However, the mechanisms of how the large cargoes can be packaged into COPII carriers are still largely unclear.
Genome-wide screening in Drosophila S2 cells led to the identification of several genes involved in transport and Golgi organization (TANGO) (Bard et al., 2006). Among them, we have recently characterized TANGO1—an integral membrane protein localized to the ER exit sites. TANGO1, also known as melanoma inhibitory activity 3 (MIA3), interacts with Sec23/24 by its C-terminal cytoplasmic side, as well as with collagen VII by the luminal SH3 domain. TANGO1 is required for collagen VII export from the ER, but it is not involved in the export of the other proteins from the ER. Thus TANGO1 appears to act as a cargo receptor for collagen VII, although this receptor itself does not seem to exit from the ER (Saito et al., 2009).
Cutaneous T-cell lymphoma–associated antigen 5 (cTAGE5), also known as meningioma-expressed antigen 6 (MEA6/MGEA6), is overexpressed in several cancer tissues, including T-cell lymphoma, meningioma, and melanoma, and thus is regarded as a tumor antigen candidate (Comtesse et al., 2002; Heckel et al., 1997; Usener et al., 2003). Here we present data that cTAGE5 localizes to the ER exit sites and interacts with TANGO1 to coordinate collagen export from the ER.
RESULTS
cTAGE5 localizes to the ER exit sites
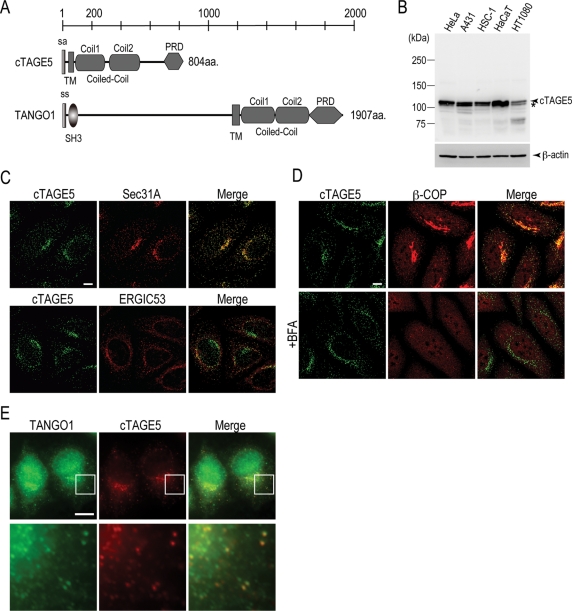
Domain search by database revealed that cTAGE5 consists of 804 amino acids containing a signal anchor motif, a single transmembrane domain, two coiled-coil motifs, and a proline-rich domain (PRD) from the N-terminus (Figure 1A). This domain organization is reminiscent of the C-terminal part of TANGO1. We raised polyclonal antibody against the first coiled-coil motif of cTAGE5 (anti–cTAGE5 CC1) and blotted the lysates from human cell lines, including HeLa, A431, HSC-1, HaCaT, and HT1080 cells, to detect the endogenous protein. The antibody recognized a doublet protein with the molecular weight of ∼110 kDa, of which the upper bands were predominant species in most of the cell lines tested (Figure 1B). Knockdown of cTAGE5 by RNA interference specifically reduced the upper bands; however, the lower bands remained unchanged (Figure 4B). These data suggest that the upper bands correspond to the cTAGE5-specific signal and the lower bands were nonspecific cross-reactive species.
FIGURE 1:
cTAGE5 localizes to the ER exit sites. (A) Schematic representation of human cTAGE5 and TANGO1 domain organization. Coiled-Coil, coiled-coil motif; PRD, proline-rich domain.; sa, signal anchor; ss, signal sequence; TM, transmembrane. (B) HeLa, A431, HSC-1, HaCaT, and HT1080 cells were collected and extracted with Triton X-100. Each cell lysate (70 μg) was resolved by SDS–PAGE and Western blotted with anti–cTAGE5 CC1 antibody. Each cell lysate (20 μg) was analyzed by SDS–PAGE and Western blotted with anti–β-actin antibody as a loading control. Asterisks indicate nonspecific signals. (C) HeLa cells were costained with anti–cTAGE5 CT antibody and either anti-Sec31A antibody or anti-ERGIC53 antibody. (D) HeLa cells were incubated with or without 5 μg/ml of BFA for 1 h at 37°C and stained with anti-cTAGE5 CT and β-COP antibodies. (E) Anti-TANGO1 antibody was directly conjugated with Alexa 488 dye. Anti–cTAGE5 CT antibody was conjugated with Alexa 555 dye. HeLa cells were fixed and stained with conjugated antibodies and analyzed by Zeiss Axio Imager M1 microscopy and processed with AxioVision software. Bottom panels show a magnified view of the boxed areas in the top panels. Bar, 10 μm.
FIGURE 4:
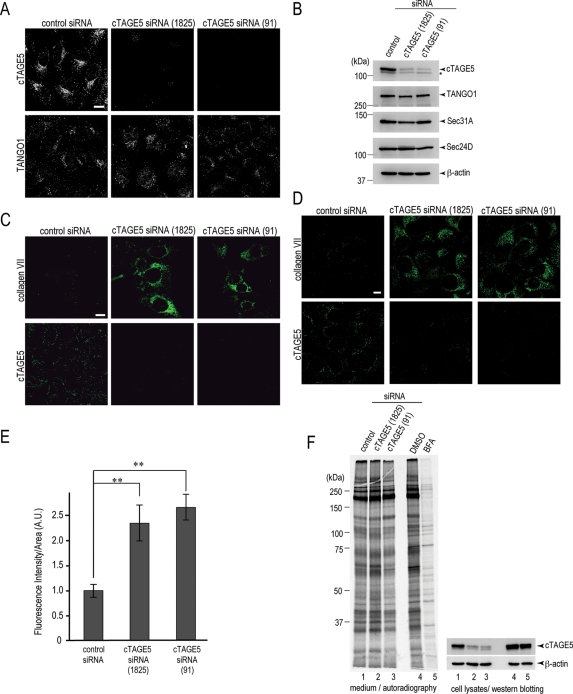
cTAGE5 is required for collagen VII secretion from the ER. (A) HeLa cells were transfected with control siRNA or cTAGE5 siRNA (1825) or cTAGE5 siRNA (91). After 70 h, the cells were fixed and stained with anti–cTAGE5 CT antibody and TANGO1 antibodies. Bars, 10 μm. (B) HeLa cells transfected with control siRNA or cTAGE5 siRNA (1825) or cTAGE5 siRNA (91) were extracted and subjected to SDS–PAGE, followed by Western blotting with anti–cTAGE5 CC1, TANGO1, Sec31A, Sec24D, and β-actin antibodies. Asterisks indicate nonspecific signals. (C) A431 cells were transfected with control siRNA or cTAGE5 siRNA and stained with anti–collagen VII or anti–cTAGE5 CT antibodies. (D) HSC-1 cells were transfected with control siRNA or cTAGE5 siRNA and stained with anti–collagen VII or anti–cTAGE5 CT antibodies. (E) Quantification of collagen VII immunofluorescence signal per cell in HSC-1 cells (A.U.). The detailed procedure is described in Materials and Methods. Error bars represent mean ± SEM; **p < 0.005 compared with control siRNA, n = 42. (F) HeLa cells were either transfected with control or cTAGE5 siRNA (1825) or cTAGE5 siRNA (91). After 46 h, cells were cultured with medium without methionine and cysteine for 1 h and then labeled with [35S]methionine for 15 min. Cells were washed and then chased for 3 h. Medium was collected and precipitated with TCA and subjected to SDS–PAGE, followed by autoradiography. Cell lysates were extracted and analyzed by SDS–PAGE, followed by Western blotting with anti–cTAGE5 CC1 and β-actin antibodies. BFA was added to the nontransfected cells and kept throughout the chase.
We next determined the localization of endogenous cTAGE5 in HeLa cells with another polyclonal antibody against the C-terminal (last 14 amino acids) of cTAGE5 (anti–cTAGE5 CT) since the former antibody was found to be unsuitable for immunofluorescence microscopy. cTAGE5 was visualized as punctuate dots scattered throughout the cytoplasm, and some accumulated at perinuclear regions (Figure 1C). These signals were reduced extensively by cTAGE5 knockdown, indicating that the antibody staining is specific (Figure 4A, top). Costaining with Sec31A suggests that cTAGE5 localizes to the ER exit sites (Figure 1C, top). The colocalization between ER-Golgi intermediate compartment 53 (ERGIC53) and cTAGE5 is less evident (Figure 1C, bottom). The localization of β-COP, which is a marker for COPI component, is separated from cTAGE5 (Figure 1D, top). To clarify the localization of cTAGE5 to the ER exit sites, the cells were treated with brefeldin A (BFA), which causes rapid dissociation of COPI components from the membranes (Orci et al., 1991). BFA treatment, as expected, relocalized COPI component (β-COP) from membranes to the cytoplasm (Figure 1D, bottom). cTAGE5 localization, however, was unchanged upon BFA treatment (Figure 1D, bottom). Taken together, these findings strongly indicate that cTAGE5 localizes to the ER exit sites.
Next we checked whether cTAGE5 and TANGO1 are localized at the same ER exit site. Because all of our antibodies available for cTAGE5 and TANGO1 are made in rabbits, it is difficult to immunostain by conventional methods. Besides, it is unsuitable to use overexpressed proteins for this study since overexpression renders both cTAGE5 and TANGO1 diffused throughout the ER even with the modestly expressed cells. To overcome this technical issue, we directly labeled antibodies with Alexa dyes. As shown in Figure 1E, some of the ER exit sites stained by Alexa 488–conjugated TANGO1 antibody could be also stained by Alexa 555–conjugated cTAGE5 CT antibody. Due to the significant background signals, however, we cannot argue for the existence of mutually exclusive ER exit sites. Nevertheless, these results indicate that at least in some conditions and/or limited areas, cTAGE5 and TANGO1 are present at the same ER exit sites.
cTAGE5 binds to TANGO1
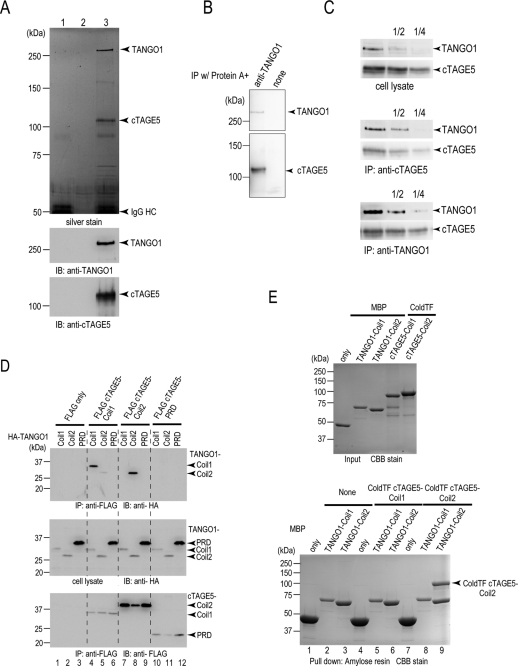
HeLa cell lysates were immunoprecipitated with anti-cTAGE5 antibody and analyzed by SDS–PAGE. As shown in Figure 2A, endogenous cTAGE5 was efficiently immunoprecipitated and detected as a 110-kDa band by both silver staining and Western blotting. Of interest, there was an additional band of upper 250 kDa in the immunoprecipitate of cTAGE5 antibody. The Western blot was reprobed with the anti-TANGO1 antibody, and the upper 250-kDa protein was identified as TANGO1 (Figure 2A). HeLa cell lysate was also immunoprecipitated with anti-TANGO1 antibody, and the immunoprecipitants were Western blotted with anti-cTAGE5 and anti-TANGO1 antibodies, respectively. The anti-TANGO1 immunoprecipitants was found to contain both TANGO1 and cTAGE5 (Figure 2B). These results strongly suggest that endogenous cTAGE5 forms a complex with TANGO1. We roughly estimated the efficiency of immunoprecipitation by Western blotting. As shown in Figure 2C, the immunoprecipitants of either anti-cTAGE5 antibody or anti-TANGO1 antibody were serially diluted and blotted with anti-cTAGE5 and anti-TANGO1 antibodies. As compared with cell lysates, the immunoprecipitants of cTAGE5 antibody contained a concentrated amount of TANGO1. On the contrary, the TANGO1 immunoprecipitants contained less cTAGE5 than did the cell lysates. These results would indicate that most of TANGO1 is in a complex with cTAGE5; however, there is a significant fraction of cTAGE5 free from the complex with TANGO1.
FIGURE 2:
cTAGE5 binds to TANGO1 at the ER exit sites. (A) Protein A beads conjugated with (lanes 1 and 3) or without (lane 2) anti–cTAGE5 CC1 antibody were incubated with (lanes 2 and 3) or without (lane 1) HeLa cell lysates. The beads were washed, and proteins retained to the beads were analyzed by SDS–PAGE, followed by silver staining or Western blotting with anti–cTAGE5 CC1 and TANGO1 antibodies. (B) Protein A beads were either untreated or conjugated with anti-TANGO1 antibody and then incubated with HeLa cell lysates. The beads were washed, and proteins retained to the beads were analyzed by SDS–PAGE, followed by Western blotting with anti–cTAGE5 CC1 and TANGO1 antibodies. (C) HeLa cell lysates were immunoprecipitated with anti–cTAGE5 CC1 antibody or anti-TANGO1 antibody, and immunoprecipitants and cell lysates were sequentially diluted and analyzed by SDS–PAGE and blotted with anti–cTAGE5 CC1 and anti-TANGO1 antibodies. (D) 293T cells were transfected with FLAG-tagged cTAGE5-Coil1 (amino acids 61–300), Coil2 (amino acids 301–650), or PRD (amino acids 651–804) with HA-tagged TANGO1-Coil1 (amino acids 1211–1440), Coil2 (amino acids 1441–1650), or PRD (amino acids 1651–1907). Cell lysates were immunoprecipitated with anti-FLAG antibody and eluted with FLAG peptide. Eluates and cell lysates were analyzed by SDS–PAGE, followed by Western blotting with anti-FLAG or anti-HA antibodies. (E) MBP, MBP-tagged TANGO1-Coil1, and TANGO1-Coil2 were expressed in E. coli and purified with Amylose resin. ColdTF-tagged cTAGE5-Coil1 and cTAGE5-Coil2 were expressed in E. coli and purified with Ni Sepharose. Purified proteins were analyzed by SDS–PAGE, followed by Coomassie brilliant blue (CBB) stain (top); MBP, MBP-tagged TANGO1-Coil1, and TANGO1-Coil2 were immobilized to amylose resin and untreated or incubated with ColdTF cTAGE5 Coil1 or ColdTF cTAGE5 Coil2. Resins were washed and eluted with maltose. Eluted proteins were subjected to SDS–PAGE followed by CBB stain (bottom).
Next we mapped the region responsible for this interaction. We made several deletion constructs and coexpressed a FLAG-tagged deletion of cTAGE5 with an HA-tagged TANGO1 deletion in 293T cells. The cell lysates were immunoprecipitated with anti-FLAG antibodies followed by elution with FLAG peptide. The N-terminal coiled-coil motif of cTAGE5, named cTAGE5-Coil1, interacted specifically with TANGO1-Coil1 (Figure 2D, lane 4). In addition, cTAGE5-Coil2 had the ability to interact with TANGO1-Coil2 (Figure 2D, lane 8). Of interest, there is specificity in the binding between coiled-coiled motifs: Coil1 binds Coil1, and Coil2 binds Coil2, of the two respective proteins (Figure 2D, lanes 4, 5, 7, and 8). In addition, proline-rich regions in cTAGE5 and TANGO1 are not involved in binding between these two proteins (Figure 2D, lanes 6, 9, and 10–12).
To further ascertain the specificity of the binding between coiled-coiled motifs of TANGO1 and cTAGE5, we expressed recombinant cTAGE5 and TANGO1 mutants in bacteria, purified the respective proteins (Figure 2E top), and tested their binding in vitro. Purified TANGO1 constructs were conjugated with beads by maltose-binding protein (MBP) epitope and incubated with cTAGE5 constructs. TANGO1-Coil2 efficiently bound to the similar amount of cTAGE5-Coil2 regions (Figure 2E bottom, lane 9), suggesting that the interaction between cTAGE5 and TANGO1 occurs stoichiometrically at the ratio of 1:1. It is intriguing that we could not observe any interaction between TANGO1-Coil1 and cTAGE5-Coil1 under the present conditions (Figure 2E, bottom, lane 5). This is not likely due to the steric hindrance raised by the comparatively huge tags that we introduced to these proteins because we still could not observe the interaction when using a tag-depleted version of cTAGE5 (unpublished data). Thus cTAGE5 and TANGO1 appear to interact directly at least via their second coiled-coil regions; their first coiled-coil regions may interact indirectly via unidentified intermediate(s).
The PRD of cTAGE5 binds Sec23/24
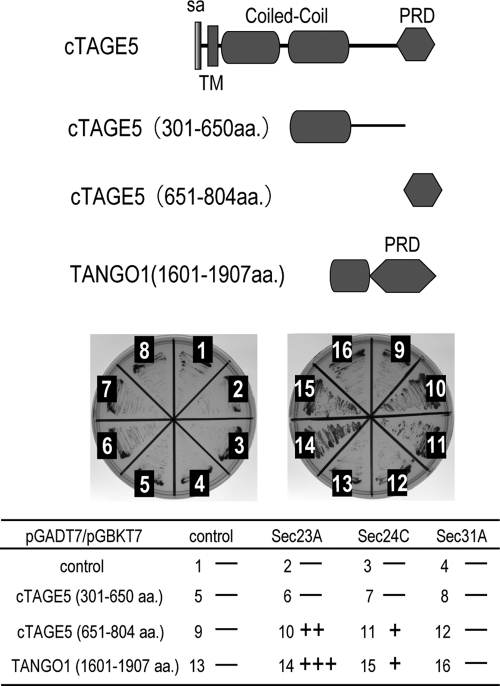
cTAGE5, as well as TANGO1, possesses a C-terminal proline-rich region. We reported previously that the C-terminus of TANGO1 interacts with Sec23/24 complex (Saito et al., 2009). Therefore we tested whether cTAGE5 can also bind Sec23/24 by yeast two-hybrid assay. As shown in Figure 3, the region of cTAGE5 amino acids 651–804 , corresponding to the proline-rich domain, interacts with both Sec23A and Sec24C (Figure 3, samples 10 and 11). Of interest, cTAGE5 interacted more potently with Sec23A than with Sec24C.
FIGURE 3:
cTAGE5 interacts with Sec23/24 through the C-terminal PRD domain. cTAGE5 and TANGO1 deletions in pGADT7 plasmids were cotransformed with pGBKT7 plasmids containing Sec23A, Sec24C, and Sec31A into AH109 yeast strains. Interactions were investigated by observing the cell growth on tryptophan-, leucine-, histidine-, and adenine-deficient plate.
cTAGE5 is required for collagen VII export from the ER
cTAGE5 knockdown was carried out by two distinct oligos, cTAGE5 (1825) and cTAGE5 (91). Both oligos efficiently reduced the expression of cTAGE5, as observed by Western blotting and immunofluorescence (Figure 4A, top, and B, top). The knockdown does not alter the expression level of other ER exit-site proteins, including TANGO1, Sec31A, and Sec24D (Figure 4B). Furthermore, there was no or, if any, minor effect on the localization of TANGO1 after siRNA treatment, and TANGO1 antibody could produce scattered punctuate staining, which is characteristic of ER exit sites (Figure 4A, bottom). These results assure that the phenotype observed by siRNA treatment is likely caused by the decrement of cTAGE5 expression level and not indirectly by the effects on other ER exit-site proteins, including TANGO1. Of interest, however, upon TANGO1 knockdown, cTAGE5 is less localized to the ER exit sites and presumably diffuses to the ER membrane (Supplemental Figure S2B), although the expression level of cTAGE5 is unaffected (Supplemental Figure S2A).
Because TANGO1 was characterized previously as a protein required for collagen VII export from the ER (Saito et al., 2009), we next tested whether cTAGE5 is also involved in this process. A431 cells were transfected with control or cTAGE5 siRNAs. After 48 h, the cells were stained with collagen VII antibody or cTAGE5 antibody. In control cells, we could detect faint staining of collagen VII inside the cells as previously described. On the other hand, a significant amount of collagen VII is accumulated within the ER in cTAGE5-knockdown cells (Figure 4C). A similar accumulation is also seen when cutaneous squamous carcinoma cell line, HSC-1, was subjected to cTAGE5 knockdown (Figure 4D). The quantification also suggests that immunofluorescent signals of collagen VII within HSC-1 cells were significantly increased upon cTAGE5 knockdown (Figure 4E). These results strongly suggest that, like TANGO1, cTAGE5 is also important for collagen VII export from the ER.
We next performed [35S]methionine pulse-chase assay to see the effect of cTAGE5 knockdown on general protein secretion. Cells were transfected with control or cTAGE5 siRNAs and further incubated with [35S]methionine for 15 min, followed by a chase for 3 h. The medium was concentrated with trichloroacetic acid (TCA) precipitation and analyzed by SDS–PAGE and autoradiography. As expected, incubation with BFA severely reduced the radiolabeled bands observed by autoradiography, suggesting that protein secretion is generally inhibited (Figure 4F, lanes 4 and 5). At the same condition, cTAGE5 knockdown did not affect overall protein secretion as compared with the control knockdown (Figure 4F, lanes 1–3). Vesicular stomatitis virus glycoprotein (VSVG) transport assay was also performed. VSVG seemed normally to traffic to the cell surface with cTAGE5 knockdown (Supplemental Figure S1). These results support the idea that cTAGE5 functions together with TANGO1 to specifically regulate collagen VII secretion.
DISCUSSION
In this study, we have identified cTAGE5 as another integral membrane protein at ER exit sites and characterized the protein as a direct binder of TANGO1. Furthermore, we have revealed that cTAGE5, as well as TANGO1, is required for collagen VII export from the ER. On the basis of the following observations, we would like to propose that cTAGE5 functions as a coreceptor of TANGO1 for collagen export. First, cTAGE5 and TANGO1 bind directly through their second coiled-coiled domain in a 1:1 stoichiometry (Figure 2). Second, the function of cTAGE5 seems not to be replaceable by TANGO1 since cTAGE5 knockdown induces collagen VII accumulation without affecting the localization and the expression of TANGO1 at the ER exit sites (Figure 4, A and B). Third, cTAGE5 does not possess the N-terminal long luminal stretch containing the SH3 domain, where collagen VII binds to TANGO1. Thus it is not likely that cTAGE5 exerts its effects on collagen VII by direct binding. Therefore cTAGE5 is not a functional homologue of TANGO1, but it rather coordinately regulates collagen VII secretion with TANGO1.
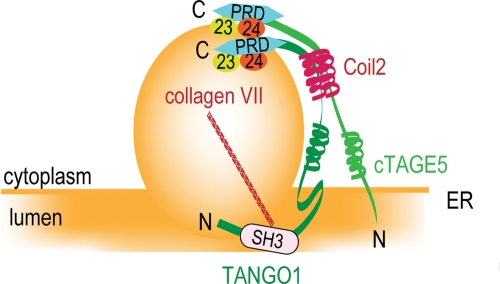
The interaction between cTAGE5 and TANGO1 is mediated via their coiled-coil domains (Figure 2, D and E). Furthermore, the C-terminus of cTAGE5 is also capable of interacting with Sec23/24 complex as in the case of TANGO1 (Figure 3). Taking these results together, we hypothesize that cTAGE5/TANGO1 complex forms a dimer, possibly shaped like the letter “y”, and that each cytoplasmic PRD of the complex interacts with Sec23/24, whereas the luminal trunk of TANGO1 interacts with cargoes such as collagen VII (Figure 5). In support of this notion, the cytoplasmic domain of cTAGE5 is almost the same length as that of TANGO1 (Figure 1A). Further investigation, particularly a structural approach, is definitely needed to validate this hypothesis.
FIGURE 5:
A dimeric model for the cTAGE5–TANGO1 complex. cTAGE5 interacts with TANGO1 through their second coiled-coil regions (Coil2). PRDs of both proteins interact with Sec23/24 complex on the COPII-coated carrier. Collagen VII bound to the SH3 domain of TANGO1 may be packaged into COPII-coated carrier by the coordinated action of cTAGE5 and TANGO1.
The immunofluorescence study suggests that cTAGE5 is localized together with TANGO1 at certain ER exit sites, supporting the complex formation as described. It is also interesting to note that estimation by immunoprecipitation implies that most of TANGO1 should be in a complex with cTAGE5; however, there would be a certain amount of cTAGE5, which is free from the complex with TANGO1. Thus there is a possibility that a certain amount of cTAGE5 acts by itself or with other oligomeric states. We should clarify this point in the future by further biochemical characterization, but we can speculate that the specificity of cargoes transported by the cTAGE5/TANGO1 complex might be regulated by the combination of these two proteins.
TANGO1 was originally identified as a protein required for protein transport by genome-wide screening in Drosophila S2 cells (Bard et al., 2006). The orthologue of TANGO1 can be found throughout metazoans, implying that TANGO1 would have a conserved roles over these species. On the contrary, we could find a cTAGE5 orthologue only from human to zebrafish but failed to identify the Drosophila counterpart. Thus the complex formation between cTAGE5 and TANGO1 may be preserved only through the vertebrates, and Drosophila may have a different structural entity for TANGO1 function. The fact that both cTAGE5 and TANGO1 are absent in the budding yeast strongly suggests that both proteins are not the minimal components required for COPII vesicle formation from the ER exit sites. Rather, these proteins might be modulators of COPII vesicle formation to coordinate the secretion of large cargoes such as collagens.
Recent reports indicated that the proteins such as Sec23A, Sec24D, Sec13, and TANGO1 are involved in the secretion of certain collagen types from the ER (Boyadjiev et al., 2006; Townley et al., 2008; Saito et al., 2009; Ohisa et al., 2010; Sarmah et al., 2010). Of interest, as for Sec13 and TANGO1, knockdown by RNAi does not impair bulk cargo secretion (Townley et al., 2008; Saito et al., 2009). In this study, we also observed similar phenotypes with cTAGE5 knockdown (see Figure 4). It is tempting to speculate that large-cargo secretion might be more tightly regulated through the coordinate action of ER exit-site proteins in higher eukaryotes. The present study has revealed that collagen VII export from the ER is driven by the cTAGE5/TANGO1 complex. The detailed molecular mechanisms of how this complex works for collagen VII secretion await further investigation.
MATERIALS AND METHODS
Antibodies
cTAGE5 polyclonal antibody for Western blotting (anti–cTAGE5 CC1) was raised in rabbits by immunization with recombinant glutathione S-transferase fusion of cTAGE5 fragment (amino acids 118–227). Raised antibody was affinity purified by the column conjugated with MBP fusion of cTAGE5 corresponding to the antigen. cTAGE5 polyclonal antibody for immunofluorescence study (anti–cTAGE5 CT) was raised in rabbits by immunization with keyhole limpet hemocyanin-conjugated peptide (C–NEPATEHPEPQQET) corresponding to the C-terminal 791–804 amino acids of cTAGE5. The antibody was affinity purified on column conjugated with the peptide (ThermoFisher Scientific, Waltham, MA). Other antibodies were used as described previously (Saito et al., 2009).
Immunoprecipitation and Western blotting
Cells extracted with extraction buffer consisting of 20 mM Tris-HCl (pH 7.4), 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, and protease inhibitors were centrifuged for 65,000 rpm for 30 min at 4°C. The cell lysate was immunoprecipitated with anti-cTAGE5 antibody conjugated with protein A Sepharose beads (GE Healthcare Bio-Sciences, Piscataway, NJ). The beads were washed with Tris-buffered saline (TBS)/0.1% Triton X-100 for five times and processed for sample preparation.
In vitro binding assay
MBP, MBP-tagged TANGO1-Coil1 (amino acids 1211–1440), and MBP-tagged TANGO1-Coil2 (amino acids 1440–1650) were expressed in Escherichia coli and purified with amylose resin. cTAGE5-deletion constructs corresponding to Coil1 (amino acids 61–300) and Coil2 (amino acids 301–650) were cloned into pColdTF vectors and purified with Ni Sepharose 6 Fast Flow (GE Healthcare). MBP fusion proteins were conjugated to amylose resin and incubated with cTAGE5 constructs. Beads were washed with TBS/0.1% Triton X-100 four times, followed by elution with maltose.
Pulse chase assay
Pulse chase assay was performed essentially as described previously (Saito et al., 2009). Control or cTAGE5 siRNA-treated or untreated HeLa cells were cultured in DMEM without l-methionine and l-cysteine for 1 h and then pulsed with 80 μCi of [35S]methionine for 15 min. Cells were washed and chased for 3 h in DMEM containing 10 mM cold methionine. Medium was collected and precipitated with TCA. The sample was resolved by SDS–PAGE, followed by autoradiography. For BFA-treated assay, 10 μg/ml of BFA was added throughout the experiments.
Immunofluorescence microscopy
Immunofluorescence microscopy was performed as described previously (Saito et al., 2009). Cells grown on cover slips were fixed with cold methanol. After blocking, cells were stained with primary antibody, followed by Alexa Fluor–conjugated secondary antibody (Invitrogen, Carlsbad, CA). Images were taken by Zeiss LSM700 confocal microscopy and processed with Zeiss Zen software (Zeiss, Jena, Germany).
Quantification of collagen VII staining
SiRNA-treated HSC-1 cells were fixed and incubated either with anti–collagen VII rabbit polyclonal antibody or rabbit control immunoglobulin G (IgG) or anti-cTAGE5 CT antibodies. Cells were washed and incubated with anti–rabbit IgG secondary antibody. Stained HSC-1 cells were analyzed by Zeiss Axio Imager M1 microscopy and processed with AxioVision software. Area calculation and intensity scanning were done by ImageJ software. The fluorescence intensity per area (A.U.) from collagen VII antibody is subtracted from that of rabbit control IgG.
siRNA oligos
Stealth select siRNAs for cTAGE5 were purchased from Invitrogen. The oligo sequences used were cTAGE5 siRNA (1825), CCGCCAGGACAAUCAUAUCCUGAUU, and cTAGE5 siRNA (91), GACCAGAUUCUAAUCUUUAUGGUUU. For control siRNA, Negative Universal Control Med #2 (Invitrogen) was used.
Cell culture and transfection
HeLa, A431, HSC-1, HaCaT, HT1080, and 293T cells were cultured in DMEM supplemented with 10% fetal bovine serum. Lipofectamine RNAiMAX (Invitrogen) was used for transfecting siRNA for HeLa and HSC-1 cells. For A431 cells, HiPerFect (Qiagen, Valencia, CA) was used. For plasmids transfection, Lipofectamine 2000 (Invitrogen) or FuGENE (Roche Diagnostics, Indianapolis, IN) was used.
Supplementary Material
Acknowledgments
We thank the members of the labs of V.M. and T.K. for valuable discussions. This work is supported in part by research grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (T.K.), the Japan Society for the Promotion of Science (K.S., T.K.), the Kanae Foundation for the Promotion of Medical Science (K.S.), the Astellas Foundation for Research on Metabolic Disorders (K.S.), the Cosmetology Research Foundation (K.S.), and the Mochida Memorial Foundation for Medical and Pharmaceutical Research (K.S.).
Abbreviations used:
- BFA
brefeldin A
- COP
coat protein
- ER
endoplasmic reticulum
- ERGIC
ER-Golgi intermediate compartment
- TCA
trichloroacetic acid
- VSVG
vesicular stomatitis virus glycoprotein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-02-0143) on April 27, 2011.
REFERENCES
- Bard F, et al. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature. 2006;439:604–607. doi: 10.1038/nature04377. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Schekman R. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 1993;365:347–349. doi: 10.1038/365347a0. [DOI] [PubMed] [Google Scholar]
- Barlowe C, d'Enfert C, Schekman R. Purification and characterization of SAR1p, a small GTP-binding protein required for transport vesicle formation from the endoplasmic reticulum. J Biol Chem. 1993;268:873–879. [PubMed] [Google Scholar]
- Bhattacharyya D, Glick BS. Two mammalian Sec16 homologues have nonredundant functions in endoplasmic reticulum (ER) export and transitional ER organization. Mol Biol Cell. 2007;18((3),):839–849. doi: 10.1091/mbc.E06-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Mancias JD, Goldberg J. Insights into COPII coat nucleation from the structure of Sec23Sar1 complexed with the active fragment of Sec31. Dev Cell. 2007;13:635–645. doi: 10.1016/j.devcel.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyadjiev SA, et al. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat Genet. 2006;38:1192–1197. doi: 10.1038/ng1876. [DOI] [PubMed] [Google Scholar]
- Comtesse N, Niedermayer I, Glass B, Heckel D, Maldener E, Nastainczyk W, Feiden W, Meese E. MGEA6 is tumor-specific overexpressed and frequently recognized by patient-serum antibodies. Oncogene. 2002;21:239–247. doi: 10.1038/sj.onc.1205005. [DOI] [PubMed] [Google Scholar]
- Connerly PL, Esaki M, Montegna EA, Strongin DE, Levi S, Soderholm J, Glick BS. Sec16 is a determinant of transitional ER organization. Curr Biol. 2005;15:1439–1447. doi: 10.1016/j.cub.2005.06.065. [DOI] [PubMed] [Google Scholar]
- Dancourt J, Barlowe C. Protein sorting receptors in the early secretory pathway. Annu Rev Biochem. 2010;79:777–802. doi: 10.1146/annurev-biochem-061608-091319. [DOI] [PubMed] [Google Scholar]
- Fromme JC, Ravazzola M, Hamamoto S, Al-Balwi M, Eyaid W, Boyadjiev SA, Cosson P, Schekman R, Orci L. The genetic basis of a craniofacial disease provides insight into COPII coat assembly. Dev Cell. 2007;13:623–634. doi: 10.1016/j.devcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromme JC, Schekman R. COPII-coated vesicles: flexible enough for large cargo? Curr Opin Cell Biol. 2005;17:345–352. doi: 10.1016/j.ceb.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Heckel D, Brass N, Fischer U, Blin N, Steudel I, Türeci O, Fackler O, Zang KD, Meese E. cDNA cloning and chromosomal mapping of a predicted coiled-coil proline-rich protein immunogenic in meningioma patients. Hum Mol Genet. 1997;6:2031–2041. doi: 10.1093/hmg/6.12.2031. [DOI] [PubMed] [Google Scholar]
- Hicke L, Yoshihisa T, Schekman R. Sec23p and a novel 105-kDa protein function as a multimeric complex to promote vesicle budding and protein transport from the endoplasmic reticulum. Mol Biol Cell. 1992;3:667–676. doi: 10.1091/mbc.3.6.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iinuma T, Shiga A, Nakamoto K, O'Brien MB, Aridor M, Arimitsu N, Tagaya M, Tani K. Mammalian Sec16/p250 plays a role in membrane traffic from the endoplasmic reticulum. J Biol Chem. 2007;282:17632–17639. doi: 10.1074/jbc.M611237200. [DOI] [PubMed] [Google Scholar]
- Ivan V, de Voer G, Xanthakis D, Spoorendonk KM, Kondylis V, Rabouille C. Drosophila Sec16 mediates the biogenesis of tER sites upstream of Sar1 through an arginine-rich motif. Mol Biol Cell. 2008;19(10):4352–4365. doi: 10.1091/mbc.E08-03-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, et al. Mutations in a Sar1 GTPase of COPII vesicles are associated with lipid absorption disorders. Nat Genet. 2003;34:29–31. doi: 10.1038/ng1145. [DOI] [PubMed] [Google Scholar]
- Lang MR, Lapierre LA, Frotscher M, Goldenring JR, Knapik EW. Secretory COPII coat component Sec23a is essential for craniofacial chondrocyte maturation. Nat Genet. 2006;38:1198–1203. doi: 10.1038/ng1880. [DOI] [PubMed] [Google Scholar]
- Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- Nakano A, Brada D, Schekman R. A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J Cell Biol. 1988;107:851–863. doi: 10.1083/jcb.107.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano A, Muramatsu M. A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J Cell Biol. 1989;109:2677–2691. doi: 10.1083/jcb.109.6.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohisa S, Inohaya K, Takano Y, Kudo A. sec24d encoding a component of COPII is essential for vertebra formation, revealed by the analysis of the medaka mutant, vbi. Dev Biol. 2010;342:85–95. doi: 10.1016/j.ydbio.2010.03.016. [DOI] [PubMed] [Google Scholar]
- Orci L, Tagaya M, Amherdt M, Perrelet A, Donaldson JG, Lippincott-Schwartz J, Klausner RD, Rothman JE. Brefeldin A, a drug that blocks secretion, prevents the assembly of non-clathrin-coated buds on Golgi cisternae. Cell. 1991;64:1183–1195. doi: 10.1016/0092-8674(91)90273-2. [DOI] [PubMed] [Google Scholar]
- Saito K, Chen M, Bard F, Chen S, Zhou H, Woodley D, Polischuk R, Schekman R, Malhotra V. TANGO1 facilitates cargo loading at endoplasmic reticulum exit sites. Cell. 2009;136:891–902. doi: 10.1016/j.cell.2008.12.025. [DOI] [PubMed] [Google Scholar]
- Salama NR, Yeung T, Schekman RW. The Sec13p complex and reconstitution of vesicle budding from the ER with purified cytosolic proteins. EMBO J. 1993;12:4073–4082. doi: 10.1002/j.1460-2075.1993.tb06091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmah S, Barrallo-Gimeno A, Melville DB, Topczewski J, Solnica-Krezel L, Knapik EW. Sec24D-dependent transport of extracellular matrix proteins is required for zebrafish skeletal morphogenesis. PloS One. 2010;5:e10367. doi: 10.1371/journal.pone.0010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg SM, LaPointe P, Razvi A, Gürkan C, Potter CS, Carragher B, Balch WE. Structural basis for cargo regulation of COPII coat assembly. Cell. 2008;134:474–484. doi: 10.1016/j.cell.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor F, Gautschi M, Geiger R, Helenius A. Bulk flow revisited: transport of a soluble protein in the secretory pathway. Traffic. 2009;10:1819–1830. doi: 10.1111/j.1600-0854.2009.00989.x. [DOI] [PubMed] [Google Scholar]
- Townley AK, Feng Y, Schmidt K, Carter DA, Porter R, Verkade P, Stephens DJ. Efficient coupling of Sec23-Sec24 to Sec13-Sec31 drives COPII-dependent collagen secretion and is essential for normal cranio-facial development. J Cell Sci. 2008;121:3025–3034. doi: 10.1242/jcs.031070. [DOI] [PubMed] [Google Scholar]
- Usener D, Schadendorf D, Koch J, Dübel S, Eichmüller S. cTAGE: a cutaneous T cell lymphoma associated antigen family with tumor-specific splicing. J Invest Dermatol. 2003;121:198–206. doi: 10.1046/j.1523-1747.2003.12318.x. [DOI] [PubMed] [Google Scholar]
- Wendeler MW, Paccaud JP, Hauri HP. Role of Sec24 isoforms in selective export of membrane proteins from the endoplasmic reticulum. EMBO Rep. 2007;8:258–264. doi: 10.1038/sj.embor.7400893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.