Our study reveals an alternative route in the SRP-dependent protein targeting pathway that includes a preassembled, membrane-bound SRP-SR complex. This alternative route is fully sufficient to maintain cell viability in the absence of a soluble SRP.
Abstract
Protein targeting by the signal recognition particle (SRP) and the bacterial SRP receptor FtsY requires a series of closely coordinated steps that monitor the presence of a substrate, the membrane, and a vacant translocon. Although the influence of substrate binding on FtsY-SRP complex formation is well documented, the contribution of the membrane is largely unknown. In the current study, we found that negatively charged phospholipids stimulate FtsY-SRP complex formation. Phospholipids act on a conserved positively charged amphipathic helix in FtsY and induce a conformational change that strongly enhances the FtsY-lipid interaction. This membrane-bound, signal sequence–independent FtsY-SRP complex is able to recruit RNCs to the membrane and to transfer them to the Sec translocon. Significantly, the same results were also observed with an artificial FtsY-SRP fusion protein, which was tethered to the membrane via a transmembrane domain. This indicates that substrate recognition by a soluble SRP is not essential for cotranslational targeting in Escherichia coli. Our findings reveal a remarkable flexibility of SRP-dependent protein targeting, as they indicate that substrate recognition can occur either in the cytosol via ribosome-bound SRP or at the membrane via a preassembled FtsY-SRP complex.
INTRODUCTION
Protein trafficking is an essential process for maintaining cellular integrity, and all living cells employ specific mechanisms for the delivery of proteins into subcellular compartments (Rapoport, 2007; Driessen and Nouwen, 2008; Cross et al., 2009). One such mechanism is the universally conserved cotranslational targeting by the signal recognition particle (SRP). In both eukaryotic and prokaryotic cells, SRP targets roughly 25–30% of newly synthesized proteins to distinct cellular compartments, i.e., to the eukaryotic endoplasmic reticulum (ER) or to the bacterial cytoplasmic membrane (Luirink et al., 1994; Koch et al., 2003; Bibi, 2010; Facey and Kuhn, 2010).
Cotranslational targeting is initiated by the early recognition of a substrate by ribosome-bound SRP (Powers and Walter, 1997; Neumann-Haefelin et al., 2000; Halic et al., 2006a; Schaffitzel et al., 2006; Grudnik et al., 2009; Janda et al., 2010; Hainzl et al., 2011) which then targets the ribosome-nascent chain (RNC) via the membrane-bound SRP receptor (SR) to the Sec translocon (Koch et al., 1999; Fulga et al., 2001; Halic et al., 2006b). After docking of the RNCs onto the Sec translocon (Cheng et al., 2005), the SRP-SR complex dissociates in a GTP-dependent reaction (Shan et al., 2009), and SRP can begin another targeting cycle. In bacteria like Escherichia coli, SRP-dependent targeting is achieved by just three components: the Ffh protein and the 4.5S RNA constitute the bacterial SRP (Poritz et al., 1990; Ribes et al., 1990), while FtsY, a homologue of the eukaryotic SRα subunit, serves as the bacterial SR (Luirink et al., 1994; Koch et al., 2003). The bacterial SRP mainly targets inner membrane proteins; secretory proteins are primarily targeted posttranslationally by the SecA pathway (Driessen and Nouwen, 2008). In contrast, eukaryotic cells employ the SRP pathway for the delivery of both secretory and membrane proteins to the ER (Rapoport, 2007; Cross et al., 2009).
Like many other cellular processes, SRP-dependent targeting requires a series of closely coordinated steps that monitor the presence of a correct substrate, the membrane, and a vacant translocon. Multiple studies have shown that this coordination is primarily achieved via subtle conformational rearrangements within the SRP-SR complex (Shan and Walter, 2005; Buskiewicz et al., 2009). The intrinsically slow complex formation between Ffh and FtsY is accelerated by the 4.5S RNA (Peluso et al., 2000, 2001; Jagath et al., 2001) and by the presence of RNCs, which expose a hydrophobic signal sequence (Bradshaw et al., 2009; Zhang et al., 2009). Recent data indicate that this acceleration is the result of a transient contact between the 4.5S RNA and FtsY upon RNC binding (Shen and Shan, 2010; Ataide et al., 2011). The presence of the substrate also increases the stability of the FtsY-SRP complex (Zhang et al., 2009), which otherwise would rapidly dissociate due to GTP hydrolysis, resulting in a half-life of <1 s in solution (Peluso et al., 2001). RNC-dependent stalling of the FtsY-SRP complex in a stable and GTPase-deficient conformation probably opens a time window for location of a vacant translocon (Zhang et al., 2009). Only upon binding to the translocon and release of the RNCs does the FtsY-SRP complex undergo additional conformational changes that finally lead to full GTPase activation, which then induces the dissociation of the complex. In agreement with this, RNCs, SRP, and SR have been shown to form a stable complex in the absence of the translocon (Song et al., 2000).
This highly ordered series of events provides the basis for the directionality of the targeting reaction and implements several proofreading steps. Nevertheless, some observations suggest a certain degree of flexibility within the targeting reaction. In mammalian cells, the large ribosomal subunit can stay attached to the ER membrane after completion of targeting (Adelman et al., 1973; Borgese et al., 1973), and thus does not need to be membrane targeted after reinitiation of translation (Seiser and Nicchitta, 2000; Potter et al., 2001). Efficient docking of these RNCs onto the Sec translocon probably still requires the SRP-SR complex, because SRP promotes RNC binding to the translocon in the presence of competing ribosomes (Neuhof et al., 1998; Raden and Gilmore, 1998; Schaletzky and Rapoport, 2006). In addition, SR appears to be required for sensing a vacant translocon (Helmers et al., 2003; Jiang et al., 2008). Thus SRP needs to recognize potential substrates not only in the cytosol but also at the membrane. In agreement with this, in eukaryotic cells ∼40% of SRP is membrane-localized, while the remaining SRP is either ribosome-associated (50%) or occurs in free form (Walter and Blobel, 1983). As a similar distribution has also been observed in bacteria (Koch et al., 1999), it appears that the recognition of substrates by membrane-bound SRP is a general feature of the SRP pathway. SRP does not seem to bind directly to the membrane, and therefore it is likely that SRP is bound to the membrane via its contact with FtsY/SR. This, however, would necessitate that the half-life of the SR-SRP complex be significantly increased not only by the presence of a substrate (Zhang et al., 2009) but, alternatively, also upon membrane contact. In agreement with this, a recent kinetic study has shown that phospholipids accelerate the formation of a stable FtsY-SRP complex (Lam et al. 2010).
In the current study, we have analyzed the significance of phospholipid-induced FtsY-SRP complex formation for cotranslational targeting. Our data demonstrate that the preformed, membrane-bound FtsY-SRP complex is able to recruit RNCs to the membrane and to subsequently transfer them to the Sec translocon. Furthermore, we show that the recognition of RNCs by cytosolic SRP is not essential for viability of E. coli. This discloses a remarkable flexibility of SRP-dependent protein targeting, because it shows that substrate recognition can occur either in the cytosol by ribosome-bound SRP or at the membrane by a preformed FtsY-SRP complex.
RESULTS
Negatively charged phospholipids stimulate FtsY-SRP complex formation
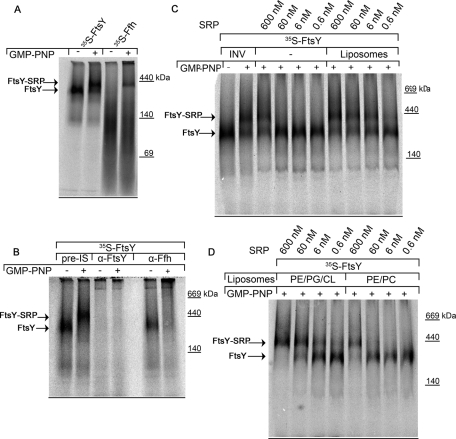
For understanding the impact of phospholipids on cotranslational targeting, we analyzed FtsY-SRP complex formation by Blue-native PAGE (BN-PAGE) analyses. Previous data had shown that in vitro synthesized FtsY (35S-FtsY) migrates at ∼250-kDa on BN-PAGE, but assembles into a 400-kDa complex in the presence of E. coli inner membrane vesicles (INV) and the nonhydrolyzable GTP analogue guanosine 5′(β,γ-imido) triphosphate (GMP-PNP; Angelini et al., 2006; Figure 1A). The 250-kDa band most likely corresponds to monomeric FtsY, which has been shown to migrate in size exclusion chromatography at ∼250 kDa, despite its predicted molecular mass of 56 kDa (Luirink et al., 1994). In a previous study, we were unable to localize Ffh in the 400-kDa complex by Western blotting (Angelini et al., 2006); however, when we synthesized Ffh in vitro (35S-Ffh) and incubated it in the presence of INV with GMP-PNP, we also observed a 400-kDa band on BN-PAGE (Figure 1A). This could indicate that the 400-kDa complex corresponds to an FtsY-SRP complex formed in the presence of membranes. This was confirmed by antibody-shift assays. In these assays, the composition of protein complexes is determined by pretreating them with antibodies before loading them on BN-PAGE (Boy and Koch, 2009). Antibodies that recognize specific components of the complex increase its molecular mass, resulting in a massive shift of the complex under the nondenaturing conditions of the BN-PAGE. In vitro synthesized FtsY was incubated with INV in the presence or absence of GMP-PNP and solubilized. Before the sample was loaded on BN-PAGE, the reaction mixture was further incubated with preimmune serum or antibodies against either FtsY or Ffh. After treatment with preimmune serum, the 250-kDa FtsY band was detectable in the absence of GMP-PNP and the 400-kDa complex was detectable in the presence of GMP-PNP (Figure 1B). Polyclonal FtsY antibodies recognized both bands and increased the molecular mass of the complexes so that they did not enter the separating gel of the BN-PAGE; this is expected when multiple epitopes are recognized by the antibodies. Treating the samples with polyclonal Ffh antibodies did not influence the migration of the 250-kDa FtsY band, but shifted the 400-kDa complex (Figure 1B). We also performed the antibody-shift assay with the 400-kDa complex observed with 35S-Ffh (see Figure 1A). Both Ffh and FtsY antibodies shifted the 400 kDa complex (Supplemental Figure S1). This further supports our conclusion that the 400-kDa complex corresponds to an FtsY-SRP complex formed in the presence of SRP- and FtsY-containing membranes and GMP-PNP.
FIGURE 1:
Anionic phospholipids stimulate FtsY-SRP complex formation. (A) In vitro synthesized FtsY and Ffh were affinity purified via metal-affinity chromatography and subsequently incubated with E. coli INV in the presence or absence of 2 mM GMP-PNP. After solubilization with DDM, the proteins were separated on a 5–15% BN-PAGE gel. (B) FtsY was incubated with INV in the presence and absence of GMP-PNP. The sample was then solubilized and incubated with preimmune serum or with the indicated antibodies and separated on a 5–10% BN-PAGE gel. (C) FtsY was incubated with either INV or buffer/liposomes together with the indicated amount of SRP. Proteins were solubilized and separated on a 5–10% BN-PAGE gel. (D) As for (C), but liposomes were prepared from synthetic lipids. PE/PG/CL (70, 25, and 5%, respectively) liposomes mimic the E. coli inner membrane lipid composition, PE/PC (65 and 35%, respectively) are zwitterionic phospholipids, which lead to the formation of neutral liposomes.
In our in vitro analyses, the formation of the 400-kDa FtsY-SRP complex on BN-PAGE was only observed in the presence of INV, which contain ∼200 nM SRP (Figure S1). Thus the in vitro analyses were performed at a final SRP concentration of ∼15–20 nM SRP. However, in previous studies, FtsY-SRP complex formation was also observed in the absence of INV (Jagath et al., 2000; Shan et al., 2004). We therefore wanted to analyze whether the SRP concentration required for observing complex formation was dependent on the presence of lipids. In vitro synthesized FtsY was incubated with different SRP concentrations in the presence or absence of liposomes. In the absence of liposomes, the 400-kDa FtsY-SRP complex was observed at high SRP concentrations (600 nM; Figure 1C), confirming the above-mentioned reports that the FtsY-SRP complex can be observed in the absence of lipids. However, reducing the SRP concentration below 600 nM prevented complex formation in the absence of liposomes. In contrast, complex formation was already observed at 6 nM SRP in the presence of liposomes (Figure 1C), demonstrating that lipids stimulate the formation of the FtsY-SRP complex. In these assays, complex formation required the addition of GMP-PNP for preventing GTPase-dependent dissociation. Although this clearly demonstrates that complex dissociation is much faster than our analyses, the data nevertheless reveal the strong effect of lipids on complex formation.
The strong stimulation of complex formation by phospholipids was further analyzed by comparing liposomes containing the endogenous lipid composition of the E. coli inner membrane (70% phosphatidylethanolamine [PE], 25% phosphatidylglycerol [PG], 5% cardiolipin [CL]) with liposomes containing only the neutral phospholipids (PE [65%]) and phosphatidylcholine (PC [35%]). Complex formation in the presence of neutral phospholipids was observed only at high SRP concentrations (600 nM; Figure 1D) and thus at SRP concentrations that also allowed a phospholipid-independent FtsY-SRP complex formation. This demonstrates that complex formation is stimulated only by negatively charged phospholipids like PG or CL and explains the important contribution of PG and CL to FtsY function, which has been observed in previous in vitro (Parlitz et al., 2007; Braig et al., 2009; Lam et al., 2010) and in vivo studies (Erez et al., 2010).
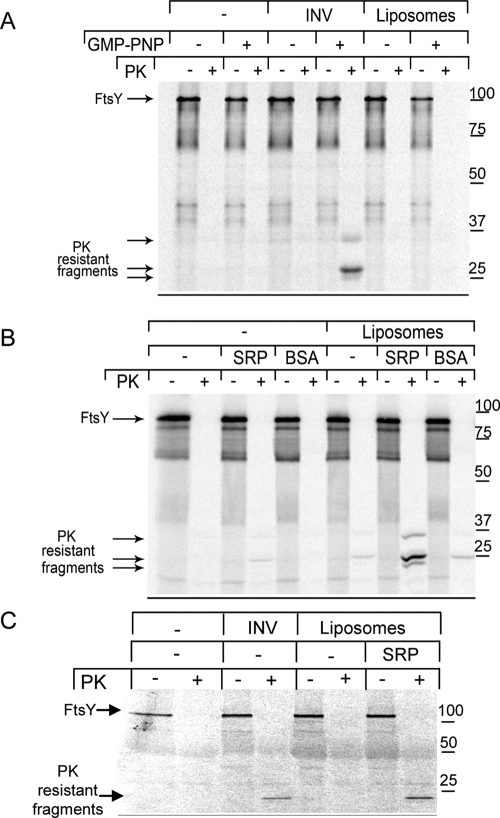
FtsY-SRP complex formation confers proteinase K resistance to FtsY
A recent flotation gradient analysis indicated that upon contacting SRP, FtsY binds more strongly to lipids (Lam et al., 2010). However, because FtsY exhibits strong lipid binding even in the absence of SRP (de Leeuw et al., 2000; Parlitz et al., 2007; Braig et al., 2009), we further tested the FtsY-lipid interaction by proteinase K (PK) protection assays. We have recently shown that after incubating FtsY with INV in the presence of GMP-PNP, two major protease-protected fragments of FtsY can be observed: a 33-kDa fragment, which over time is further proteolysed into two fragments running at 25 and 24 kDa (Angelini et al., 2006; Figure 2A). The 33-kDa fragment was shown to lack the complete A domain but to contain the complete NG domain (Angelini et al., 2006), which is responsible for SRP binding (Egea et al., 2004; Focia et al., 2004). Since the A domain is responsible for the anomalous migration of FtsY (Weiche et al., 2008), the size of the 33-kDa fragment is in agreement with the predicted molecular mass of the NG domain. The 25- and 24-kDa fragments correspond to C-terminally truncated NG domains (Angelini et al., 2006). Protease protection of FtsY was not observed in the presence of GMP-PNP and liposomes containing the endogenous lipid composition of the E. coli inner membrane (Figure 2A). This demonstrates that the FtsY–lipid contact is not sufficient to render FtsY PK resistant. We therefore analyzed whether the PK resistance of FtsY was dependent on SRP. In the presence of liposomes, SRP, and GMP-PNP, FtsY was largely protease protected (Figure 2B), but failed to become PK resistant in the absence of either liposomes or SRP. The addition of bovine serum albumin (BSA) had only a minor effect on PK protection of FtsY. This indicates that FtsY undergoes a conformational change upon interacting with SRP that protects the NG domain against PK cleavage. PK protection of FtsY was also observed when the in vitro synthesized FtsY was first purified via metal-affinity chromatography and then incubated with purified SRP and liposomes (Figure 2C). Thus the conformational change does not require the presence of the translocon and or the presence of ribosomes or RNCs.
FIGURE 2:
FtsY acquires a PK-resistant conformation upon interaction with SRP and lipids. (A) FtsY was in vitro synthesized and then incubated in the absence or presence of INV or liposomes with GMP-PNP (2 mM) or INV buffer and treated with PK (0.5 mg/ml for 20 min at 25°C). Samples were precipitated with trichloroacetic acid (TCA, 5% final concentration), separated on 13% SDS–PAGE, and visualized on a phosphorimager. (B) PK resistance was tested as in (A), but after preincubation with SRP (0.1 μM) or BSA (8 μM). (C) FtsY was in vitro synthesized and purified via metal-affinity chromatography before PK resistance testing.
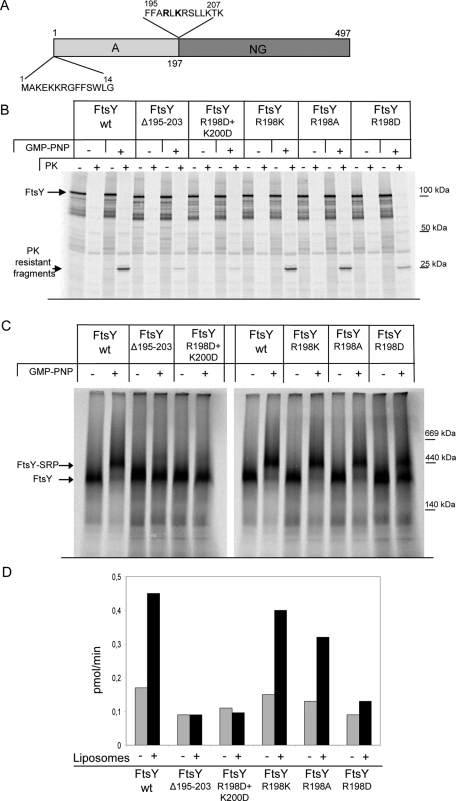
For correlating the PK-resistant state of FtsY (Figure 2) with the occurrence of the 400-kDa FtsY-SRP complex (Figure 1), we tested several FtsY mutants. FtsY contains two autonomous lipid-binding helices (Figure 3A; Parlitz et al., 2007; Weiche et al., 2008; Braig et al., 2009). One is located at the very N-terminus of FtsY, but is not essential for function, although deleting it reduces membrane binding of FtsY (Weiche et al., 2008; Braig et al., 2009). The second helix is located at the interface between the A domain and the conserved NG domain and is absolutely essential for FtsY function and, therefore, for cell viability (Bahari et al., 2007; Parlitz et al., 2007; Braig et al., 2009; Grudnik et al., 2009). When this second helix was deleted (FtsYΔ195–203), PK protection of FtsY was drastically reduced (Figure 3B) and a similar effect was observed when two conserved positively charged residues were replaced by negatively charged aspartate residues (R198D + K200D; Figure 3B). Replacing R198 with a positively charged lysine or a neutral alanine did not impair protease protection, but replacing it with aspartate drastically reduced protease protection (Figure 3B). The same pattern was observed when FtsY-SRP complex formation was determined by BN-PAGE analyses (Figure 3C): The 400-kDa complex was observed only for the FtsY(R198K) and the FtsY(R198A) mutants. Complex formation was significantly reduced with the R198D mutant, and no 400-kDa complexes were observed with the double-mutant FtsY(R198D+K200D) or for the FtsY mutant that completely lacked the second lipid-binding helix. Thus the 400-kDa complex and the PK-resistant state of FtsY most likely depict the same conformational organization of the FtsY-SRP complex.
FIGURE 3:
A positively charged helix of FtsY is crucial for complex formation with SRP. (A) Drawing showing the domain structure of E. coli FtsY. The localization of the two lipid-binding helices and their amino acid sequences are also shown. (B) Wild-type (wt) FtsY and FtsY derivatives carrying mutations within the second lipid-binding helix were in vitro synthesized and affinity purified via a C-terminal His tag. PK resistance of the mutants was analyzed in the presence of INV as described in Figure 2. (C) As in (B), but the samples were incubated with INV, solubilized, and separated on a 5–10% BN-PAGE gel. (D). The FtsY mutant proteins were expressed in E. coli C43 (DE3) and purified via metal-affinity purification. The GTPase activity of the FtsY mutants was analyzed in a 20 μl reaction mixture containing 0.5 μM FtsY. The reaction was started by the addition of GTP (200 μM GTP and 2.5 μM [γ-32P]-GTP). When indicated, 2 μl of liposomes (70% PE, 25% PG and 5% CL, as in Figure 1D) was added.
To exclude the possibility that the mutations/deletions had a more global effect on the FtsY structure, we determined the basal GTPase activity of the FtsY derivatives. In comparison to wild-type FtsY, the GTPase activity of the mutants was only slightly reduced (Figure 3D), demonstrating that GTP binding and hydrolysis was not significantly impaired by the mutations. Importantly, when we tested the effects of liposomes on the GTPase activity, we observed in agreement with previous reports (de Leeuw et al., 2000; Bahari et al., 2007) an increase for wild-type FtsY that was not observed for the deletion mutant or the double mutant (Figure 3D). Of the single mutants, only the R198K and R198A mutants displayed increased GTPase activity.
In summary, these mutant analyses highlight the importance of lipid binding for FtsY-SRP complex formation and demonstrate that the interaction between negatively charged lipids and the positively charged lipid-binding helix of FtsY stimulates complex formation. The data also explain why the lipid-binding helix at the interface of the A and NG domains of FtsY is universally conserved and essential for cotranslational targeting.
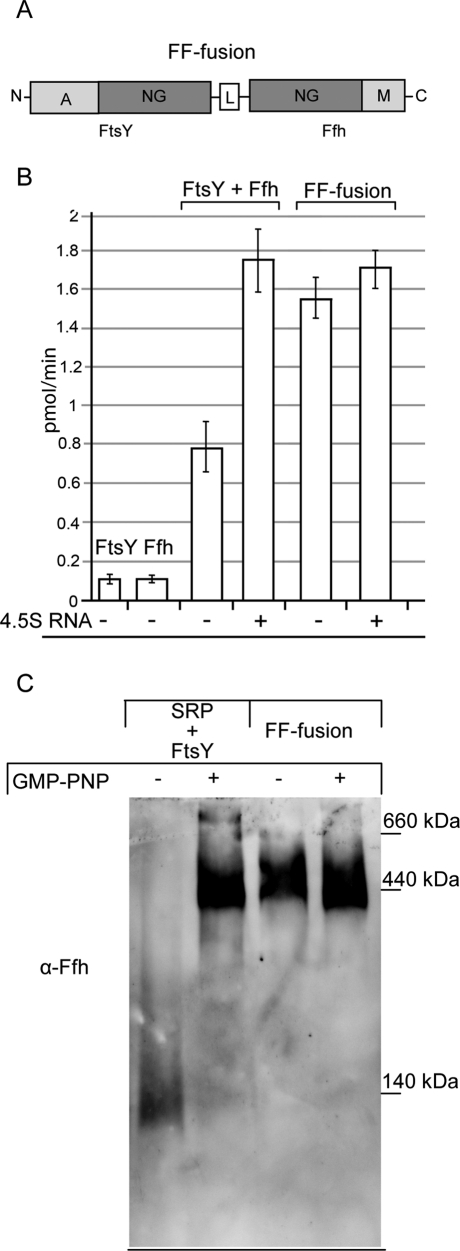
A FtsY-Ffh fusion protein forms a 400-kDa complex and is capable of GTP hydrolysis
The stimulatory effect of phospholipids on FtsY-SRP complex formation was comparable to the stimulation that occurs in solution when RNCs are added (Zhang et al., 2009). Whether the preformed, membrane-bound FtsY-SRP complex was capable of recruiting RNCs to the membrane and transferring them to the SecY translocon was analyzed by constructing an FtsY-Ffh fusion protein. In this construct, the respective NG domains of FtsY and Ffh were genetically linked by a 20-amino-acid-long flexible linker, and the corresponding fusion protein was termed FF-fusion (Figure 4A). During the course of our study, a similar FtsY-Ffh fusion protein was constructed for CryoEM studies (Estrozi et al., 2011) but not tested for its functionality and membrane binding.
FIGURE 4:
An FtsY-Ffh fusion protein forms a 400-kDa complex on BN-PAGE and hydrolyzes GTP. (A) The FtsY-Ffh-fusion protein (FF-fusion) was constructed by genetically fusing the C-terminal end of the FtsY NG domain to the N-terminal end of the Ffh NG domain via a 20-amino-acid-long, flexible linker (L). Label A indicates the A domain of FtsY and label M the signal sequence–binding domain of Ffh. (B) The GTPase activity of the FF-fusion was analyzed in a 20 μl reaction mixture containing 0.05 μM FF-fusion. The reaction was started by the addition of GTP (200 μM GTP and 2.5 μM [γ-32P]-GTP). FtsY and Ffh were also used at a final concentration of 0.05 μM each and, when indicated, 0.5 μg 4.5S RNA was added. The mean values and SD of at least three independent experiments are shown. (C) 4 μM FtsY was incubated with 1.5 μM Ffh and 0.1 mg/ml 4.5S RNA in the presence or absence of 2 mM GMP-PNP; alternatively, 1.5 μM of the FtsY-Ffh fusion protein was incubated with 0.1 mg/ml 4.5S RNA in the presence or absence of GMP-PNP and then separated on a 5–15% BN-PAGE gel. After transfer to a nitrocellulose membrane, the protein complexes were detected by α-Ffh antibodies.
For determining the properties of the FF-fusion, we first analyzed whether the purified FF-fusion would be able to bind and hydrolyze GTP. FtsY and Ffh display only low intrinsic GTPase activities, which are significantly stimulated if both proteins interact in the presence of the 4.5S RNA. When purified Ffh and FtsY were incubated together, we observed significant GTP hydrolysis, which was not observed for the individual proteins (Figure 4B). The addition of 4.5S RNA further stimulated GTP hydrolysis, which is in agreement with the previous observation that 4.5S RNA stabilizes the FtsY-SRP complex formation (Neher et al., 2008; Shen and Shan, 2010; Ataide et al., 2011). The FF-fusion displayed a GTPase activity that was comparable to the GTP hydrolysis measured in the presence of FtsY, Ffh, and 4.5S RNA, and was only weakly stimulated by the 4.5S RNA; this would be expected if 4.5S RNA is required for stabilizing the FtsY-SRP complex (Shen and Shan, 2010).
We next tested whether the purified FF-fusion would be detectable as a 400-kDa complex on BN-PAGE. When micromolar amounts of purified FtsY and purified SRP were incubated in the presence of GMP-PNP, the 400-kDa FtsY-SRP complex was easily detectable on BN-PAGE by Western blotting using α-Ffh antibodies (Figure 4C). The use of purified components also demonstrated that the 400-kDa complex does not contain stoichiometric amounts of any other component and the reduced migration on BN-PAGE is most likely due to the well-documented aberrant migration behavior of FtsY (Luirink et al., 1994; de Leeuw et al., 2000; Weiche et al., 2008). In addition, the presence of the SRP-RNA and binding of detergent and Coomassie further reduces the mobility on BN-PAGE (Kulajta et al., 2006). Because SRP and FtsY were present in concentrations above 600 nM, complex formation in this assay did not require the presence of lipids. The very same 400-kDa complex was detected when the purified FF-fusion was separated on BN-PAGE and analyzed by Western blotting with α-Ffh antibodies (Figure 4C). We did not detect dimerization of the FF-fusion in the presence of GMP-PNP, which suggested that the FtsY and Ffh parts of the FF-fusion interacted primarily with each other and not with their cognate partner on a second FF-fusion molecule. In summary, these data further verify that the 400-kDa complex represents an FtsY-SRP complex.
The preformed FtsY-SRP complex is functional in cotranslational targeting
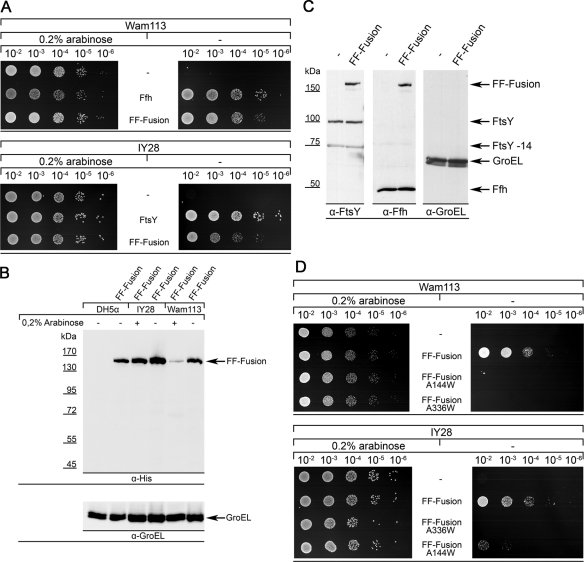
The functionality of the FF-fusion was tested in vivo by expressing it in the conditional E. coli Ffh mutant Wam113 (Phillips and Silhavy, 1992), which expresses the endogenous Ffh under the arabinose promoter. In the absence of arabinose this strain is not viable (Figure 5A), however, expressing either wild-type Ffh or the FF-fusion protein in this strain allowed cell growth even in the absence of arabinose. This demonstrates that the Ffh part of the FF-fusion protein is functional. Performing the same experiment with the conditional E. coli FtsY mutant strain IY28 (Bürk et al., 2009; Erez et al., 2010) also demonstrated that the FtsY part within the fusion protein is functional.
FIGURE 5:
The FtsY-Ffh fusion protein is functional in vivo. (A) Plasmid-borne copies of FtsY, Ffh, or of the FF-fusion protein were expressed in the conditional Ffh mutant WAM113 or the conditional FtsY mutant strain IY28. In both strains, the respective gene is under the control of the arabinose promoter and growth requires the addition of 0.2% arabinose. Overnight cultures were grown in LB medium in the presence of arabinose to an OD600 of 1.0, and serially diluted on LB plates containing either arabinose or fructose. The complementation did not require the presence of IPTG, which demonstrates that the basal expression level is sufficient. (B) Western blot analyses of the FF-fusion expressed in either wild-type DH5α cells or in IY28 and WAM113 cells, grown either in the presence of arabinose or fructose. Antibodies against the C-terminal His tag of the FF-fusion revealed no detectable cleavage of the fusion protein. As control, the same samples were also analyzed for the presence of the bacterial Hsp60 protein GroEL. (C) The expression level of the FF-fusion in wild-type DH5α cells was compared with the endogenous FtsY/Ffh content by Western blotting using antibodies against FtsY and Ffh. FtsY-14 corresponds to an N-terminally truncated FtsY-derivative, which lacks the first 14 amino acids. Note that FtsY has a predicted molecular mass of 56 kDa but runs at ∼100-kDa on SDS–PAGE due to its highly charged N-terminal A domain. As control, antibodies against GroEL were used. (D) For excluding the possibility that the functionality of the FF-fusion is due to proteolytic cleavage within the linker region, two FF-fusion derivatives were constructed that contained either an inactive Ffh-part (FF-fusion A144W) or an inactive FtsY part (FF-fusion A336W). The complementation assay was performed as in (A).
In principle, the FF-fusion could support cotranslational targeting in vivo if it were proteolytically cleaved into the individual proteins at the linker region. However, in both the absence and the presence of arabinose, we detected by Western blotting using antibodies against the C-terminal His tag of the FF-fusion only the full-size FF-fusion in wild-type DH5α, in IY28, and in WAM113 cells (Figure 5B). The amount of the bacterial Hsp60 protein GroEL was determined as a loading control in these experiments. We noticed for this series of experiments that the expression of the FF-fusion was lower when the endogenous FtsY and, in particular, the endogenous Ffh were present (Figure 5B, arabinose-containing cells). However, the reasons for this observation were not further analyzed in this study. To exclude the possibility that the in vivo functionality of the FF-fusion was due to unphysiologically high expression, we compared its expression in wild-type cells with the endogenous FtsY and Ffh levels (Figure 5C). In wild-type cells, FtsY is present as full-size FtsY running at ∼100 kDa and as an N-terminally truncated version running at 75 kDa (FtsY-14; Luirink et al., 1994; Weiche et al., 2008). Western blotting revealed that the expression of the FF-fusion was comparable to the endogenous FtsY level, and that there was also no significant difference in the endogenous Ffh content. GroEL was analyzed as the control in these experiments. These results demonstrate that the in vivo complementation activity of the FF-fusion was not the result of a higher expression or a significant proteolytic cleavage of the FF-fusion into two functional proteins.
Nevertheless, cleavage of even a small portion of the FF-fusion could be sufficient for supporting growth of WAM113 or IY28. It was also possible that the FF-fusion was functional because the Ffh part interacted with the endogenous FtsY in Wam113 and the FtsY part with the endogenous Ffh in IY28. To exclude both possibilities, we constructed two additional FF-fusions, in which either the FtsY or the Ffh part was inactivated due to single amino acid substitutions. We chose the A144W mutation in the GTPase domain of Ffh and the A336W mutation in FtsY, because both mutations had been shown to inactivate Ffh or FtsY, respectively (Shan et al., 2007). The rationale of this experiment was that if the in vivo complementation was due to cleavage or due to interaction with endogenous FtsY or Ffh in the corresponding strains, then FF-fusion (A144W) should still complement the FtsY-depletion strain IY28, because the FtsY part in the fusion was unaltered. Likewise, the FF-fusion (A336W) should complement the Ffh-depletion strain WAM113. However, both half-inactivated FF-fusions failed to complement WAM113 or IY28 (Figure 5D), which demonstrates that the FF-fusion is not cleaved and that the Ffh and FtsY parts of the FF-fusion interacted exclusively with each other.
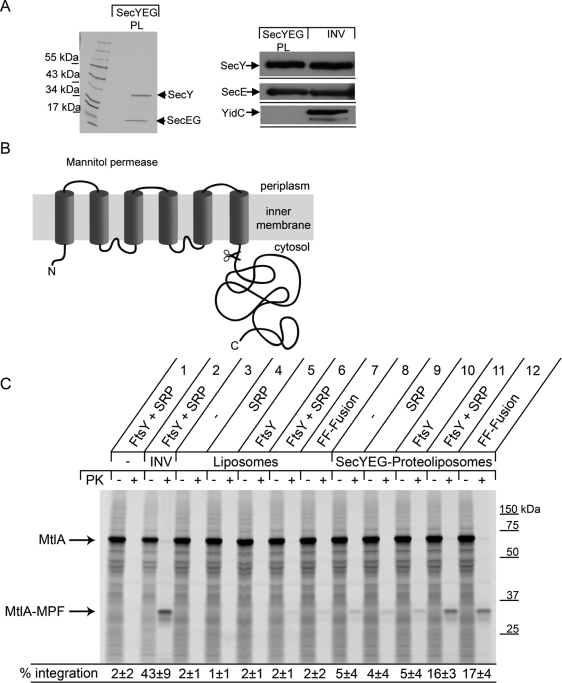
The in vivo complementation data strongly indicate that a preformed FtsY-SRP complex is capable of recruiting RNCs and transferring them to the translocon. This was further verified by an in vitro transport assay using SecYEG proteoliposomes. The purity of the components used in these assays was first determined by Coomassie staining and Western blotting (Figure 6A), which revealed no major contaminating band in the purified protein preparations. In particular, this completely reconstituted transport assay contained no FtsY or SRP, unless added (Koch et al., 1999), and also no YidC, which most likely cooperates with the SecYEG translocon to facilitate folding of membrane proteins (Facey and Kuhn, 2010). We tested the integration of the polytopic membrane protein mannitol permease (MtlA), a typical E. coli SRP substrate (Koch et al., 1999). When MtlA was synthesized in vitro in the presence of INV, we observed a membrane-protected fragment (MtlA-MPF) after PK treatment, which corresponds to the membrane-integral part of MtlA (Figure 6, B and C [lane 2]). In the absence of INV, MtlA was completely degraded by PK (Figure 6C, lane 1). When MtlA was synthesized in vitro in the presence of liposomes, it was also completely degraded by PK, independently of whether the individual components of the SRP pathway or the FtsY-Ffh fusion protein were present (Figure 6C, lanes 3–7). In SecYEG-proteoliposomes, we observed a weak background integration even in the absence of the SRP-components (Figure 6C, lane 8), which is expected considering the intrinsic affinity of the ribosome for the SecYEG translocon (Prinz et al., 2000). The addition of either SRP or FtsY did not stimulate MtlA integration in SecYEG proteoliposomes (Figure 6C, lanes 9 and 10). However, when FtsY and SRP were added simultaneously, we observed a significant stimulation of MtlA integration (Figure 6C, lane 11), and importantly, the same stimulation was also detected by adding the FtsY-Ffh-fusion protein (Figure 6C, lane 12). This demonstrates that a preformed FtsY-SRP complex is capable of recruiting RNCs to the membrane and transferring them to the SecYEG translocon. These data also show that MtlA integration does not strictly require the presence of YidC; however, it is important to note that the SecYEG content in our proteoliposomes is higher than the SecYEG content in INV (see Figure 6A). Whether a coreconstitution of YidC and SecYEG would further increase MtlA integration was not further analyzed in this study.
FIGURE 6:
The FtsY-Ffh fusion protein is capable of targeting ribosome nascent chains to the SecYEG translocon in a reconstituted in vitro transport system. (A) Coomassie staining and immune detection of SecYEG-proteoliposomes. SecYEG proteoliposomes were loaded for Coomassie staining (4 μl) and immune detection (0.2 μl). INV (3 μl) served as control. SecE and SecG have approximately the same size and are not well separated; in addition, Coomassie Brilliant Blue hardly stains SecG. (B) The SRP-dependent polytopic membrane protein MtlA consists of six transmembrane domains and a long C-terminal cytoplasmic domain, which is cleaved off by PK treatment (scissors), leaving behind a membrane-protected 30-kDa fragment (MtlA-MPF) that corresponds to the membrane-integral part of MtlA. (C) MtlA was in vitro synthesized in an E. coli transcription–translation system that does not contain FtsY, SRP, or INV (Koch et al., 1999) unless added. One-half of the reaction mixture was subsequently precipitated with TCA, whereas the second half was further digested with PK. MtlA was visualized by autoradiography after separation on a 13% SDS–polyacrylamide gel. INV correspond to E. coli inner membrane vesicles. The lipid composition of liposomes reflects the E. coli cytoplasmic membrane. SecYEG-proteoliposomes were generated as described in Material and Methods. When indicated, INV (0.5 μl), liposomes (4 μl), SecYEG proteoliposomes (4 μl), FtsY (1 μg), SRP (0.4 μg Ffh + 0.7 μg 4.5S RNA), or FF-fusion (1.5 μg FF-fusion + 0.7 μg 4.5S RNA) was added. The integration rate was calculated considering the loss of methionines for the MtlA-MPF and is the mean value of at least three independent experiments. The SD is indicated.
Substrate recognition by a membrane-bound FtsY-Ffh fusion
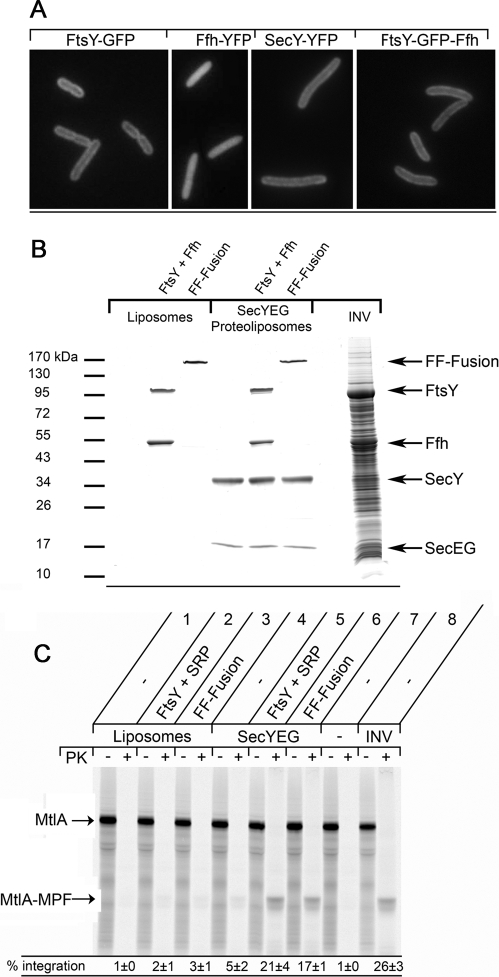
In the in vitro and in vivo assays described above, we cannot exclude that the FF-fusion shuttles between the membrane and the cytosol, that is, it binds to RNCs in the cytosol and only then targets them to the membrane. We therefore analyzed the cellular localization of the FF-fusion in living E. coli cells by fluorescence microscopy using a green fluorescent protein (GFP)-tagged FF-fusion. Fluorescently labeled FtsY has been shown to be predominantly membrane-localized in vivo (Mircheva et al., 2009), which we also show here (Figure 7A). The localization of FtsY-GFP was basically identical to the localization of the integral membrane protein SecY-YFP. In comparison, fluorescently labeled Ffh was evenly partitioned between the membrane and the cytosol (Figure 7A), supporting previous biochemical assays that have localized ∼40% of SRP to the membrane and ∼60% to the cytosol (Walter and Blobel, 1983; Koch et al., 1999). Importantly, the GFP-tagged FF-fusion, like FtsY, was predominantly membrane-localized (Figure 7A), suggesting that the FF-fusion functions at the membrane in vivo. The predominant membrane localization of the FF-fusion was observed in all E. coli cells, independent of the growth phase.
FIGURE 7:
The FF-fusion protein functions exclusively at the membrane. (A) The localization of the FF-fusion was analyzed in vivo by fluorescence microscopy and compared with fluorescently labeled FtsY, Ffh, and SecY. FtsY, Ffh, and SecY were C-terminally fused to either GFP or yellow fluorescent protein (YFP). For labeling of the FF-fusion, GFP was inserted into the linker region that connects FtsY and Ffh (see Figure 4A). (B) Liposomes or SecYEG-proteoliposomes were preincubated with FtsY/Ffh or FF-fusion in the presence of 4.5S RNA, isolated by centrifugation, and resuspended in buffer A (50 mM triethanolamine acetate, pH 8.0; 250 mM sucrose); for concentrations see legend to Figure 6. The resuspended liposomes/proteoliposomes were separated on a 5–20% SDS–polyacrylamide gel and stained with Coomassie Brilliant Blue. (C) The preincubated liposomes/proteoliposomes shown in (B) were used for MtlA in vitro transport assays as described in Figure 6. INV were used as control.
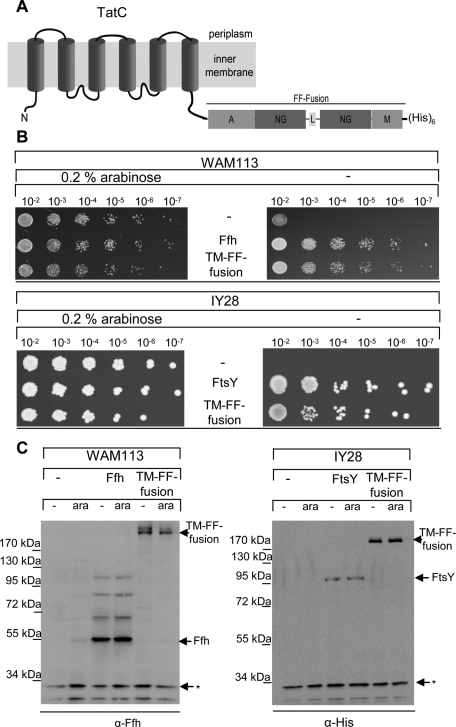
However, in the GFP construct, the small linker connecting FtsY and Ffh was replaced by the larger GFP moiety, and it was therefore possible that the presence of GFP favored membrane localization. We therefore used additional assays for demonstrating that the membrane-bound FF-fusion was functional in targeting. In a first assay, SecYEG proteoliposomes were preincubated with either FtsY/Ffh or the FF-fusion and isolated by centrifugation. Coomassie staining revealed that the FtsY/Ffh and the FF-fusion were efficiently bound to both liposomes and SecYEG-proteoliposomes (Figure 7B). Using these preincubated liposomes/proteoliposomes for in vitro transport assays demonstrated that MtlA was efficiently integrated into SecYEG proteoliposomes that only contained the membrane-bound FF-fusion (Figure 7C). Finally, we created a membrane-integral FF-fusion by attaching it to the C-terminus of TatC, a polytopic membrane protein containing six transmembrane domains (Figure 8A). The resulting TM-FF-fusion was like the FF-fusion capable of complementing WAM113 and IY28 cells (Figure 8B). We tested with Western blotting whether the complementation was due to a proteolytic release of the FF-fusion from the membrane. In WAM113 cells expressing the TM-FF-fusion, α-Ffh antibodies detected only the full-length TM-FF-fusion, but no cleavage product (Figure 8C). We also observed no cleavage in IY28 cells expressing the TM-FF-fusion (Figure 8C). However, here we used antibodies against the C-terminal His tag, because the low specificity of the polyclonal α-FtsY antibody prevented its use for whole cells. In summary, these data demonstrate that the membrane-bound FtsY-Ffh complex is sufficient for cotranslational targeting in E. coli and that soluble SRP is not absolutely required.
FIGURE 8:
A membrane-integral FtsY-Ffh-complex is functional. (A) The FF-fusion was fused to the C-terminus of the integral membrane protein TatC to generate TM-FF-fusion. (B) The functionality of the TM-FF-fusion was determined by complementation assays as described in Figure 5. (C) Expression and integrity of the TM-FF-fusion was analyzed by Western blotting using antibodies against Ffh or against the C-terminal His tag of the TM-FF-fusion. IY28 or WAM113 cells (approx. 0.5 × 108 cells) grown in the absence (−) or presence (ara) of arabinose and expressing the indicated constructs were directly precipitated by TCA (10% final concentration) and separated on 5–15% SDS–PAGE. The band running below the 34-kDa marker band, which is labeled with (*), was unspecifically recognized by both antibodies and served as an internal loading control.
DISCUSSION
Phospholipids do not merely provide a passive scaffold for membrane-integral or membrane-associated proteins, but they also determine the topology of membrane proteins (Bogdanov et al., 2008; Romantsov et al., 2008) and influence their stability. This has been shown for respiratory complexes (Wenz et al., 2009) and for the SecYEG translocon (Gold et al., 2010), which both require the presence of CL. This recent observation underscores the important link between phospholipids and protein transport, initially observed for SecA, which is stimulated by negatively charged phospholipids (de Vrije et al., 1988; Lill et al., 1990). Negatively charged phospholipids are also important for SRP-dependent protein targeting, because they provide a binding site for FtsY (de Leeuw et al., 2000; Parlitz et al., 2007; Braig et al., 2009).
Whether the contribution of phospholipids to SRP-dependent protein targeting goes beyond providing a binding site for FtsY was unknown. It was recently shown that negatively charged phospholipids activate FtsY, which subsequently accelerates FtsY-SRP complex formation (Lam et al. 2010). However, whether a phospholipid-induced FtsY-SRP complex is able to recruit RNCs to the membrane and to transfer them to the SecYEG translocon was not addressed in that study. Here we confirm the substrate-independent, phospholipid-induced FtsY-SRP complex formation by using a different experimental setup, and we also demonstrate that the preformed FtsY-SRP complex at the membrane is functional in cotranslational targeting both in vivo and in vitro. This reveals an unexpected plasticity of the SRP-dependent protein targeting, because it demonstrates that substrate recognition can occur either in the cytosol by ribosome-bound SRP or by a preassembled FtsY-SRP complex at the membrane.
This stimulatory effect of phospholipids on FtsY-SRP complex formation is similar to the stimulatory effect of RNCs on complex formation in solution (Zhang et al., 2009). It has been suggested that RNCs stall the FtsY-SRP complex in a conformation, where the complex displays only low GTPase activity. In agreement with this, the presence of RNCs delays GTP hydrolysis of the FtsY-SRP complex ∼10-fold (Zhang et al., 2009). If negatively charged phospholipids stabilize the FtsY-SRP complex in the same conformational state as RNCs, then the phospholipid-induced complex should also display reduced GTPase activity. In support of this assumption, the 100-fold stimulation of complex formation observed here and in a recent kinetic study (Lam et al., 2010) is accompanied by only a two- to fivefold increase in GTPase activity when FtsY is incubated with liposomes and SRP (Bahari et al., 2007; Marty et al., 2009). This indicates that both RNCs and negatively charged phospholipids stabilize the FtsY-SRP complex by delaying GTP hydrolysis. Stalling the FtsY-SRP complex in this early conformational state (Shan et al., 2009; Lam et al., 2010; Estrozi et al., 2011), which has reduced GTPase activity, probably provides a time window that allows for the location of vacant translocons.
In vivo, more than 80% of FtsY is bound to the E. coli membrane (Mircheva et al., 2009), mainly via contact to the head groups of PG and CL (Braig et al., 2009). However, these electrostatic interactions (Reinau et al., 2010) do not render FtsY protease-resistant. Only when lipid-bound FtsY is in contact with SRP does the NG domain of FtsY become protease-protected. FtsY contains an amphipathic lipid-binding helix at the interface between the conserved NG domain and the N-terminal A domain of FtsY (Parlitz et al., 2007; Egea et al., 2008; Braig et al., 2009; Marty et al., 2009), which is essential for function. We now show that this lipid-binding helix is required for lipid-induced FtsY-SRP complex formation and the enhanced stability of the FtsY-lipid contact. This helix is located immediately upstream of the αN1-helix of FtsY, which is delocalized upon SRP interaction (Neher et al., 2008). It is possible that this delocalization inserts the downstream lipid-binding helix more deeply between the phospholipid head groups, allowing for additional hydrophobic interactions and resulting in protease protection of FtsY.
Our data using the FtsY-Ffh fusion protein demonstrate that a preassembled FtsY-SRP complex is able to recruit RNCs to the membrane and to transfer them to the SecY translocon both in vitro and in vivo. The presence of a stable FtsY-SRP complex at the membrane would also explain why in both eukaryotes and prokaryotes ∼40% of the SRP are found at the membrane (Walter and Blobel, 1983; Koch et al., 1999) and why membrane-bound SRP is sufficient for targeting of RNCs (Bornemann et al., 2008). The RNC-independent formation of an FtsY-SRP complex at the membrane raises the question about its physiological function. Because FtsY can bind directly to SecY (Angelini et al., 2005, 2006; Kuhn et al., 2011), it is conceivable that a membrane-bound FtsY-SRP complex simply enhances the efficiency of targeting by increasing the local concentration of SRP close to an available SecY translocon. Increasing the local concentration appears to be a fundamental principle in living systems and has been observed for many biological processes (Oehler and Muller-Hill, 2010).
In addition, a preformed FtsY-SRP complex might be important beyond the efficiency aspect. Previous studies have shown that ribosomes remain bound to the ER membrane after nascent chain release (Adelman et al., 1973) and do not easily dissociate from the membrane (Potter and Nicchitta, 2000). These ribosomes can reinitiate the translation of SRP substrates at the membrane, and thus would not depend on cytosolic SRP (Potter and Nicchitta, 2000). Transfer of these RNCs to the SecYEG translocon would probably still require SRP, because SRP gives RNCs a competitive advantage over nontranslating ribosomes in translocon binding (Neuhof et al., 1998; Raden and Gilmore, 1998; Schaletzky and Rapoport, 2006). The preassembled FtsY-SRP complex would be perfectly suited to facilitate translocon binding of nascent chains initiated on membrane bound ribosomes. Finally, it has been suggested that mRNAs may contain targeting information that localizes them to the right compartment without the need of protein synthesis or the SRP pathway (Palazzo et al., 2007; Pyhtila et al., 2008; Kraut-Cohen and Gerst, 2010). Because mRNAs encoding for bacterial membrane proteins have significantly higher uracil content than mRNAs for cytosolic proteins, the existence of a specific mRNA targeting has also been proposed for bacteria (Prilusky and Bibi, 2009). In agreement with this, a recent study has demonstrated that mRNAs that encode inner membrane proteins can localize to the membrane by a translation-independent mechanism (Nevo-Dinur et al., 2011). After translation initiation of these alternatively targeted mRNAs, the preassembled membrane-bound FtsY-SRP complex could recognize the signal-anchor sequence and ensure the delivery of the RNC to the Sec translocon.
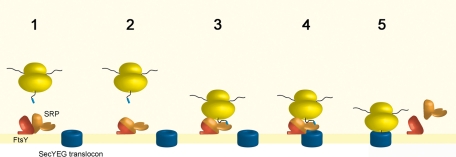
In summary, our data indicate that a second targeting pathway exists (Figure 9) in addition to the canonical targeting by the SRP-pathway that involves the recognition of the substrate in the cytosol and its subsequent targeting to the membrane-bound translocon. This pathway involves a preformed membrane-bound FtsY-SRP complex (Figure 9, steps 1 and 2) that recognizes RNCs at the membrane (step 3) and transfers them to the SecYEG translocon (step 4). Subsequently the FtsY-SRP complex can dissociate due to GTP hydrolysis (step 5) or can accept another RNC. It is difficult to determine the extent to which this alternative pathway is utilized in bacterial cells. The ability of the TM-FF-fusion to support growth of E. coli in the absence of a soluble SRP indicates that the membrane-bound FtsY-SRP complex has at least the minimal activity required for viability. This indicates that soluble SRP is not absolutely essential for cotranslational targeting in E. coli.
FIGURE 9:
Model for RNC targeting by a preformed FtsY-SRP-complex. A preassembled FtsY-SRP complex at the membrane (FtsY, red; SRP, orange) is able to recruit RNCs to the membrane and to transfer them to the SecYEG translocon (blue). It is only after contact of the FtsY-SRP complex with the translocon that the stimulation of GTPase activity induces the dissociation of the complex and allows docking of the RNCs onto the translocon. The dissociated SRP can either replenish the pool of cytosolic SRP or assemble again in a phospholipid-dependent manner with FtsY.
MATERIAL AND METHODS
Strains and plasmids
The following E. coli strains were used: MC4100 (Koch et al., 1999), DH5α (Hanahan, 1983), BL21 (DE3) pLysS (Novagen, Bad Soden, Germany), TY0 (Matsumoto et al., 2000), IY28 (Burk et al., 2009), and Wam113 (Phillips and Silhavy, 1992).
The FtsY-Ffh-fusion protein (FF-fusion) was constructed by amplifying FtsY from plasmid pTP37 (Powers and Walter, 1997) using primer FtsY_fusion_f and FtsY_fusion_r. The PCR product was digested with NcoI and EcoRI. Ffh was amplified from plasmid pET19b-Ffh (Eisner et al., 2003) using primer Ffh-fusion_f and Ffh_fusion_r. The PCR product was digested with EcoRI and HindIII. Subsequently the FtsY and Ffh fragments were ligated into NcoI and HindIII-digested pTrc99a, resulting in pTrc99a-FF-fusion. For in vitro synthesis of the fusion protein, the NcoI/HindIII FtsY-Ffh-fusion fragment was cloned into pET22b vector to allow T7-dependent expression. pTrc99a-Ffh was constructed by digesting pET19b-Ffh (Eisner et al. 2003) with NcoI and BamHI and ligating the resulting Ffh fragment into NcoI/BamHI-digested pTrc99a. pBAD33-Ffh-YFP was constructed as follow: Ffh-EYFP was amplified from plasmid pES118-Ffh-EYFP (kindly provided by Victor Sourjik, University of Heidelberg, Germany) using the primer Ffh_YFP_XbaI and Ffh_YFP_pBad33_rev digested with XbaI and ligated into XbaI-digested pBAD33. For construction of pTrc99a-FtsY-GFP-Ffh-fusion, the GFP gene was amplified from pTrc99a-FtsY-GFP (Mircheva et al. 2009) using primer GFP_EcoRI_for and GFP_EcoRI_rev and subsequently digested with EcoRI. The resulting fragment was ligated into EcoRI-digested pTrc99a-FtsY-Ffh-fusion. FF-fusion A144W and A336W were constructed by site-directed mutagenesis using Phusion DNA-Polymerase (NEB, Biolabs, Frankfurt, Germany) and the appropriate oligonucleotides (Table S1).
The TM-FF-fusion was constructed by amplifying tatC from plasmid p8737 (Alami et al., 2002) using the primer fw-NcoI-TatC and rev-NcoI-TatC. The PCR product was subsequently digested with NcoI and cloned into NcoI-digested pTrc99a-FF-fusion, resulting in pTrc99a-TM-FF-fusion. The orientation of tatC and its correct fusion to the FF-fusion protein was verified by restriction digestion and sequencing.
For the complementation analyses of strain WAM113 (see In vivo complementation activity and localization of the FtsY-Ffh fusion protein), the ampicillin cartridge of pTrc99a-FF-fusion and pTrc99a-TM-FF-fusion had to be replaced by a chloramphenicol cassette. The ampicillin cartridge was deleted by inverse PCR using the primer pTrc99a-Amp-for and pTrc99a-Amp-rev. The chloramphenicol cassette was amplified from vector pBAD33 using the primer CM_p15A-pBad-for and CM_p15A-pBad-rev and blunt-end ligated to the inverse PCR product of the pTrc99a-FF-fusion, resulting in plasmid pTrc99a-FF-fusion(Cm). The same strategy was used for generating pTrc99a-TM-FF-fusion(Cm).
The sequences of the oligonucleotides used in this study are listed in Table S1.
In vitro transcription–translation assays and PK protection
The composition of the reconstituted transcription–translation system of E. coli, the purification of its components, the preparation of INV, and the proteinase protection assay followed previously described protocols (Koch et al., 1999).
BN-PAGE analysis
In vitro synthesis and purification of 35S-FtsY, solubilization of FtsY-containing membrane complexes with subsequent BN-PAGE analysis has been described previously (Angelini et al., 2006). This protocol was also used for in vitro synthesis and purification of Ffh and the FtsY-Ffh-fusion protein. FtsY-SRP complex formation in the presence of liposomes was performed as follows. Radiolabeled samples: 35S-FtsY was incubated with the indicated amount of Ffh in the presence of 35 μg/ml 4.5S RNA (Koch et al., 1999) and 0.2 mg/ml liposomes composed of either synthetic lipids (Braig et al., 2009) or E. coli lipids in the presence or absence of 2 mM GMP-PNP for 20 min at 37°C. Further processing of the samples was identical to INV-containing samples. Samples were separated on either 5–10% or 5–15% BN-PAGE gels. For nonradioactive BN-PAGE, FtsY (4 μM final concentration) was incubated with Ffh (1.5 μM final concentration) and 0.1 mg/ml 4.5S RNA in the presence or absence of 2 mM GMP-PNP. Alternatively, the FtsY-Ffh fusion (1.5 μM final concentration) was incubated with 0.1 mg/ml 4.5S RNA in the presence or absence of 2 mM GMP-PNP. Sample preparation for BN-PAGE was identical to sample preparation for the radioactively labeled samples. Proteins were subsequently separated on a 5–15% BN-PAGE gel. After blotting on a nitrocellulose membrane, the protein complexes were detected by α-Ffh antibodies.
Expression and purification of proteins, GTPase assays
pTrc99a-FtsY (Braig et al., 2009), pTrc99a-Ffh, and pTrc99a-FF-fusion were transformed into E. coli DH5α cells (Hanahan, 1983). Cells were grown at 37°C, induced with 1 mM IPTG after reaching an OD600 of 0.6–0.8, and harvested 4h after induction. Proteins were affinity purified via their His tags using an Äkta chromatography system (GE Healthcare, Waukesha, WI) and a 1 ml HisTrap FF crude nickel column (GE Healthcare). The equilibration/wash buffer contained 50 mM HEPES KOH, pH 7.6, 1 M NH4Ac, 10 mM MgAc2, 1 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 30 mM imidazole, pH 7.8, and 10% Glycerol. The elution buffer contained 50 mM HEPES KOH, pH 7.6, 1 M NH4Ac, 10 mM MgAc2, 1 mM DTT, 0.5 mM PMSF, Roche Complete Inhibitor Cocktail Tablets, 400 mM imidazole, pH 7.8, and 10% glycerol. Subsequently, the buffer was exchanged to 2 × HT buffer (100 mM HEPES KOH, pH 7.6, 200 mM KAc pH 7.5, 20 mM MgAc2, 2 mM DTT) using PD10 columns (GE Healthcare). Ffh and the FF-fusion protein were stored at −20°C in HT buffer supplemented with 50% glycerol; FtsY was stored at −70°C in HT buffer supplemented with 10% glycerol. The FtsY derivatives carrying mutations in the second lipid-binding helix were constructed by PCR mutagenesis using pTP37 as a template (Powers and Walter, 1997). The reversed primer Dav_Hr was used in all PCR reactions. The forward primers used were Dav_Hf (for FtsY(delta195-203)), K198_Dav_f (for FtsY(R198K)), D198_Dav_f (for FtsY(R198D)), A198_Dav_f (for FtsY(R198A)), or R198D+K200D_f_Dav (for FtsY(R198D+K200D)), respectively. The mutated FtsY derivatives were expressed in E. coli CE43 (DE3) and purified via Talon (Clontech, Mountain View, CA) metal-affinity chromatography.
GTPase assays were performed at 25°C in a total volume of 20 μl and contained 0.05–0.5 μM Ffh, FtsY, or FF-fusion. The reaction was performed in HT buffer (50 mM HEPES, pH 7.6; 100 mM KOAc, pH 7.5; 10 mM MgOAc; and 1 mM DTT) and started by the addition of GTP (200 μM GTP + 2.5 μM [γ-P32]GTP [approx. 2.5 μCi]). When indicated, 0.5 μg 4.5S RNA was added. Aliquots were removed at frequent time intervals. The reaction was stopped on ice by the addition of 800 μl charcoal suspension (10% in 20 mM phosphoric acid) and the liberated phosphate in the supernatant after centrifugation was determined using a scintillation counter.
In vivo complementation activity and localization of the FtsY-Ffh fusion protein
The conditional FtsY mutant strain IY28 and the conditional Ffh mutant strain Wam113 have been described previously (Phillips and Silhavy, 1992; Burk et al., 2009). Complementation of IY28 was achieved by transforming the strain with pTrc99a vector harboring the appropriate genes or the empty vector as negative control. Strains were grown to an OD600 of 1.0 following exponential dilution of the culture in lysogeny broth (LB) medium. The diluted cultures were plated on LB-agar with and without 0.2% arabinose.
Complementation of WAM113 was achieved by transforming the strain with pTrc99a(Cm) vector harboring the appropriate genes or the empty pTrc99a(Cm) vector as control. Growth of the mutant Wam113 strains was examined identical to IY28.
Fluorescence microscopy was performed exactly as described in Mircheva et al., (2009) on an Olympus BX51 microscope (Olympus, Hamburg, Germany) at 100× magnification with a numerical aperture of 1.4. Images were acquired with a charge-coupled device camera (F-View, Olympus), driven by the cell*F software (Olympus Soft Imaging Solutions, Münster, Germany). Cells were immobilized on a microscope slide with low-melting agarose.
Integration of MtlA into SecYEG proteoliposomes
SecYEG was purified from E. coli TY0 expressing pBAD-SecYEHisG (Collinson et al., 2001). Cells were grown at 37°C and induced with 0.5% arabinose after reaching an OD600 of 0.5, and harvested 2 h after induction. After cell breakage using a French Press (Thermo Fisher Scientific, Schwerte, Germany), a crude membrane fraction was isolated and solubilized with S1 buffer for 1 h at 4°C (S1 buffer: 20 mM Tris/HCl, pH 7.5; 300 mM NaCl; 5 mM MgCl2; 10% glycerol; 1% dodecyl maltoside [DDM]; 5 mM imidazole; complete protease inhibitor [Roche, Basel, Switzerland]). After centrifugation for 25 min at 30.000 rpm in a Ti50.2 rotor, the supernatant was added to preequilibrated Talon-beads (Clontech) and incubated for 1 h at 4°C. After washing with S1 buffer containing 0.03% DDM, SecYEG was eluted with S1 buffer containing 0.03% DDM and 200 mM imidazole. Reconstitution of purified SecYEG into liposomes and in vitro integration of MtlA with subsequent PK digestion as described previously (Nishiyama et al., 2006) was used with slight modifications. SecYEG was reconstituted in E. coli polar phospholipids (Avanti Polar Lipids, Alabaster, AL) supplemented with 5% diacylglycerol. Integration of MtlA into SecYEG proteoliposomes (100 nM SecYEG; 0.1 mg/ml lipids) was analyzed in the presence of in vitro synthesized 4.5 S RNA (final concentration 15 μg/ml; Koch et al. 1999), Ffh (150 nM), FtsY (750 nM), and FF-fusion (150 nM) when indicated.
Supplementary Material
Acknowledgments
We gratefully acknowledge Eitan Bibi, Weizmann Institute, Rehovot, Israel, for providing E. coli IY28 and Victor Sourjik, ZMBH Heidelberg, Germany, for providing plasmid pES118-Ffh-EYFP. This work was supported by grants from the DFG-Forschergruppe 967 to R.B. and H.G.K., by an Innovation Fellowship of the University of Freiburg to D.B., by a DAAD Fellowship to M.M., and by a long-term fellowship from the Human Frontiers Science Program to E.O.v.d.S.
Abbreviations used:
- BN-PAGE
Blue-native PAGE
- BSA
bovine serum albumin
- CL
cardiolipin
- DAG
diacylglycerol
- DDM
dodecyl maltoside
- DTT
dithiothreitol
- FF-fusion
FtsY-Ffh-fusion
- GFP
green fluorescent protein
- GMP-PNP
guanosine 5′(β,γ-imido) triphosphate
- INV
inner membrane vesicles
- IPTG
isopropyl-β-d-thiogalactopyranoside
- LB
lysogeny broth
- MtlA
mannitol permease
- MtlA-MPF
, mannitol permease membrane-protected fragment
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- PK
proteinase K
- PMSF
phenylmethylsulfonyl fluoride
- RNC
ribosome-nascent chain
- SR
SRP receptor
- SRP
signal recognition particle
- TCA
trichloroacetic acid
- TEA
triethanolamine acetate
- TM-FF-fusion
transmembrane FF-fusion
- YFP
yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-02-0152 on May 5, 2011.
REFERENCES
- Adelman MR, Sabatini DD, Blobel G. Ribosome-membrane interaction. Nondestructive disassembly of rat liver rough microsomes into ribosomal and membranous components. J Cell Biol. 1973;56:206–229. doi: 10.1083/jcb.56.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alami M, Trescher D, Wu L-F, Müller M. Separate analysis of twin-arginine translocation (Tat)-specific membrane binding and translocation in Escherichia coli. J Biol Chem. 2002;277:20499–20503. doi: 10.1074/jbc.M201711200. [DOI] [PubMed] [Google Scholar]
- Angelini S, Boy D, Schiltz E, Koch HG. Membrane binding of the bacterial signal recognition particle receptor involves two distinct binding sites. J Cell Biol. 2006;174:715–724. doi: 10.1083/jcb.200606093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini S, Deitermann S, Koch HG. FtsY, the bacterial signal-recognition particle receptor, interacts functionally and physically with the SecYEG translocon. EMBO Rep. 2005;6:476–481. doi: 10.1038/sj.embor.7400385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataide SF, Schmitz N, Shen K, Ke A, Shan S-o, Doudna J, Ban N. The crystal structure of the signal recognition particle in complex with its receptor. Science. 2011;331:881–886. doi: 10.1126/science.1196473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahari L, Parlitz R, Eitan A, Stjepanovic G, Bochkareva ES, Sinning I, Bibi E. Membrane targeting of ribosomes and their release require distinct and separable functions of FtsY. J Biol Chem. 2007;282:32168–32175. doi: 10.1074/jbc.M705429200. [DOI] [PubMed] [Google Scholar]
- Bibi E. Early targeting events during membrane protein biogenesis in Escherichia coli. Biochim Biophys Acta. 2010;1808:841–850. doi: 10.1016/j.bbamem.2010.07.025. [DOI] [PubMed] [Google Scholar]
- Bogdanov M, Xie J, Heacock P, Dowhan W. To flip or not to flip: lipid-protein charge interactions are a determinant of final membrane protein topology. J Cell Biol. 2008;182:925–935. doi: 10.1083/jcb.200803097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese D, Blobel G, Sabatini DD. In vitro exchange of ribosomal subunits between free and membrane-bound ribosomes. J Mol Biol. 1973;74:415–438. doi: 10.1016/0022-2836(73)90037-5. [DOI] [PubMed] [Google Scholar]
- Bornemann T, Jockel J, Rodnina MV, Wintermeyer W. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat Struct Mol Biol. 2008;15:494–499. doi: 10.1038/nsmb.1402. [DOI] [PubMed] [Google Scholar]
- Boy D, Koch HG. Visualization of distinct entities of the SevYEG translocon during translocation and integration of bacterial proteins. Mol Biol Cell. 2009;20:1804–1815. doi: 10.1091/mbc.E08-08-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw N, Neher SB, Booth DS, Walter P. Signal sequences activate the catalytic switch of SRP RNA. Science. 2009;323:127–130. doi: 10.1126/science.1165971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig D, Bär C, Thumfart JO, Koch HG. Two cooperating helices constitute the lipid-binding domain of the bacterial SRP receptor. J Mol Biol. 2009;390:401–413. doi: 10.1016/j.jmb.2009.04.061. [DOI] [PubMed] [Google Scholar]
- Bürk J, Weiche B, Wenk M, Boy D, Nestel S, Heimrich B, Koch HG. Depletion of the signal recognition particle receptor inactivates ribosomes in Escherichia coli. J Bacteriol. 2009;191:7017–7026. doi: 10.1128/JB.00208-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskiewicz IA, Jockel J, Rodnina MV, Wintermeyer W. Conformation of the signal recognition particle in ribosomal targeting complexes. RNA. 2009;15:44–54. doi: 10.1261/rna.1285609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Jiang Y, Mandon EC, Gilmore R. Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J Cell Biol. 2005;168:67–77. doi: 10.1083/jcb.200408188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross BC, Sinning I, Luirink J, High S. Delivering proteins for export from the cytosol. Nat Rev Mol Cell Biol. 2009;10:255–264. doi: 10.1038/nrm2657. [DOI] [PubMed] [Google Scholar]
- Collinson I, Breyton C, Duong F, Tziatzios C, Schubert D, Or E, Rapoport T, Kühlbrandt W. Projection structure and oligomeric properties of a bacterial core protein translocase. EMBO J. 2001;20:2462–2471. doi: 10.1093/emboj/20.10.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw E, te Kaat K, Moser C, Menestrina G, Demel R, de Kruijff B, Oudega B, Luirink J, Sinning I. Anionic phospholipids are involved in membrane association of FtsY and stimulate its GTPase activity. EMBO J. 2000;19:531–541. doi: 10.1093/emboj/19.4.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vrije T, de Swart RL, Dowhan W, Tommassen J, de Kruijff B. Phosphatidylglycerol is involved in protein translocation across Escherichia coli inner membranes. Nature. 1988;334:173–175. doi: 10.1038/334173a0. [DOI] [PubMed] [Google Scholar]
- Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- Egea PF, Shan SO, Napetschnig J, Savage DF, Walter P, Stroud RM. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004;427:215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- Egea PF, Tsuruta H, de Leon GP, Napetschnig J, Walter P, Stroud RM. Structures of the signal recognition particle receptor from the archaeon Pyrococcus furiosus: implications for the targeting step at the membrane. PLoS One. 2008;3:e3619. doi: 10.1371/journal.pone.0003619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisner G, Koch HG, Beck K, Brunner J, Muller M. Ligand crowding at a nascent signal sequence. J Cell Biol. 2003;163:35–44. doi: 10.1083/jcb.200306069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erez E, Stjepanovic G, Zelazny AM, Brugger B, Sinning I, Bibi E. Genetic evidence for functional interaction of the E. coli SRP-receptor with acidic lipids in vivo. J Biol Chem. 2010;285:40508–40514. doi: 10.1074/jbc.M110.140921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrozi LE, Boehringer D, Shan So, Ban N, Schaffitzel C. Cryo-EM structure of the E. coli translating ribosome in complex with SRP and its receptor. Nature Struct Mol Biol. 2011;18:88–90. doi: 10.1038/nsmb.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facey SJ, Kuhn A. Biogenesis of bacterial inner-membrane proteins. Cell Mol Life Sci. 2010;67:2343–2362. doi: 10.1007/s00018-010-0303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM. Heterodimeric GTPase core of the SRP targeting complex. Science. 2004;303:373–377. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulga TA, Sinning I, Dobberstein B, Pool MR. SRβ coordinates signal sequence release with ribosome binding to the translocon. EMBO J. 2001;20:2338–2347. doi: 10.1093/emboj/20.9.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold VA, Robson A, Bao H, Romantsov T, Duong F, Collinson I. The action of cardiolipin on the bacterial translocon. Proc Natl Acad Sci USA. 2010;107:10044–10049. doi: 10.1073/pnas.0914680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grudnik P, Bange G, Sinning I. Protein targeting by the signal recognition particle. Biol Chem. 2009;390:775–782. doi: 10.1515/BC.2009.102. [DOI] [PubMed] [Google Scholar]
- Hainzl T, Huang S, Meriläinen G, Brannström K, Sauer-Erikson AE. Structural basis of signal sequence recognition by the signal recognition particle. Nature Struct Mol Biol. 2011;18:389–391.. doi: 10.1038/nsmb.1994. [DOI] [PubMed] [Google Scholar]
- Halic M, Blau M, Becker T, Mielke T, Pool MR, Wild K, Sinning I, Beckmann R. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006a;444:507–511. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- Halic M, Gartmann M, Schlenker O, Mielke T, Pool MR, Sinning I, Beckmann R. Signal recognition particle receptor exposes the ribosomal translocon binding site. Science. 2006b;312:745–747. doi: 10.1126/science.1124864. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Helmers J, Schmidt D, Glavy JS, Blobel G, Schwartz T. The beta-subunit of the protein-conducting channel of the endoplasmic reticulum functions as the guanine nucleotide exchange factor for the beta-subunit of the signal recognition particle receptor. J Biol Chem. 2003;278:23686–23690. doi: 10.1074/jbc.C300180200. [DOI] [PubMed] [Google Scholar]
- Jagath JR, Matassova NB, de Leeuw E, Warnecke JM, Lentzen G, Rodnina MV, Luirink J, Wintermeyer W. Important role of the tetraloop region of 4.5S RNA in SRP binding to its receptor FtsY. RNA. 2001;7:293–301. doi: 10.1017/s1355838201002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagath JR, Rodnina MV, Wintermeyer W. Conformational changes in the bacterial SRP receptor FtsY upon binding of guanine nucleotides and SRP. J Mol Biol. 2000;295:745–753. doi: 10.1006/jmbi.1999.3427. [DOI] [PubMed] [Google Scholar]
- Janda CY, Li J, Oubridge C, Hernandez H, Robinson CV, Nagai K. Recognition of a signal peptide by the signal recognition particle. Nature. 2010;465:507–510. doi: 10.1038/nature08870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Cheng Z, Mandon EC, Gilmore R. An interaction between the SRP receptor and the translocon is critical during cotranslational protein translocation. J Cell Biol. 2008;180:1149–1161. doi: 10.1083/jcb.200707196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch HG, Hengelage T, Neumann-Haefelin C, MacFarlane J, Hoffschulte HK, Schimz KL, Mechler B, Muller M. In vitro studies with purified components reveal signal recognition particle (SRP) and SecA/SecB as constituents of two independent protein-targeting pathways of Escherichia coli. Mol Biol Cell. 1999;10:2163–2173. doi: 10.1091/mbc.10.7.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch HG, Moser M, Muller M. Signal recognition particle-dependent protein targeting, universal to all kingdoms of life. Rev Physiol Biochem Pharmacol. 2003;146:55–94. doi: 10.1007/s10254-002-0002-9. [DOI] [PubMed] [Google Scholar]
- Kraut-Cohen J, Gerst JE. Addressing mRNAs to the ER: cis sequences act up. Trends Biochem Sci. 2010;35:459–469. doi: 10.1016/j.tibs.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Kuhn P, Weiche B, Sturm L, Sommer E, Drepper F, Warscheid B, Sourjik V, Koch HG. The bacterial SRP receptor, SecA and the ribosome use overlapping binding sites on the SecY translocon. Traffic. 2011;12:563–578. doi: 10.1111/j.1600-0854.2011.01167.x. [DOI] [PubMed] [Google Scholar]
- Kulajta C, Thumfart JO, Haid S, Daldal F, Koch HG. Multi-step assembly pathway of the cbb3-type cytochrome c oxidase complex. J Mol Biol. 2006;355:989–1004. doi: 10.1016/j.jmb.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Lam VQ, Akopian D, Rome M, Henningsen D, Shan SO. Lipid activation of the signal recognition particle receptor provides spatial coordination of protein targeting. J Cell Biol. 2010;190:623–635. doi: 10.1083/jcb.201004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- Luirink J, ten Hagen-Jongman CM, Van Der Weijden CC, Oudega B, High S, Dobberstein B, Kusters R. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO J. 1994;13:2289–2296. doi: 10.1002/j.1460-2075.1994.tb06511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty NJ, Rajalingam D, Kight AD, Lewis NE, Fologea D, Kumar TK, Henry RL, Goforth RL. The membrane-binding motif of the chloroplast signal recognition particle receptor (cpFtsY) regulates GTPase activity. J Biol Chem. 2009;284:14891–14903. doi: 10.1074/jbc.M900775200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto G, Homma T, Mori H, Ito K. A mutation in secY that causes enhanced SecA insertion and impaired late functions in protein translocation. J Bacteriol. 2000;182:3377–3382. doi: 10.1128/jb.182.12.3377-3382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mircheva M, Boy D, Weiche B, Hucke F, Graumann P, Koch HG. Predominant membrane localization is an essential feature of the bacterial signal recognition particle receptor. BMC Biol. 2009;7:76. doi: 10.1186/1741-7007-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher SB, Bradshaw N, Floor SN, Gross JD, Walter P. SRP RNA controls a conformational switch regulating the SRP-SRP receptor interaction. Nat Struct Mol Biol. 2008;15:916–923. doi: 10.1038/nsmb.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhof A, Rolls MM, Jungnickel B, Kalies KU, Rapoport TA. Binding of signal recognition particle gives ribosome/nascent chain complexes a competitive advantage in endoplasmic reticulum membrane interaction. Mol Biol Cell. 1998;9:103–115. doi: 10.1091/mbc.9.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann-Haefelin C, Schäfer U, Müller M, Koch HG. SRP-dependent co-translational targeting and SecA-dependent translocation analyzed as individual steps in the export of a bacterial protein. EMBO J. 2000;19:6419–6426. doi: 10.1093/emboj/19.23.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O. Translation-independent localization of mRNA in E. coli. Science. 2011;331:1081–1084. doi: 10.1126/science.1195691. [DOI] [PubMed] [Google Scholar]
- Nishiyama K, Ikegami A, Moser M, Schiltz E, Tokuda H, Muller M. A derivative of lipid A is involved in signal recognition particle/SecYEG-dependent and -independent membrane integrations. J Biol Chem. 2006;281:35667–35676. doi: 10.1074/jbc.M608228200. [DOI] [PubMed] [Google Scholar]
- Oehler S, Muller-Hill B. High local concentration: a fundamental strategy of life. J Mol Biol. 2010;395:242–253. doi: 10.1016/j.jmb.2009.10.056. [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Springer M, Shibata Y, Lee CS, Dias AP, Rapoport TA. The signal sequence coding region promotes nuclear export of mRNA. PLoS Biol. 2007;5:e322. doi: 10.1371/journal.pbio.0050322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlitz R, Eitan A, Stjepanovic G, Bahari L, Bange G, Bibi E, Sinning I. Escherichia coli signal recognition particle receptor FtsY contains an essential and autonomous membrane-binding amphipathic helix. J Biol Chem. 2007;282:32176–32184. doi: 10.1074/jbc.M705430200. [DOI] [PubMed] [Google Scholar]
- Peluso P, Herschlag D, Nock S, Freymann DM, Johnson AE, Walter P. Role of 4.5S RNA in assembly of the bacterial signal recognition particle with its receptor. Science. 2000;288:1640–1643. doi: 10.1126/science.288.5471.1640. [DOI] [PubMed] [Google Scholar]
- Peluso P, Shan SO, Nock S, Herschlag D, Walter P. Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry. 2001;40:15224–15233. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- Phillips GJ, Silhavy TJ. The E. coli ffh gene is necessary for viability and efficient protein export. Nature. 1992;359:744–746. doi: 10.1038/359744a0. [DOI] [PubMed] [Google Scholar]
- Poritz MA, Bernstein HD, Strub K, Zopf D, Wilhelm H, Walter P. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990;250:1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- Potter MD, Nicchitta CV. Regulation of ribosome detachment from the mammalian endoplasmic reticulum membrane. J Biol Chem. 2000;275:33828–33835. doi: 10.1074/jbc.M005294200. [DOI] [PubMed] [Google Scholar]
- Potter MD, Seiser RM, Nicchitta CV. Ribosome exchange revisited: a mechanism for translation-coupled ribosome detachment from the ER membrane. Trends Cell Biol. 2001;11:112–115. doi: 10.1016/s0962-8924(00)01905-x. [DOI] [PubMed] [Google Scholar]
- Powers T, Walter P. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 1997;16:4880–4886. doi: 10.1093/emboj/16.16.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prilusky J, Bibi E. Studying membrane proteins through the eyes of the genetic code revealed a strong uracil bias in their coding mRNAs. Proc Natl Acad Sci USA. 2009;106:6662–6666. doi: 10.1073/pnas.0902029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz A, Behrens C, Rapoport TA, Hartmann E, Kalies KU. Evolutionarily conserved binding of ribosomes to the translocation channel via the large ribosomal RNA. EMBO J. 2000;19:1900–1906. doi: 10.1093/emboj/19.8.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyhtila B, Zheng T, Lager PJ, Keene JD, Reedy MC, Nicchitta CV. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA. 2008;14:445–453. doi: 10.1261/rna.721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raden D, Gilmore R. Signal recognition particle-dependent targeting of ribosomes to the rough endoplasmic reticulum in the absence and presence of the nascent polypeptide-associated complex. Mol Biol Cell. 1998;9:117–130. doi: 10.1091/mbc.9.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Reinau ME, Thogersen IB, Enghild JJ, Nielsen KL, Otzen DE. The diversity of FtsY-lipid interactions. Biopolymers. 2010;93:595–606. doi: 10.1002/bip.21404. [DOI] [PubMed] [Google Scholar]
- Ribes V, Romisch K, Giner A, Dobberstein B, Tollervey D. E. coli 4.5S RNA is part of a ribonucleoprotein particle that has properties related to signal recognition particle. Cell. 1990;63:591–600. doi: 10.1016/0092-8674(90)90454-m. [DOI] [PubMed] [Google Scholar]
- Romantsov T, Stalker L, Culham DE, Wood JM. Cardiolipin controls the osmotic stress response and the subcellular location of transporter ProP in Escherichia coli. J Biol Chem. 2008;283:12314–12323. doi: 10.1074/jbc.M709871200. [DOI] [PubMed] [Google Scholar]
- Schaffitzel C, Oswald M, Berger I, Ishikawa T, Abrahams JP, Koerten HK, Koning RI, Ban N. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature. 2006;444:503–506. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- Schaletzky J, Rapoport TA. Ribosome binding to and dissociation from translocation sites of the endoplasmic reticulum membrane. Mol Biol Cell. 2006;17:3860–3869. doi: 10.1091/mbc.E06-05-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiser RM, Nicchitta CV. The fate of membrane-bound ribosomes following the termination of protein synthesis. J Biol Chem. 2000;275:33820–33827. doi: 10.1074/jbc.M004462200. [DOI] [PubMed] [Google Scholar]
- Shan SO, Chandrasekar S, Walter P. Conformational changes in the GTPase modules of the signal reception particle and its receptor drive initiation of protein translocation. J Cell Biol. 2007;178:611–620. doi: 10.1083/jcb.200702018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan SO, Schmid SL, Zhang X. Signal recognition particle (SRP) and SRP receptor: a new paradigm for multistate regulatory GTPases. Biochemistry. 2009;48:6696–6704. doi: 10.1021/bi9006989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan SO, Stroud RM, Walter P. Mechanism of association and reciprocal activation of two GTPases. PLoS Biol. 2004;2:e320. doi: 10.1371/journal.pbio.0020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan SO, Walter P. Co-translational protein targeting by the signal recognition particle. FEBS Lett. 2005;579:921–6. doi: 10.1016/j.febslet.2004.11.049. [DOI] [PubMed] [Google Scholar]
- Shen K, Shan SO. Transient tether between the SRP RNA and SRP receptor ensures efficient cargo delivery during cotranslational protein targeting. Proc Natl Acad Sci USA. 2010;107:7698–7703. doi: 10.1073/pnas.1002968107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Raden D, Mandon EC, Gilmore R. Role of Sec61alpha in the regulated transfer of the ribosome-nascent chain complex from the signal recognition particle to the translocation channel. Cell. 2000;100:333–343. doi: 10.1016/s0092-8674(00)80669-8. [DOI] [PubMed] [Google Scholar]
- Walter P, Blobel G. Subcellular distribution of signal recognition particle and 7SL-RNA determined with polypeptide-specific antibodies and complementary DNA probe. J Cell Biol. 1983;97:1693–1699. doi: 10.1083/jcb.97.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiche B, Burk J, Angelini S, Schiltz E, Thumfart JO, Koch HG. A cleavable N-terminal membrane anchor is involved in membrane binding of the Escherichia coli SRP receptor. J Mol Biol. 2008;377:761–773. doi: 10.1016/j.jmb.2008.01.040. [DOI] [PubMed] [Google Scholar]
- Wenz T, Hielscher R, Hellwig P, Schagger H, Richers S, Hunte C. Role of phospholipids in respiratory cytochrome bc1 complex catalysis and supercomplex formation. Biochim Biophys Acta. 2009;1787:609–616. doi: 10.1016/j.bbabio.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Zhang X, Schaffitzel C, Ban N, Shan SO. Multiple conformational switches in a GTPase complex control co-translational protein targeting. Proc Natl Acad Sci USA. 2009;106:1754–1759. doi: 10.1073/pnas.0808573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.