Abstract
Chromosomal double-strand breaks (DSBs) in mammalian cells are repaired by either homology-directed repair (HDR), using a homologous sequence as a repair template, or nonhomologous end-joining (NHEJ), which often involves sequence alterations at the DSB site. To characterize the interrelationship of these two pathways, we analyzed HDR of a DSB in cells deficient for NHEJ components. We find that the HDR frequency is enhanced in Ku70−/−, XRCC4−/−, and DNA-PKcs−/− cells, with the increase being particularly striking in Ku70−/− cells. Neither sister-chromatid exchange nor gene-targeting frequencies show a dependence on these NHEJ proteins. A Ku-modulated two-ended versus one-ended chromosome break model is presented to explain these results.
Keywords: Homologous recombination, DNA double-strand break, nonhomologous end-joining, Ku protein, DNA-PK, sister-chromatid exchange, gene targeting
Double-strand breaks (DSBs) are potentially catastrophic lesions that if not repaired will lead to loss of genetic information and mutagenesis or cell death. In mammalian cells, two major pathways exist to repair DSBs—homologous recombination and nonhomologous end-joining (NHEJ; Liang et al. 1998). NHEJ, the rejoining of DNA ends with the use of little or no sequence homology, involves the processing of ends such that nucleotides are often deleted or inserted at the break site prior to ligation (Jeggo 1998). Such modifications are likely central to the ability of mammalian cells to rejoin DNA ends with a variety of structures. Homology-directed repair (HDR) of a DSB, in contrast, requires significant lengths of sequence homology so that a DNA end from one molecule can invade a homologous sequence and prime repair synthesis (Pâques and Haber 1999). Processing of DNA ends also occurs with HDR; however, repair is typically precise, because a homologous sequence templates the repair event.
Several processes exist in which repair of a DSB is restricted to either NHEJ or HDR. For example, DSBs introduced by the RAG proteins to generate antigen receptor diversity during V(D)J rearrangement are repaired by the NHEJ pathway (Jeggo 1998), whereas those introduced during meiosis by the Spo11 protein are repaired by the HDR pathway (Keeney 2001). The restriction in type of DSB repair raises the question as to how pathway choice is regulated. Several studies point to cell cycle phase as one factor that modulates repair pathway choice. In chicken cells, HDR has been found to play a dominant role in repairing radiation-induced DSBs in late S/G2 phase, whereas NHEJ is preferentially used during G1/early S phase (Takata et al. 1998). Consistent with this in mammalian cells, the preferred homologous template for HDR—the sister chromatid (Johnson and Jasin 2001)—is present only during the S/G2 phase of the cell cycle. Despite a cell cycle preference, HDR and NHEJ can nevertheless be coupled for the repair of a single DSB in mammalian cells (Richardson and Jasin 2000), indicating that the two repair pathways are not completely restricted to different cell cycle phases and that other factors influence pathway choice.
Based on in vitro studies of DNA end-binding proteins such as RAD52, it has been suggested that end-binding proteins may direct entry into alternative DSB repair pathways (Van Dyck et al. 1999). However, no evidence yet exists in vivo to support this model, and mutation of the Rad52 gene in mouse does not confer a cellular DSB repair phenotype (Rijkers et al. 1998). NHEJ proteins, that is, the DNA-dependent protein kinase, composed of the Ku70/Ku80 heterodimer and the catalytic subunit DNA-PKcs, and the XRCC4/DNA Ligase IV complex, have also been characterized to interact with DNA ends (Jeggo 1998; Smith and Jackson 1999; Chen et al. 2000). The Ku heterodimer in particular has been postulated to be an alignment factor to hold DNA ends in an appropriate configuration for end-processing and subsequent ligation (see, e.g., Feldmann et al. 2000).
To begin to understand the factors that modulate the choice of DSB repair pathway in mammalian cells, we investigated homologous recombination in NHEJ mutants. We examined repair of a DSB generated at a defined site in the genome of NHEJ mutants by in vivo expression of the I-SceI endonuclease. We find that HDR of the DSB is enhanced in frequency in NHEJ mutant cells, with a particularly striking sixfold increase in Ku70−/− cells. However, other events involving homologous recombination, that is, sister-chromatid exchange (SCE) and gene targeting, were not increased in frequency in the NHEJ mutants. These results suggest that DSB repair can be modulated by components of repair pathways, in particular by Ku, although the structure of the initiating lesion may be important in determining whether repair is modulated.
Results
Sister-chromatid exchange is not altered in frequency in NHEJ mutants
To begin to understand the factors that modulate homologous recombination pathways, we measured rates of SCE, a common indicator of homologous recombination (Sonoda et al. 1999; Dronkert et al. 2000), in cell lines mutant for representative components of the NHEJ complexes. We compared the spontaneous SCE frequency in mouse embryonic stem (ES) cell lines containing targeted mutations in the Ku70 (Gu et al. 1997), DNA-PKcs (Gao et al. 1998a), or XRCC4 (Gao et al. 1998b) genes with the wild-type J1 ES cell line. We found no significant differences in SCE formation between any of these lines (Table 1), suggesting that the NHEJ machinery is neither required for nor competitive with the generation of SCEs.
Table 1.
SCE frequencies in NHEJ mutant ES cell lines
| Cell line
|
No. of metaphases scored
|
No. chromosomes per metaphase (avg.)
|
Mean no. SCEs per metaphase ± S.D.
|
|---|---|---|---|
| wild type (J1) | 62 | 40.2 | 8.6 ± 4.0 |
| Ku70−/− | 36 | 40.2 | 8.4 ± 3.6 |
| XRCC4−/− | 36 | 40.6 | 7.7 ± 4.3 |
| DNA–PKcs−/− | 23 | 40.0 | 8.2 ± 4.4 |
Gene-targeting efficiency in NHEJ mutants
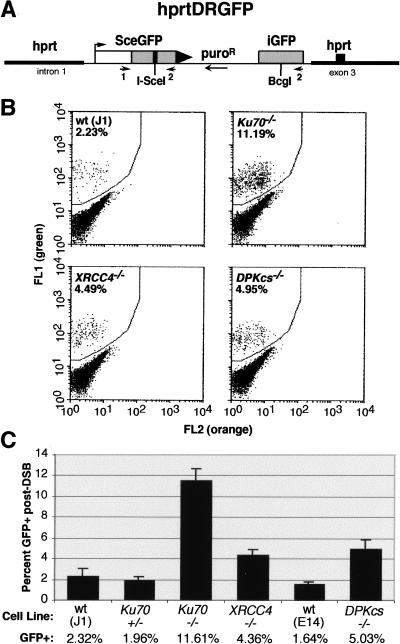
We next examined homologous and random integration of a transfected DNA fragment. As well as providing insight into DNA repair pathways, a shift in the relative frequencies of these events could lead to the identification of cell lines with more favorable gene-targeting efficiencies. The transfected fragment, hprtDRGFP, has homology to the hypoxanthine phosphoribosyl transferase (hprt) locus and contains the selectable puromycin resistance gene (puroR) as well as the HDR reporter substrate DR-GFP (see below; Fig. 1A). Cells that integrate this fragment at a random genomic location are resistant to puromycin, whereas cells in which the reporter is successfully targeted to the hprt locus are additionally resistant to 6-thioguanine (6-TG).
Figure 1.
HDR is elevated in NHEJ mutant cell lines. (A) The hprtDRGFP targeting vector contains homologous hprt targeting arms, for integration of the HDR reporter substrate DR-GFP into the hprt locus, and a dominant selectable puroR marker. In the DR-GFP substrate, the iGFP gene has 0.8 kb of sequence homology to direct repair of an I-SceI-cleaved SceGFP gene, to restore functionality to the GFP gene. (B) Flow cytometric analysis of ES cells containing the targeted hprtDRGFP reporter after electroporation with an I-SceI expression vector. The GFP-positive population is shifted “greenward” from the GFP-negative population. (DPKcs) DNA-PKcs. (C) Summary of the percentage of GFP-positive cells from each of the electroporated cell lines. Bars represent the average of three independently isolated hprtDRGFP subclones for each cell line. Error bars are ±1 S.D. (N = 3).
Cells were electroporated with the hprtDRGFP fragment and selected with puromycin alone or in combination with 6-TG. Clones that were doubly resistant (i.e., puroR/6-TGR clones) were readily obtained in all cell lines and verified by Southern blot analysis to have undergone gene targeting at the hprt locus (data not shown). Although variability in the absolute frequency of integrations was seen between experiments, within each experiment no significant difference in gene-targeting efficiency (i.e., homologous vs. total integration) was seen in the Ku70−/− and XRCC4−/− lines relative to the control J1 line (Table 2). Although not isogenic with the other lines, similar results were obtained with the DNA-PKcs−/− cell line (data not shown). Therefore, NHEJ mutants do not have a more favorable gene-targeting efficiency.
Table 2.
Homologous and random integration of a transfected fragment in NHEJ mutants
| Cell line
|
Frequency of integration (×10−6)a
|
|||||
|---|---|---|---|---|---|---|
| Experiment 1
|
Experiment 2
|
Experiment 3
|
||||
|
puroR/6-TGR
|
puroR
|
puroR/6-TGR
|
puroR
|
puroR/6-TGR
|
puroR
|
|
| wild type (J1) | 2.4 | 207 | 7.4 | 505 | 1.2 | 38 |
| Ku70+/− | 1.5 | 197 | 5.2 | 372 | 1.2 | 49 |
| Ku70−/− | 0.9 | 198 | 8.8 | 499 | 1.3 | 94 |
| XRCC4−/− | ND | ND | ND | ND | 2.1 | 222 |
Number of homologous (puroR/6-TGR) or total (puroR) integrants divided by the number of cells electroporated with the hprtDRGFP fragment.
ND, Not determined.
NHEJ mutants have enhanced HDR of a chromosomal DSB
NHEJ proteins are known to be involved in the repair of chromosomal DSBs such as those generated by ionizing radiation. We therefore investigated whether HDR of a DSB would be affected by mutation of NHEJ genes, by examining repair of a defined chromosomal DSB in the mutant cell lines. For this, we used the DR-GFP reporter, integrated at the hprt locus, which contains the cleavage site for the rare-cutting I-SceI endonuclease (Fig. 1A). The I-SceI site is placed in one of two defective GFP gene repeats, such that an HDR event by a noncrossover gene conversion will restore a functional GFP gene (Pierce et al. 1999). To determine the frequency of this HDR event in the NHEJ mutants, an I-SceI expression vector was electroporated into three independently hprt-targeted clones for each of six different ES cell lines: wild-type ES cell lines J1 and E14, and the targeted Ku70+/−, Ku70−/−, XRCC4−/−, and DNA-PKcs−/− ES cell lines. As the DR-GFP reporter is integrated at the same locus in each of these cell lines, possible position effects on cleavage or repair that may arise from random integration should be abrogated (Liang and Jasin 1996).
In the absence of I-SceI expression, few cells were GFP-positive in any of the cell lines; however, after transient I-SceI expression, GFP-positive cells were readily detected, indicating DSB-induced gene conversion. Approximately 2% of cells from the two wild-type ES lines or the heterozygous Ku70 line were GFP-positive (Fig. 1B,C). In contrast, in cell lines deficient for either of the NHEJ proteins XRCC4 or DNA-PKcs, 4%–5% of transfected cells were GFP-positive, more than twofold higher than the control lines.
Strikingly, an even greater number of recombinants were obtained with the Ku70−/− cells, such that almost 12% of the transfected cell population was GFP-positive. We found a similar five- to sixfold increase in recombinants in Ku70−/− cells containing a related HDR substrate in which the GFP repeats are in opposite orientation (data not shown). GFP-positive cells derived from the Ku70−/− mutant were sorted by flow cytometry and then analyzed by Southern blotting to determine the nature of the repair event (Pierce et al. 1999). As with the J1 control cells, the DR-GFP reporter in the GFP-positive Ku70−/− cells had the structure expected for a noncrossover gene conversion event (data not shown). Complementation of the Ku70 mutation was also performed, by cotransfecting a Ku70 expression vector with the I-SceI expression vector. Levels of GFP-positive cells were reduced to that of the J1 cell line (H. Lee, A.P, and M.J., unpubl.), confirming that HDR is reduced with Ku expression.
To compare transfection efficiency, each of the cell lines was electroporated with a vector in which GFP is expressed from the same regulatory elements as I-SceI. A similar number of GFP-positive cells was found for each of the cell lines, indicating that the increased frequency of GFP-positive cells in the NHEJ mutants was not attributable to a higher transfection efficiency (data not shown). The increased number of GFP-positive cells in the NHEJ mutants after I-SceI expression also could not be accounted for by a generalized loss of cells with an unrepaired DSB, as cell counts 24 or 48 h after electroporation did not differ for the various cell lines. These results show, therefore, that each of the NHEJ mutants has an increased frequency of HDR in response to a DSB, but that the Ku70 mutant is distinct, with a much more pronounced increase than the XRCC4 or DNA-PKcs mutants.
Physical analysis of DSB repair
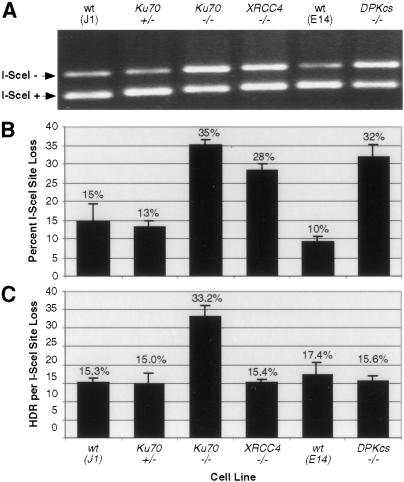
To compare HDR to other DSB repair processes, we amplified the genomic region surrounding the I-SceI site after expression of the endonuclease in cells. PCR primers flanked the I-SceI site (Fig. 1A) so as to amplify products from several types of repair: HDR, precise and imprecise NHEJ, and homologous deletion from single-strand annealing at the GFP repeats. To determine how much of the amplified fragment had a physically detectable repair product, we cleaved the fragment with I-SceI. For the wild-type and Ku70+/− cell lines, the I-SceI site was lost in 10%–15% of the amplified fragment, whereas for the NHEJ mutant lines it was lost in roughly 30%–35% of the amplified fragment (Fig. 2A,B). Because I-SceI generates precisely ligatable 4-base overhangs, the lower amount of the I-SceI+ PCR fragment suggests that the NHEJ mutants are impaired for precise NHEJ of a chromosomal DSB. Although we cannot definitively rule out that the I-SceI site was cleaved more efficiently in the NHEJ mutant cells, the similar transfection efficiency for each of the cell lines is evidence against such differences.
Figure 2.
NHEJ mutants show increased I-SceI site loss after DSB repair. (A) PCR assay to detect DSB repair. The genomic region surrounding the I-SceI break site was amplified from cell lines transfected with the I-SceI expression vector using primers shown in Figure 1A. PCR products were digested with I-SceI and resolved on agarose gels. (B) Fraction of I-SceI site loss from transfected cell populations. I-SceI+ and I-SceI− PCR products were quantified, with the fraction of amplified product from each cell population no longer digestible by I-SceI indicated. Note that because the smaller band incorporates proportionately less ethidium bromide, band intensities were normalized for product length. Error bars are ±1 S.D. (N = 3). (C) Fraction of I-SceI− PCR product from each population derived from HDR. The fraction of GFP-positive cells was normalized to the fraction of I-SceI− PCR product from each transfected cell population. Error bars are ±1 S.D. (N = 3).
We compared the levels of HDR and precise ligation by determining the proportion of GFP-positive cells relative to the percentage of the amplified fragment that had lost the I-SceI site (Fig. 2C). The XRCC4−/− and DNA-PKcs−/− lines had no more HDR relative to I-SceI site loss than the wild-type and Ku70+/− control cell lines, whereas the Ku70 mutant had more than a twofold increase. Therefore, although the increased HDR in the XRCC4−/− and DNA-PKcs−/− lines can be attributed to simple competition between the ligation and HDR pathways, the additional increase in HDR seen in the Ku70−/− line cannot be fully explained in this way.
Discussion
Our results show that lines deficient for NHEJ proteins have significantly increased levels of HDR of a chromosomal DSB, with a Ku70 mutant having a particularly striking increase compared to XRCC4 and DNA-PKcs mutants. Therefore, Ku has a distinct role from other NHEJ proteins in modulating DSB repair pathway choice in mammalian cells. Presumably, Ku80 and LigIV mutants would give similar results to the Ku70 and XRCC4 mutants, respectively, because the stability of Ku80 and LigIV are compromised by mutation of the other protein in the respective complex (Gu et al. 1997; Bryans et al. 1999). In contrast to HDR, SCE events and gene targeting, however, were not elevated in the NHEJ mutants. Using a physical assay, we also found that mammalian NHEJ proteins promote precise rejoining of a chromosomal DSB.
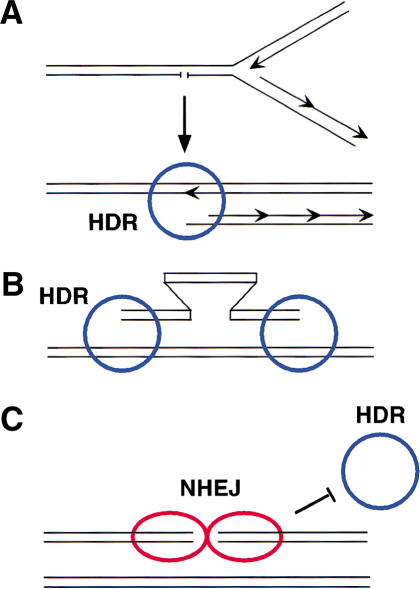
Although SCE, gene targeting, and HDR each involve homologous recombination, only HDR is elevated in the NHEJ mutants. What is notable is that the specific geometry of the DNA ends is likely to be different for each of these processes (Fig. 3), which we propose may account for the differential effects of NHEJ mutation. The etiology of spontaneous SCE events in mammalian cells is unknown; however, in Escherichia coli, DNA damage arising during replication is repaired by homologous recombination (Cox et al. 2000; Michel 2000), suggesting a mechanism for how SCEs might arise. When a replication fork encounters a nicked template, the fork will collapse and give rise to a one-ended DSB and an intact chromosome (Fig. 3A). Strand invasion of the one-ended DSB into the intact chromosome reconstitutes the collapsed fork, leading to the formation of a single Holliday junction, which can be resolved to give exchange or nonexchange sister chromatids. Exchange events (i.e., SCEs) are specifically predicted from an encounter with a nick on the lagging strand (Cromie and Leach 2000). Ku may not be involved in the regulation of this DSB repair process, because there is no other end with which the one-ended partially replicated sister could be joined.
Figure 3.
DNA ends and homologous recombination: one-ended versus two-ended DSBs. (A) Sister chromatid exchange. Replication fork collapse leads to a partially replicated chromosome with a one-ended DSB, which can invade the intact sister to restart the fork. As there is only one double-stranded end per fork collapse, ligation mediated by NHEJ proteins would not be predicted to be involved in its repair, and Ku would not block entry of the HDR machinery (blue circle). (B) Gene targeting. A linearized gene-targeting substrate involves the homologous invasion of a one-ended DSB with an intact chromosome, similar to SCE formation in A, but in gene targeting there is a one-ended DSB on each side of the targeting fragment. NHEJ-mediated ligation does not appear to be competitive with this reaction in mammalian cells, possibly because Ku is nonblocking for HDR proteins at these one-ended DSBs. (C) Classical DSB with two ends. I-SceI endonuclease generates a two-ended DSB, with the ends in close proximity. The NHEJ proteins (red ovals) mediate a ligation reaction that can compete with the HDR machinery. Even in the absence of an ability to ligate the ends (e.g., by loss of XRCC4 or DNA-PKcs), Ku can interact with the two-ended DSB and block entry by HDR proteins.
Likewise in the omega configuration of a gene-targeting substrate, as used in this study, alignment of the targeting arms with homologous sequences in the genome gives rise to one-ended DNA intermediates that can invade the chromosome (Fig. 3B). Thus, the reaction involves two one-ended DSBs that are inappropriately configured for NHEJ. Although it is possible for two noncomplementary ends of a targeting molecule (e.g., SacI and KpnI overhangs in this study) to be joined to each other, intramolecular rejoining events occur in only a fraction of transfected plasmids in mammalian cells (Liang and Jasin 1996), such that by itself this event is unlikely to be in competition with gene targeting, which requires only a single linear molecule.
In contrast to one-ended intermediates generated during SCE and gene targeting, a DSB in our repair substrate generated by I-SceI cleavage has two ends that are in close proximity to each other (Fig. 3C). That this two-ended DSB is a substrate for precise ligation mediated by the NHEJ machinery is shown by Figure 2B, where a greater amount of I-SceI site loss occurs in the NHEJ mutants than in wild-type lines. Although NHEJ and HDR have been shown to genetically interact to promote cell survival, as after radiation damage (Takata et al. 1998; Essers et al. 2000), our molecular analysis shows that both pathways compete for the repair of a defined lesion. Based on our data, we propose that Ku binding to both ends of a two-ended DSB inhibits access by the homologous recombination machinery (Fig. 3C), such that when Ku is absent, HDR is enhanced. The XRCC4 and DNA-PKcs mutant lines would therefore not be expected to show the same degree of enhanced HDR as the Ku mutant, because Ku acts upstream of the other NHEJ proteins and is still present in these cells to bind to the I-SceI-generated ends.
Physical studies have shown that Ku binds to DNA ends with a directed orientation (Yoo et al. 1999) and that Ku heterodimers interact on DNA (Cary et al. 1997). Possibly, binding to both ends of a two-ended DSB stabilizes contacts between Ku heterodimers, tethering the DNA ends and preventing access by the HDR machinery. The recent crystal structure of the Ku heterodimer (Walker et al. 2001) suggests how this could be achieved. Ku cradles the DNA on one face of the helix and could be predicted to block access of the DNA ends to Rad51 or to other recombination proteins, either directly or indirectly, in the latter case by preventing access to nucleases that would process DNA ends in preparation for HDR. Genetic and physical data would be consistent with such a role in preventing end-processing, as DNA ends are degraded to a greater extent in Ku mutants than in wild-type cells (Liang and Jasin 1996; Smith and Jackson 1999; Pellicioli et al. 2001). In the absence of stabilized heterodimeric complexes, as when Ku binds to a one-ended DSB, Ku may translocate internally from the single end, and not impede access by the HDR machinery. As NHEJ of two one-ended DSBs generated during replication by collapse of multiple forks would have the potential for translocations and other rearrangements, this may be a mechanism to protect the genome from deleterious NHEJ events.
In summary, the work presented here provides strong evidence that the two major DSB repair pathways in mammalian cells can be alternative pathways for repair of the same DSB, and even compete for repair of the same DSB. Because gross chromosomal rearrangements are observed in cells mutant for NHEJ or HDR components and each repair pathway appears to have tumor-suppressor function (Pierce et al. 2001), it will be important to determine how altered DSB repair pathway use compromises genetic integrity to promote tumor formation.
Materials and methods
SCE assays
Subconfluent ES cells cultured on 100-mm plates were incubated with BrdU (Sigma) at 20 μM for 48 h. Colcemid (Life Technology) was then added at a final concentration of 0.15 μg/mL for 1 h, after which time cells were harvested for metaphase preparation. Slides were aged for 3 d and stained with 10−4 M 33258 Hoechst (Sigma) for 10 min. After washing in water, slides were mounted and then placed under a 120-W Plant Lite (General Electric, Inc.) at a distance of 10 cm for 2 h. Slides were then washed and stained in 2% Giemsa (LabChem, Inc.) in Gurr's buffer (Bio/medical Specialties, Inc.) for 20 min, and then washed again and examined under an Olympus microscope mounted with a Sony CCD camera.
DNA manipulations and cell transfections
The DR-GFP reporter was cloned into the mouse hprt targeting vector pGPD351 (a kind gift of Greg Donoho; Donoho et al. 1998). The direction of transcription of the SceGFP gene is the same as that for hprt. After linearization with SacI/KpnI and subsequent purification, 70 μg of hprtDRGFP was electroporated into cells suspended in phosphate-buffered saline in a 0.4-cm cuvette at 800 V, 3 μF. Puromycin selection was applied 24 h later at 1 μg/mL for all lines except DNA-PKcs−/−, for which the final concentration was 2 μg/mL. After 6 d of puromycin selection, 6-thioguanine was added at a final concentration of 10 μg/mL for an additional 7 d, at which time surviving colonies were isolated. For HDR assays, 50 μg of the I-SceI expression vector pCBASce (Richardson et al. 1998) was electroporated into log-phase ES-cells suspended in 650 μL of PBS at 250 V, 1000 μF in a 0.4-cm cuvette. Flow cytometric analysis was performed on a Beckton Dickinson FACScan. To compare transfection efficiency, 15 μg of the GFP expression vector (pNZE-CAG; Pierce et al. 1999) was electroporated similarly, with the result that approximately half of the population became GFP-positive for each of the cell lines tested.
PCR assays
PCR reactions were performed with 2 μg of genomic DNA digested with SpeI. Primer sequences were DRGFP1, 5′-AGGGCGGGGTTCGGCT TCTGG; and DRGFP2, 5′-CCTTCGGGCATGGCGGACTTGA. Following amplification, PCR products were digested with I-SceI (Roche) and resolved on 1.2% agarose gels. Following staining with ethidium bromide, bands were quantified using a BioRad ChemiDoc System with rolling disc background subtraction. Results were normalized for product length and I-SceI cutting efficiency as measured on genomic DNA from untransfected cells (>95%). This method was verified to give a quantitative linear response over the ranges of I-SceI site loss described in this work by performing this assay using defined mixed amounts of genomic DNA from untransfected cells with genomic DNA from a pure population of transfected cells that express functional GFP, isolated on this basis by fluorescence-activated cell sorting.
Acknowledgments
We thank Jeremy Stark for helpful comments on the manuscript, and Kevin Mills, Yansong Gu, and Fred Alt for providing materials. This work was funded by NIH grant GM54688 (M.J.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL m-jasin@ski.mskcc.org; FAX (212) 717-3317.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.946401.
References
- Bryans M, Valenzano MC, Stamato TD. Absence of DNA ligase IV protein in XR-1 cells: Evidence for stabilization by XRCC4. Mutat Res. 1999;433:53–58. doi: 10.1016/s0921-8777(98)00063-9. [DOI] [PubMed] [Google Scholar]
- Cary RB, Peterson SR, Wang J, Bear DG, Bradbury EM, Chen DJ. DNA looping by Ku and the DNA-dependent protein kinase. Proc Natl Acad Sci. 1997;94:4267–4272. doi: 10.1073/pnas.94.9.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Sung P, Tomkinson AE. Interactions of the DNA ligase IV–XRCC4 complex with DNA ends and the DNA-dependent protein kinase. J Biol Chem. 2000;275:26196–26205. doi: 10.1074/jbc.M000491200. [DOI] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- Cromie GA, Leach DR. Control of crossing over. Mol Cell. 2000;6:815–826. doi: 10.1016/s1097-2765(05)00095-x. [DOI] [PubMed] [Google Scholar]
- Donoho G, Jasin M, Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol Cell Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dronkert ML, Beverloo HB, Johnson RD, Hoeijmakers JH, Jasin M, Kanaar R. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol Cell Biol. 2000;20:3147–3156. doi: 10.1128/mcb.20.9.3147-3156.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J, van Steeg H, de Wit J, Swagemakers SM, Vermeij M, Hoeijmakers JH, Kanaar R. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J. 2000;19:1703–1710. doi: 10.1093/emboj/19.7.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: Implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2896. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Chaudhuri J, Zhu C, Davidson L, Weaver DT, Alt FW. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity. 1998a;9:367–376. doi: 10.1016/s1074-7613(00)80619-6. [DOI] [PubMed] [Google Scholar]
- Gao Y, Sun Y, Frank KM, Dikkes P, Fujiwara Y, Seidl KJ, Sekiguchi JM, Rathbun GA, Swat W, Wang J, et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998b;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- Gu Y, Jin S, Gao Y, Weaver DT, Alt FW. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc Natl Acad Sci. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo PA. DNA breakage and repair. Adv Genet. 1998;38:185–218. doi: 10.1016/s0065-2660(08)60144-3. [DOI] [PubMed] [Google Scholar]
- Johnson RD, Jasin M. Double-strand-break-induced homologous recombination in mammalian cells. Biochem Soc Trans. 2001;29:196–201. doi: 10.1042/0300-5127:0290196. [DOI] [PubMed] [Google Scholar]
- Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- Liang F, Jasin M. Ku80 deficient cells exhibit excess degradation of extrachromosomal DNA. J Biol Chem. 1996;271:14405–14411. doi: 10.1074/jbc.271.24.14405. [DOI] [PubMed] [Google Scholar]
- Liang F, Han M, Romanienko PJ, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B. Replication fork arrest and DNA recombination. Trends Biochem Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- Pâques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol Cell. 2001;7:293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes & Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Stark JM, Araujo FD, Moynahan ME, Berwick M, Jasin M. Double-strand breaks and tumorigenesis. Trends Cell Biol. 2001;11:S52–S59. doi: 10.1016/s0962-8924(01)02149-3. [DOI] [PubMed] [Google Scholar]
- Richardson C, Jasin M. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol Cell Biol. 2000;20:9068–9075. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: Suppression of chromosomal translocations. Genes & Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijkers T, Van Den Ouweland J, Morolli B, Rolink AG, Baarends WM, Van Sloun PP, Lohman PH, Pastink A. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GC, Jackson SP. The DNA-dependent protein kinase. Genes & Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol Cell Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyck E, Stasiak AZ, Stasiak A, West SC. Binding of double-strand breaks in DNA by human Rad52 protein. Nature. 1999;398:728–731. doi: 10.1038/19560. [DOI] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- Yoo S, Kimzey A, Dynan WS. Photocross-linking of an oriented DNA repair complex. Ku bound at a single DNA end. J Biol Chem. 1999;274:20034–20039. doi: 10.1074/jbc.274.28.20034. [DOI] [PubMed] [Google Scholar]