Abstract
Zebrafish (Danio rerio) is a model organism to study the mechanisms and pathways of human disorders. Many dysfunctions in neurological, development and neuromuscular systems are due to glycosylation deficiencies, but the glycoproteins involved in zebrafish embryonic development have not been established. In this study, a mass spectrometry-based glycoproteomic characterization of zebrafish embryos was performed to identify the N-linked glycoproteins and N-linked glycosylation sites. To increase the number of glycopeptides, proteins from zebrafish were digested with two different proteases, chymotrypsin and trypsin, into peptides of different length. The N-glycosylated peptides of zebrafish were then captured by the solid phase extraction of N-linked glycopeptides (SPEG) method and the peptides were identified with an LTQ OrbiTrap Velos mass spectrometer. From 265 unique glycopeptides, including 269 consensus NXT/S glycosites, we identified 169 different N-glycosylated proteins. The identified glycoproteins were highly abundant in proteins belonging to the transporter, cell adhesion, and ion channel/ion binding categories which are important to embryonic, organ, and central nervous system development. This proteomics data will expand our knowledge about glycoproteins in zebrafish and may be used to elucidate the role glycosylation plays in cellular processes and disease. The glycoprotein data are available through the GlycoFish database (http://betenbaugh.jhu.edu/GlycoFish) introduced in this paper.
Keywords: N-linked glycoproteins, glycosylation, glycomics, glycoproteomics, proteomics, zebrafish (Danio rerio) embryos, Solid Phase Extraction of N-linked glycopeptides (SPEG), Mass spectrometry (MS), GlycoFish
Introduction
Zebrafish (Danio rerio) has become widely used as a model organism in recent years for the understanding of vertebrate development because of a number of advantageous features.1–2 This freshwater fish can produce 100–200 embryos in 2 or 3 months providing opportunities to create transgenic systems and to rapidly screen for the effects of different mutations at different developmental stages.3–4 Furthermore, the optically transparent fertilized zebrafish embryos develop externally which allows monitoring of the developmental processes such as cardiac and vascular systems, angiogenesis, and axon guidance.1, 5 Extensive genetic screens of zebrafish have also demonstrated the functional and morphological similarity of zebrafish to humans6–7 and studies over the past two decades have shown conserved pathways between zebrafish and humans.8 Indeed, as many of the mutations in the zebrafish resemble human disorders, zebrafish represent a promising model system to explore mechanisms and pathways in human diseases and therapy.7–8 Zebrafish has been used to understand cardiovascular, skeletal, kidney disorders and Huntington’s, Alzheimer’s, and Parkinson’s diseases.6, 9–11 In addition, zebrafish is used in drug discovery studies to evaluate potential therapeutics for human diseases since some of the pharmacological agents have exhibited similar effects on zebrafish and humans.12–13
Glycosylation is one of the most important post translational modifications since it can be critical for the stability, conformation, and function of proteins.14 One of two major protein glycan modifications, N- glycosylation is the attachment of an oligosaccharide moiety to the amino group of an asparagine residue of the N-X-S/T polypeptide sequence where X is any amino acid other than proline. N-glycosylated proteins are highly involved in the cell membrane and extracellular matrix processes because of their crucial roles in cell-cell and cell-matrix signaling interactions.14–15 Many of the proteins in the central nervous system (CNS) or peripheral nervous system (PNS) are N-glycosylated and a glycosylation deficiency in the CNS may lead to a neurological disorder. The existence of complex type N-glycan structures in the zebrafish3 and the conservation of proteins between zebrafish and mammals12 suggest that N-linked glycosylation of the proteins in zebrafish can be important to cellular function and potentially relevant to human diseases. Zebrafish is widely used as a model for many human related diseases and drugs, the genome of the zebrafish is almost complete, and there are studies to establish a complete proteome profile15–16 yet there is no data available on the N-glycosylation of proteins of the zebrafish. The identification of N-glycosylation sites will help to provide a glycoproteome profile for zebrafish and may enhance our knowledge on the role of glycosylation and glycoproteins in disease states.
Therefore, our aim in this project was to identify specific N-glycosylated proteins and the N-linked glycosites of zebrafish using mass spectrometry. To achieve that goal, solid phase extraction of N-linked glycopeptides (SPEG)17–18 was used to enrich the N-glycosylated proteins. SPEG is a technique for identification of N-glycosites developed by our group.17–20 SPEG technique uses hydrazide chemistry to immobilize sugar containing peptides or proteins on hydrazide beads. The formation of chemical hydrazone bonds between the glycans and beads enables glycosylated peptides to be separated from the non-glycosylated peptides. Then the captured peptides can be hydrolyzed with glycosidases followed by analysis using mass spectrometry. Recent studies have shown the efficiency of chemical immobilization for the N-linked glycopeptide capture.21 Previously, SPEG coupled LC/MS methods have been successfully used to identify the N-glycosylated proteins of plasma, tissue, and cancer cell lines.22 Furthermore, this study introduces the newly established GlycoFish database, http://betenbaugh.jhu.edu/GlycoFish,23 as a public resource for housing the experimentally identified N-linked glycoproteins and glycopeptides of zebrafish through its web interface.
Experimental Procedures
Materials
Instant Ocean for zebrafish pools was purchased from Marinelabs (Columbus, OH). Sequencing grade trypsin and chymotrypsin were purchased from Promega (Madison, WI). Affiprep hydrazide resin beads and sodium periodate were from Bio-Rad (Hercules, CA). PNGaseF was from New England Biolabs (Ipswich, MA). Trifluoroethanol (TFE) was purchased from Sigma-Aldrich (Milwaukee, WI) and iodoacetamide was purchased from Sigma-Aldrich (St. Louis, MO). Tris (2-carboxyethyl) phosphine (TCEP) was from Pierce (Rockford, IL), BCA protein assay kit was from Thermo Scientific Pierce (Rockford, IL), and the 1 mL C18 Sep-Pak cartridges were from Waters (Milford, MA). Furthermore, C18 column (75 μm × 10 cm, 5 μm, 120Å, Magic C18) (MicromBioresources, Auburn, Ca), LTQ Orbitrap Velos mass spectrometer (Thermo Scientific-Germany), Mass spectrometer emitter tip (1 μm) (New Objective, Woburn, MA), and Mascot Software (Matrix Science, Boston, MA) were used in this experiment. All other chemicals used in this study were purchased from Sigma-Aldrich (St. Louis, MO).
Zebrafish Preparation
Wild type zebrafish embryos were collected at 0 hour post fertilization and kept at 28°C in embryo water (60 μg/mL Instant Ocean in water) for optimum growth of embryos. Embryos were frozen 48 hour post fertilization at −80°C until processing for glycopeptides capture.
Enzymatic Digestion
Thirty milligrams of zebrafish embryos were homogenized in PBS buffer containing 1% SDS. In order to extract all the proteins, the solution was mixed, sonicated, and then heated at 65°C for 10 minutes. The cuticle debris was removed and the solubilized protein concentration was determined to be 6370 μg/mL using BCA assay. Urea and TCEP were used for the denaturation and reduction of the proteins, respectively. Urea was added until the concentration was 8M and TCEP was added until the concentration was 10 mM and the samples were incubated with gentle shaking at 60°C for two hours. The proteins were alkylated with 12 mM iodoacetamide in the dark at room temperature for 30 minutes. Then three 800 μg protein sample replicates were prepared for each trypsin and chymotrypsin digestion. Firstly, the SDS percentage of the solutions was reduced to 0.1% for trypsin reaction and 0.025% for chymotrypsin digestion by diluting with trypsin buffer (100 mM KHPO4 (pH 7.6–8) and CaCl2 to a final concentration of 1 mM). The trypsin/chymotrypsin to protein ratio was adjusted to 1 to 50 and the pH of the solution was adjusted to 7–8. The enzymatic reaction took place overnight at 37 °C with gentle shaking. In order to ensure the completeness of the enzymatic reaction, the samples from before and after trypsin/chymotrypsin addition were run on the SDS-PAGE gel using the silver staining method. Then the digested peptides were desalted using C18 SepPak columns. The C18 SepPak column was first prewashed with 1 mL of 80% ACN in 0.1% TFA and then washed twice with 0.1% TFA. The pH of the samples was adjusted to below 3 and then the samples were slowly loaded into the column. Before eluting the samples, the column was washed three times with 1 mL of 0.1% TFA. Then the samples were eluted with 700–800 μL of 80% ACN in 0.1% TFA. After the desalting procedure, the samples were vacuum-dried stored at −20 °C.
Chemical Immobilization and Glycosidase Digestion
The dried peptides were dissolved in 0.1% trifluoroacetic acid (TFA) and 5% acetonitrile (ACN) solution. The proteins were oxidized with 10 mM sodium periodate at room temperature in the dark for 1 hour. The oxidized samples were immediately desalted with a C18 column and at the last step of C18 desalting; peptides were eluted with 800 μL of 80% ACN and 0.1% TFA. 50 μL of (50% slurry) hydrazide resins were prepared for each coupling reaction. The resins were first spun down at 3000 rpm for 30 seconds and then washed with 1 mL of deionized water. After this, the aldehydes were immobilized on the solid hydrazide support overnight at room temperature with gentle shaking. Next, the non-glycosylated peptides were removed by washing the immobilized resins with 800 μL of 1.5 M NaCl and water.
In order to release the N-glycosylated peptides from the immobilized support, the resins were resuspended in 50 μL of G7 reaction buffer (50 mM sodium phosphate at pH 7.5) with 2 μL PNGaseF at 37 °C and overnight shaking. The supernatant was transferred into a glass vial and the resins were washed twice with 100 μL of water. The washes and the supernatant were then combined in a glass vial and its pH was adjusted with hydrochloric acid (HCl) to below 3. The contents of the glass vial were applied to a C18 SepPak column and the eluted samples were dried in a SpeedVac.
LTQ OrbiTrap Velos Mass Spectrometer Analysis
Samples were injected into a 10 μL volume of starting mobile phase (2% CAN in 0.1% formic acid) with a nanoaquity liquid chromatography (LC) system containing a 75 μm × 15 cm C18 column at 750 nL/min. Samples were loaded for 15 minutes before switching the sample loop out of the flowpath and decreasing the flow to 300 nL/min. The peptides were then separated by gradient elution at 300 nL/min over 90 minutes and injected onto the Velos Orbitrap electrospray ionization mass spectrometer running at a resolution of 30,000 for precursors and 15,000 for fragment ions. The data was collected in data dependent mode with the top ten precursors selected for MS/MS analyses. MS/MS spectra were acquired in higher-energy collision dissociation (HCD) mode with normalized collision energy of 35%. Dynamic exclusion was set for 30 seconds with a 20 ppm window and monoisotopic precursor selection (MIPS) was enabled.24 Each sample was analyzed in the LTQ OrbiTrap Velos mass spectrometer three times to increase the confidence of peptide and glycosite identification and assess the reproducibility.
The MS/MS data were searched against the NCBI Danio Rerio (zebrafish) database using Mascot software. The enzyme was set to trypsin or chymotrypsin. Deamidation (NQ) and oxidation (M) were set as variable modifications and carbamidomethylation (C) was set as a fixed modification. These modifications were necessary to identify the glycopeptides because of the chemical reactions that can occur during the SPEG method. Carbamidomethylation modification was possible due to the alkylation reaction that occurred during the iodoacetamide incubation. Furthermore, PNGaseF was used to cleave captured peptides from the hydrazide resin. This cleavage leads to a 1 Da increase because of the conversion of asparagine to aspartic acid, which is equivalent chemically to deamidation of the asparagine. Such modifications have been included in the informatics analysis in order to identify the glycosylated peptides. The data files were then uploaded to Scaffold3 for viewing 24.
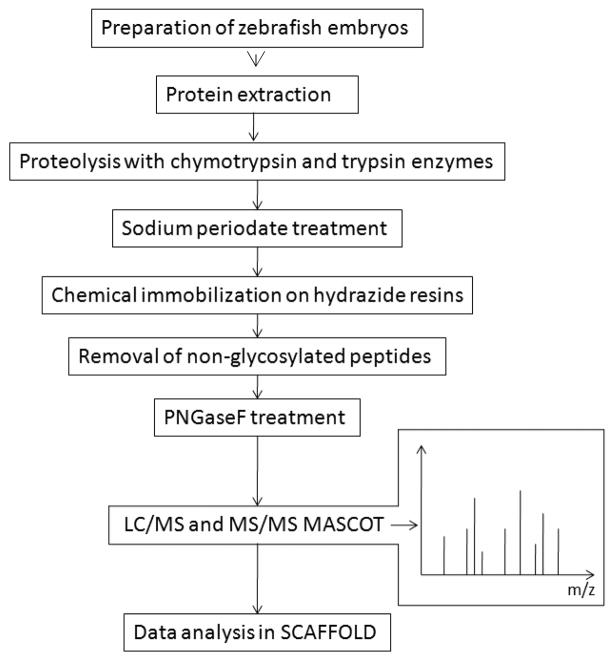
Results
Zebrafish embryos were first homogenized and then the proteins were extracted, denatured, and reduced in urea, alkylated with TCEP, and iodoactemide. In order to increase the identification of N-glycosylated peptides, the denatured zebrafish proteins were proteolyzed with two different enzymes, either chymotrypsin or trypsin. The digested peptides were oxidized with sodium periodate to convert the cis-diol groups of the carbohydrates to aldehydes, which were then immobilized on hydrazide beads by a coupling reaction. Next, the nonglycosylated peptides were washed away and the glycosylated peptides were cleaved from the glycans and beads by treating with PNGaseF. The eluted N-glycosylated peptides were analyzed with LC-MS/MS in a LTQ OrbiTrap Velos Mass Spectrometer three times.24 A schematic representation of the SPEG method used in this study is shown in Figure 1. The proteins that were identified in all three repeats for chymotrypsin or trypsin digested samples are listed in the Supporting information Table S-2 and online database.
Figure 1.
Schematic representation of SPEG coupled LC/MS method for zebrafish glycopeptide analysis.
The MS/MS spectra were used to search against the NCBI database using MASCOT software. The data was compiled in a Scaffold file for viewing and statistical analysis. A total of 855 peptide identifications from trypsin and chymotrypsin digestion were assigned. Six hundred seventy-six of these peptide identifications included the consensus N-X-S/T glycosylation sites, showing the capability of SPEG method to enrich the N-linked glycopeptides containing consensus N-linked glycosylation sites. While a number of peptides were observed multiple times, two hundred sixty-five of these six hundred seventy-six glycopeptide identifications were unique glycopeptide sequences. Shown in Table 1 is detailed information on peptides and proteins detected from the chymotrypsin and trypsin digests.
Table 1.
Total number of peptide identifications, number of identified proteins and peptides from trypsin and chymotrypsin digested zebrafish samples.
| # | Total number of identifications for peptides or proteins | NXT/S motif containing identifications for peptides or proteins | Percentage of NXT/S motif containing peptides/proteins | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Trypsin digestion | Chymotrypsin digestion | Total | Trypsin digestion | Chymotrypsin digestion | Total | Trypsin digestion | Chymotrypsin digestion | Total | |
| Identified Peptides | 724 | 131 | 855 | 563 | 113 | 676 | 78% | 86% | 79% |
| Unique Peptides | 324 | 50 | 374 | 229 | 36 | 265 | 71% | 72% | 71% |
| Identified Proteins | 193 | 34 | 227 | 154 | 31 | 185* | 80% | 91% | 82% |
Note that 16 of 185 glycosylated proteins identified in both chymotrypsin and trypsin digestions were identical.
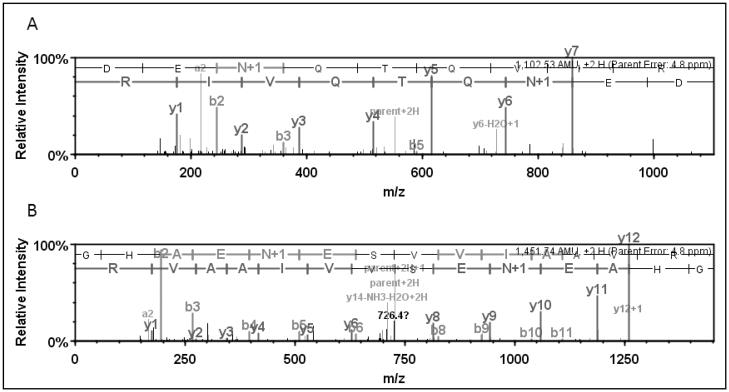
Two hundred twenty-nine different glycopeptides were uniquely detected from trypsin digests whereas only 36 unique glycopeptides were detected from the chymotrypsin digests. A total of 185 N-glycosylated proteins containing consensus glycosylation sites were specified from 265 unique glycopeptides. Interestingly, 16 of 31 glycoproteins identified in chymotrypsin digests were also found in the trypsin digest together with an additional 138 unique glycoproteins. As a result, a total of 169 different N-glycosylated proteins were identified by either chymotrypsin digestion, trypsin digestion or both (Figure 2). Some of these glycoproteins such as nicastrin included multiple different N-glycopeptides. The MS/MS spectra of the two unique glycopeptides identified in nicastrin following trypin digestion are shown in Figure 3.
Figure 2.
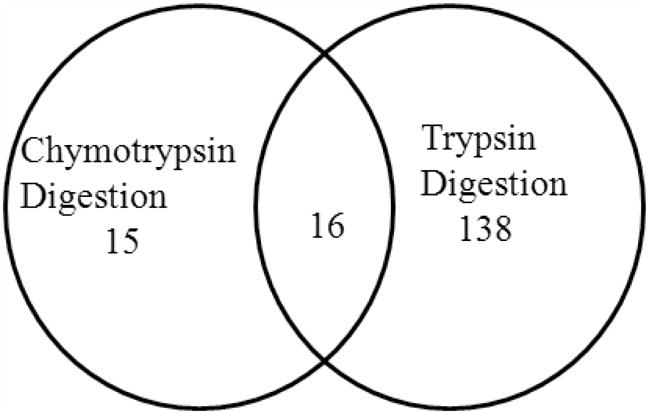
The number of commonly or uniquely detected N-glycosylated proteins in zebrafish embryos from chymotrypsin and trypsin digests.
Figure 3.
MS/MS spectra of identified N-glycosylated peptides of nicastrin A) The deamidated form of the glycopeptide (K)DEnQTQVIR(K) (top panel) and B) the deamidated form of glycopeptide (R)GHAEnESVVIAAVR(L) (bottom panel) were identified following trypsin digestion.
Although the peptide sequences were only 10 or 11 amino acids long, four of the unique glycoproteins contained 2 different N-glycosylation sites in their detected peptide sequences. The other 165 proteins had one N-glycosylation site in each of their peptides, giving a total of 269 different consensus N-glycosites as illustrated in Table S-1 in supporting information. One hundred eighty three of these peptides contained the NXT motif whereas only 86 of them contained the NXS motif.
In order to obtain a better understanding of the zebrafish glycoproteins, the detected proteins were searched using the zebrafish model organism database (ZFIN - http://zfin.org).25 The putative molecular and biological functions were specified for each of the glycoproteins. Detailed information on these proteins, including peptides detected, N-glycosylation sites, and functions, is listed in Table S-2 of the supporting information. Moreover, these findings for each zebrafish glycoprotein are catalogued in the GlycoFish Database freely accessible online (http://betenbaugh.jhu.edu/GlycoFish).23
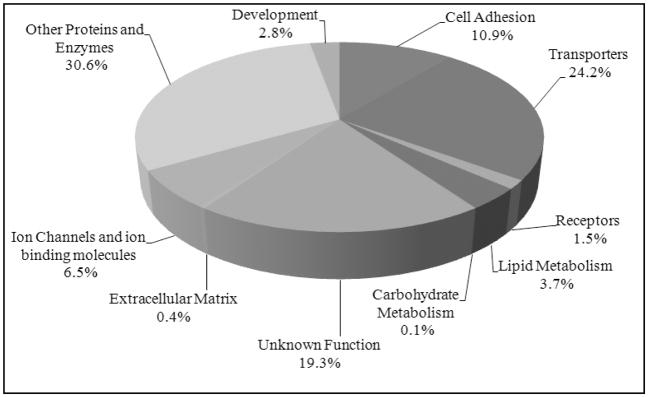
The Scaffold program allows mass spectrometrists to manage large amounts of proteomic data, including quantification of the spectral counts of each protein. In order to estimate the distribution of glycoproteins collected from the zebrafish embryos, the number of spectral counts for each glycoprotein was determined. The ratios of spectral counts of each protein divided by the total counts indicates the abundance of a glycoprotein. We categorized the glycoproteins into 10 groups based on their putative functions. The distribution diagram of the N-glycosylated proteins according to their functional category in the zebrafish embryo is shown in Figure 4.
Figure 4.
Distribution of the N-glycosylated proteins of zebrafish embryos based on their putative functions.
The identified proteins were categorized into transporters (24.2%), cell adhesion (10.9%), ion channels and ion binding (6.5%), lipid metabolism (3.7%), development (2.8%), and receptors (1.5%). The ability of the method to capture ion channels, cell adhesion molecules, and receptors demonstrates the capacity of the methodology to enrich membrane and related proteins. Other molecules having functions different than the specified categories were grouped as other proteins and enzymes (30.6%). A large number of proteins were not previously characterized, so those were categorized in the unknown function group (19.3%). All of these proteins, their molecular and biological function and their captured peptide sequences are listed in Supporting Information Table S-2 under their functional category and in the GlycoFish Database as described below.
GlycoFish Database
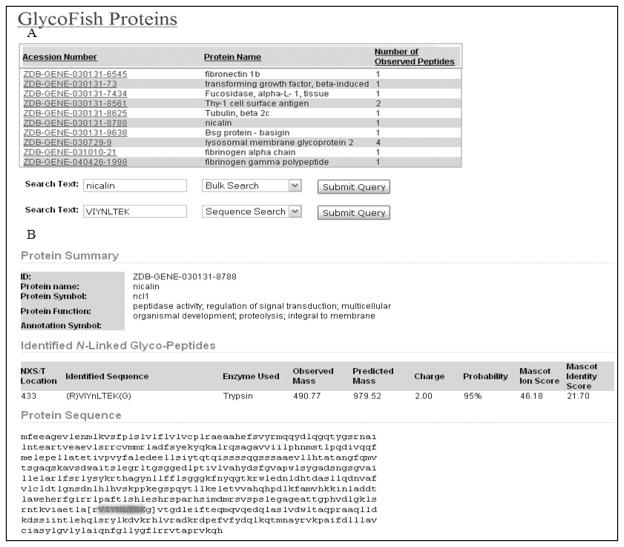
The GlycoFish database (http://betenbaugh.jhu.edu/GlycoFish)23 was established to provide public access to the glycoproteomics data of zebrafish embryos. Php coding in a mySQL database was used to program this website.26 All the experimentally detected glycopeptides and glycoproteins and their specificities can be found using the web interface of this database. Example outputs from the website are shown in Figure 5A and 5B.
Figure 5.
An example output for an identified N-glycopeptides from the GlycoFish database (http://betenbaugh.jhu.edu/GlycoFish)23. A) The accession number, protein name, and the number of observed glycosylated peptides are presented on the main page of the website. B) After clicking on the protein of interest, detailed information on the protein can be seen. The protein name, ZFIN ID, protein symbol, and protein function are presented in the protein summary section of the website. The detected N-glycosylated peptides with their observed and predicted mass and also confidence score are given in the second section. The third section gives the protein sequence highlighted with the experimentally detected N-glycopeptides and N-glycosites.
The detected glycoproteins, their accession numbers from the Zebrafish Model Organism (ZFIN) database25 and the number of observed glycopeptides for each protein are listed on the main page of the website as shown in Figure 5A. In addition to the protein list, bulk and sequence search functions are available on the main page. The protein of interest can be found by searching protein name, protein symbol, accession number or protein function in the Bulk Search feature. Furthermore, a protein of interest can be found by searching any peptide or protein sequence up to 256 amino acids using the sequence search feature of the website. Once the proteins are listed that meet the search criteria, one can click on the link for the protein of interest to learn more detailed information.24
For each protein entry, information available for that protein is summarized in three sections, as shown in Figure 5B for an example membrane protein, nicalin. These three sections include a protein summary, identified N-linked glycopeptides and the protein sequence. In the protein summary, general information on the protein such as its ZFIN ID (http://zfin.org);25 name, symbol, and a functional category is provided. As illustrated in Figure 5B, nicalin is a glycoprotein that functions in multicellular organism development. In the second section, features of the identified N-linked glycos are listed in nine columns. The NXS/T location of the captured N-glycan site in the protein sequence is given in the first column. The captured peptide sequence is given in the second column. The next column lists the proteolytic enzyme (i.e. chymotrypsin or trypsin) that was used to capture that specific peptide. The observed mass of the captured peptide in the mass spectrometer is given in the fourth column. Once the observed mass is known, the actual (average) mass can be predicted using:
where M stands for the actual/predicted mass, z is the charge of the ion, and H is the number of hydrogen atoms.24 The mascot program calculates the predicted mass of the peptides, which can be viewed using the Scaffold program. The predicted mass of the peptide calculated by the Mascot program is provided in the fifth column. The charge of the peptide is given in the sixth column and the seventh column provides the confidence score of the analysis given as a probability. A majority (>90%) of 265 N-linked glycopeptides have been identified with greater than 95% confidence. The identified N-linked glycopeptides with lower than 95% confidence (20 of N-linked glycopeptides) are also included in the database with their probability values to provide additional information for the readers. The Mascot ion score and Mascot identity score are provided in the eighth and ninth columns, respectively. In the third section, the protein sequence is shown, including the experimentally determined N-glycosylated peptide, which is highlighted to be easily distinguishable (see protein sequence in Figure 5B). The GlycoFish database will allow the integration of additional zebrafish glycoproteins to the website by contributing laboratories. Thus, GlycoFish is designed to serve as a continually updated public resource for the N-glycosylated proteins of zebrafish.
Discussion
Zebrafish embryos are appropriate model organisms for both developmental and central nervous system studies because of their functional and morphological similarities to humans.6–7 Glycosylation can play a critical role in cell-cell and protein-protein communication, and signaling,14 and for this reason, glycosylated proteins are often involved in developmental processes,27 as well as in the central and peripheral nervous system.28 Zebrafish central nervous system development is almost complete by 48 hours and interestingly many of the pathways in the nervous system are conserved between zebrafish and humans.12 Although, there are many ongoing efforts involving genomic and proteomic studies of Danio rerio, glycoprotein characterization of the zebrafish system has not been well studied. Glycosylation of some individual proteins has been evaluated before,29–30 however, there has not been previous glycoproteomics analysis of zebrafish to our knowledge. In order to understand the potential role of glycosylation on the biological processes, the glycoproteins and their respective glycosylation sites need to be elucidated first. Different strategies exist to fractionate the N-glycosylated proteins of the organisms. These strategies can be coupled with proteomics to investigate a broad spectrum of proteins. Lectin affinity chromatography and solid phase extraction of (N-linked) glycopeptides (SPEG) are two common methods to analyze glycoproteins. SPEG can be advantageous both for small sample sizes and for enrichment of low abundance glycoproteins from complex mixtures.21, 24 Thus, SPEG has been coupled to an LC/MS method in this study to detect and identify the N-linked glycosites of zebrafish embryos. Zebrafish embryos were chosen because they are widely used as a model system for understanding developmental processes especially the neuronal system.12 Further, glycosylation and glycoproteins are often associated with neural and organ development.3, 31
Chymotrypsin and trypsin are two proteolytic enzymes which have different specific cleavage sites. In this study, both of these enzymes were used to generate different populations of N-glycosylated peptides. Although chymotrypsin has more cleavage sites compared to trypsin, the number of glycopeptides detected from the trypsin digests was almost five times higher (Table 1). There are several possible reasons for fewer glycoproteins detected by chymotrypsin digestion. The optimal m/z ratio for detection with our mass spectrometric analysis was in the range of 500 to 5000. Peptides cleaved by trypsin (at K or R) were generally in this range, whereas the peptides cleaved by chymotrypsin (at Y, W, or L) were often shorter or longer. A second possible cause is that the probability of detection by mass spectrometry of lysine (K) or arginine (R) amino acids at the C-terminus of trypsin digested peptides is higher than other amino acid. In addition, the efficiency of chymotrypsin digestion may need to be further optimized or alternatively other proteolytic enzymes can be considered.
One hundred sixty-nine unique zebrafish proteins were identified as N-glycosylated in this glycoproteomics study.23 Identifications of several previously known zebrafish glycoproteins, such as vitellogenin, sarcoglycan, and others in this study strengthen the reliability of the findings by the SPEG method.29–30 Vitellogenin, the major yolk protein of zebrafish, has been shown to be glycosylated and the mass of its N-glycosylated peptides were estimated by western blot studies.29 Also, Guyon et al. studied the glycosylation and glycosites of sarcoglycans in zebrafish embryos.30 Significantly, vitellogenin and sarcoglycan were both identified as glycosylated from a very few embroyos in the current study to authenticate the sensitivity and effectiveness of the SPEG coupled MS method at least for these two glycoproteins.
The fraction of the proteins having the consensus NXS/T motif was 82% of all the proteins identified from the trypsin and chymotrypsin digests which indicates the effectiveness of the SPEG method for capturing the N-linked glycopeptides. Sixty-eight percent of the digested peptides had a NXT motif whereas only 32% of them included the NXS motif. The abundance of the NXT motif may be explained by the conformational compatibility of threonine for N-glycosylation when compared to the serine amino acid.32 Alternatively, proteins without either consensus NXS/T glycosylation sites represented 18% of the total that may or may not be glycosylated. These peptide sequences included some atypical motifs such as N-X-V motif which were designated as glycosylated in this study. Interestingly, these motifs are also conserved in a number of species including Oryzias latipes, Homo sapiens, and other fish species. In addition, these atypical motifs are also observed as glycosylation sites in Archaea in previous studies,33 strengthening their probability of being glycosylated. However, these proteins have not been included in the glycosylated protein database because of the lack of knowledge concerning atypical sequence motifs. Further analysis is needed on the individual proteins for a full characterization of their glycosylation status. The identification of peptides that did not have any potential glycosylation sites including either NXS/T or NXV may result from two sources: 1) Nonspecific binding of proteins, 2) Incorrectly assigned peptide sequences (false positives) based on the MS/MS spectra.22
The analysis of the zebrafish embryo bodies resulted in a broad spectrum of glycoproteins, so the identified N-glycosylated proteins were grouped into 10 general categories based on their putative or known biological functions. The majority of these proteins were classified as transporters, cell adhesion molecules, and ion channel/ion binding molecules. Vitellogenin, the a yolk glycophosphoprotein, had the highest spectral count ratio.29 Many of the other identified proteins in this study have roles either in the central nervous system, development, or the muscular system indicating the importance of glycosylation for these systems. For example, laminin alpha 1 is a cell adhesion molecule with roles in axon guidance, the central nervous system, axonogenesis, neuron migration, and optic nerves.34–37 Cadherin 2 (N-Cadherin) is a calcium ion binding protein important for synapse assembly, neural tube development, the peripheral nervous system, and brain development.38–40 Two unique deamidated glycopeptides were found for the nicastrin protein. Nicastrin is one of the essential transmembrane proteins involved in the activity and integrity of the γ-Secretase complex, which proteolyzes the β-amyloid precursor protein. The accumulation of some proteolyzed amyloid-β peptides can be neurotoxic and initiate Alzheimer’s disease.41–43 The secretase complex also proteolyzes notch, which is critical for developmental processes.41–43 Interestingly, nicastrin is the only glycosylated protein in this complex44 and its glycosylation is associated with execution of γ-Secretase cleavage.45 In addition, one N-glycosite was identified in this study on nicalin, a nicastrin-like-protein. The importance of nicalin and nicastrin in cell fate and development system decisions is well known.43 The potential role glycosylation plays for these proteins could be elucidated further using zebrafish as a model system to obtain potential insights into Alzheimer’s disease and development. Furthermore, both the sarcoglycan and sarcolemmal membrane related proteins, sarcalumenin and sarcoma amplified sequence, identified as glycosylated in this study, are important to the dystrophic pathology of skeletal and cardiac muscles. The dystrophin-associated sarcoglycan localizes in the sarcolemmal membrane of muscle cells and plays a critical role in muscle development.46 Guyon et al. showed that mutations in the sarcoglycan of zebrafish caused muscle disorganization and uninflation of swim bladders, leading to limb girdle muscular dystrophy.30 It is also known that various types of dystrophies are due to defects in glycosylation47–48 and interestingly the transmembrane region and N-glycosites of sarcoglycan are highly conserved between humans and zebrafish.30 The characterization of these glycosites on these proteins in the current study may be useful for a better understanding of the role of glycosylation in muscular dystrophies.
Furthermore, the characterization of the glycosylation sites on these 169 proteins can indicate a role for glycosylation in protein function. Indeed, a number of proteins detected in the current study (53 of 169) do not have any reported function in the literature. Experimentally proving that these proteins are N-glycosylated in the zebrafish embryos is a first step towards beginning to elucidate their potential functions. Indeed, these glycoproteins may have homologs in other organisms including humans. Characterizing glycosites of the proteins can be useful to comprehending their function and a possible role in associated human diseases.
Conclusion
Although zebrafish is a common model organism, profiling of its glycoproteins has not been established. This study has served to identify numerous N-glycosylated proteins and their glycosylatsites in zebrafish embryos. This glycoproteomic data is also available online at the newly established GlycoFish website for public use. Many of the proteins play significant roles in the neuromuscular system and embryonic development and their N-glycosylation status will provide insights into understanding the potential role glycosylation plays in neurological, muscular, and developmental processes and disorders.
Supplementary Material
Acknowledgments
This work was supported by National Scientific Foundation by Grant NSF-EFRI: 0736000. This work was also supported by federal fund from the National Cancer Institute, National Institutes of Health, by Early Detection Research Network (NIH/NCI/EDRN) grant U01CA152813. We thank Johns Hopkins University School of Medicine (JHUSOM) Mass Spectrometry and Proteomics Facility for their effort in this study.
Footnotes
Supporting Information Available
Supplementary Tables: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Neumann CJ. Semin Cell Dev Biol. 2002;13(6):469. doi: 10.1016/s108495210200099x. [DOI] [PubMed] [Google Scholar]
- 2.Grunwald DJ, Eisen JS. Nat Rev Genet. 2002;3(9):717–24. doi: 10.1038/nrg892. [DOI] [PubMed] [Google Scholar]
- 3.Guerardel Y, Chang LY, Maes E, Huang CJ, Khoo KH. Glycobiology. 2006;16(3):244–57. doi: 10.1093/glycob/cwj062. [DOI] [PubMed] [Google Scholar]
- 4.Detrich HW, WM, Zon LI. The Zebrafish: Biology. Vol. 59. Academic Press; San Diego, California, USA: 1999. [Google Scholar]
- 5.Muller B, Grossniklaus U. J Proteomics. 2010;73(11):2054–63. doi: 10.1016/j.jprot.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Tay TL, Lin Q, Seow TK, Tan KH, Hew CL, Gong Z. Proteomics. 2006;6(10):3176–88. doi: 10.1002/pmic.200600030. [DOI] [PubMed] [Google Scholar]
- 7.Shin JT, Fishman MC. Annu Rev Genomics Hum Genet. 2002;3:311–40. doi: 10.1146/annurev.genom.3.031402.131506. [DOI] [PubMed] [Google Scholar]
- 8.Abramsson A, Westman-Brinkmalm A, Pannee J, Gustavsson M, von Otter M, Blennow K, Brinkmalm G, Kettunen P, Zetterberg H. Zebrafish. 2010;7(2):161–8. doi: 10.1089/zeb.2009.0644. [DOI] [PubMed] [Google Scholar]
- 9.Karlovich CA, John RM, Ramirez L, Stainier DY, Myers RM. Gene. 1998;217(1–2):117–25. doi: 10.1016/s0378-1119(98)00342-4. [DOI] [PubMed] [Google Scholar]
- 10.Son OL, Kim HT, Ji MH, Yoo KW, Rhee M, Kim CH. Biochem Biophys Res Commun. 2003;312(3):601–7. doi: 10.1016/j.bbrc.2003.10.163. [DOI] [PubMed] [Google Scholar]
- 11.Leimer U, Lun K, Romig H, Walter J, Grunberg J, Brand M, Haass C. Biochemistry. 1999;38(41):13602–9. doi: 10.1021/bi991453n. [DOI] [PubMed] [Google Scholar]
- 12.Guo S. Expert Opin Drug Discov. 2009;4(7):715–726. doi: 10.1517/17460440902988464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pichler FB, Laurenson S, Williams LC, Dodd A, Copp BR, Love DR. Nat Biotechnol. 2003;21(8):879–83. doi: 10.1038/nbt852. [DOI] [PubMed] [Google Scholar]
- 14.Jones J, Krag SS, Betenbaugh MJ. Biochim Biophys Acta. 2005;1726(2):121–37. doi: 10.1016/j.bbagen.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 15.De Souza AG, MacCormack TJ, Wang N, Li L, Goss GG. Zebrafish. 2009;6(3):229–38. doi: 10.1089/zeb.2009.0591. [DOI] [PubMed] [Google Scholar]
- 16.Singh SK, Sundaram CS, Shanbhag S, Idris MM. Zebrafish. 2010;7(2):169–77. doi: 10.1089/zeb.2010.0657. [DOI] [PubMed] [Google Scholar]
- 17.Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. Nat Protoc. 2007;2(2):334–9. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Li XJ, Martin DB, Aebersold R. Nat Biotechnol. 2003;21(6):660–6. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y, Aebersold R, Zhang H. Anal Chem. 2007;79(15):5826–37. doi: 10.1021/ac0623181. [DOI] [PubMed] [Google Scholar]
- 20.Tian Y, Kelly-Spratt KS, Kemp CJ, Zhang H. Clin Proteomics. 2008;4(3–4):117–136. doi: 10.1007/s12014-008-9014-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H. Curr Protoc Protein Sci. 2007;Chapter 24(Unit 24):3. doi: 10.1002/0471140864.ps2403s48. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Loriaux P, Eng J, Campbell D, Keller A, Moss P, Bonneau R, Zhang N, Zhou Y, Wollscheid B, Cooke K, Yi EC, Lee H, Peskind ER, Zhang J, Smith RD, Aebersold R. Genome Biol. 2006;7(8):R73. doi: 10.1186/gb-2006-7-8-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.GlycoFish: A Database for Zebrafish Glycoproteins. http://betenbaugh.jhu.edu/GlycoFish.
- 24.Baycin Hizal D, Tian Y, Akan I, Jacobson E, Clark D, Chu J, Palter K, Zhang H, Betenbaugh M. Journal of Proteome Research. 2011 doi: 10.1021/pr200004t. in publish. [DOI] [PubMed] [Google Scholar]
- 25.ZFIN: The Zebrafish Model Organism Database. http://zfin.org/
- 26.MySQL. http://www.mysql.com/
- 27.Haltiwanger RS, Lowe JB. Annu Rev Biochem. 2004;73:491–537. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi Y. Biochim Biophys Acta. 2002;1573(3):369–76. doi: 10.1016/s0304-4165(02)00405-1. [DOI] [PubMed] [Google Scholar]
- 29.Fan X, Klein M, Flanagan-Steet HR, Steet R. J Biol Chem. 2010;285(43):32946–53. doi: 10.1074/jbc.M110.158295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guyon JR, Mosley AN, Jun SJ, Montanaro F, Steffen LS, Zhou Y, Nigro V, Zon LI, Kunkel LM. Exp Cell Res. 2005;304(1):105–15. doi: 10.1016/j.yexcr.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 31.Hwang H, Zhang J, Chung KA, Leverenz JB, Zabetian CP, Peskind ER, Jankovic J, Su Z, Hancock AM, Pan C, Montine TJ, Pan S, Nutt J, Albin R, Gearing M, Beyer RP, Shi M. Mass Spectrometry Reviews. 2010;29(1):79–125. doi: 10.1002/mas.20221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koles K, Lim JM, Aoki K, Porterfield M, Tiemeyer M, Wells L, Panin V. Glycobiology. 2007;17(12):1388–403. doi: 10.1093/glycob/cwm097. [DOI] [PubMed] [Google Scholar]
- 33.Yurist-Doutsch S, Chaban B, VanDyke DJ, Jarrell KF, Eichler J. Mol Microbiol. 2008;68(5):1079–84. doi: 10.1111/j.1365-2958.2008.06224.x. [DOI] [PubMed] [Google Scholar]
- 34.Sittaramane V, Sawant A, Wolman MA, Maves L, Halloran MC, Chandrasekhar A. Dev Biol. 2009;325(2):363–73. doi: 10.1016/j.ydbio.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulus JD, Halloran MC. Dev Dyn. 2006;235(1):213–24. doi: 10.1002/dvdy.20604. [DOI] [PubMed] [Google Scholar]
- 36.Wolman MA, Sittaramane VK, Essner JJ, Yost HJ, Chandrasekhar A, Halloran MC. Neural Dev. 2008;3(6) doi: 10.1186/1749-8104-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biehlmaier O, Makhankov Y, Neuhauss SC. Invest Ophthalmol Vis Sci. 2007;48(6):2887–94. doi: 10.1167/iovs.06-1212. [DOI] [PubMed] [Google Scholar]
- 38.Jontes JD, Emond MR, Smith SJ. J Neurosci. 2004;24(41):9027–34. doi: 10.1523/JNEUROSCI.5399-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiellette E, Grinblat Y, Austen M, Hirsinger E, Amsterdam A, Walker C, Westerfield M, Sive H. Genesis. 2004;40(4):231–40. doi: 10.1002/gene.20090. [DOI] [PubMed] [Google Scholar]
- 40.Kerstetter AE, Azodi E, Marrs JA, Liu Q. Dev Dyn. 2004;230(1):137–43. doi: 10.1002/dvdy.20021. [DOI] [PubMed] [Google Scholar]
- 41.Kaether C, Haass C, Steiner H. Neurodegener Dis. 2006;3(4–5):275–83. doi: 10.1159/000095267. [DOI] [PubMed] [Google Scholar]
- 42.Haffner C, Haass C. Neurodegener Dis. 2006;3(4–5):284–9. doi: 10.1159/000095268. [DOI] [PubMed] [Google Scholar]
- 43.Haffner C, Haass C. Neurodegener Dis. 2004;1(4–5):192–5. doi: 10.1159/000080985. [DOI] [PubMed] [Google Scholar]
- 44.Morais VA, Brito C, Pijak DS, Crystal AS, Fortna RR, Li T, Wong PC, Doms RW, Costa J. Biochim Biophys Acta. 2006;1762(9):802–10. doi: 10.1016/j.bbadis.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 45.Yang DS, Tandon A, Chen F, Yu G, Yu H, Arawaka S, Hasegawa H, Duthie M, Schmidt SD, Ramabhadran TV, Nixon RA, Mathews PM, Gandy SE, Mount HT, St George-Hyslop P, Fraser PE. J Biol Chem. 2002;277(31):28135–42. doi: 10.1074/jbc.M110871200. [DOI] [PubMed] [Google Scholar]
- 46.Hack AA, Groh ME, McNally EM. Microsc Res Tech. 2000;48(3–4):167–80. doi: 10.1002/(SICI)1097-0029(20000201/15)48:3/4<167::AID-JEMT5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 47.Kawahara G, Guyon JR, Nakamura Y, Kunkel LM. Hum Mol Genet. 2010;19(4):623–33. doi: 10.1093/hmg/ddp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brockington M, Blake DJ, Prandini P, Brown SC, Torelli S, Benson MA, Ponting CP, Estournet B, Romero NB, Mercuri E, Voit T, Sewry CA, Guicheney P, Muntoni F. Am J Hum Genet. 2001;69(6):1198–209. doi: 10.1086/324412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.