Abstract
The immediate response to skin injury is the release of inflammatory signals. It is shown here, by use of cultures of primary keratinocytes from wild-type and PPARβ/δ−/− mice, that such signals including TNF-α and IFN-γ, induce keratinocyte differentiation. This cytokine-dependent cell differentiation pathway requires up-regulation of the PPARβ/δ gene via the stress-associated kinase cascade, which targets an AP-1 site in the PPARβ/δ promoter. In addition, the pro-inflammatory cytokines also initiate the production of endogenous PPARβ/δ ligands, which are essential for PPARβ/δ activation and action. Activated PPARβ/δ regulates the expression of genes associated with apoptosis resulting in an increased resistance of cultured keratinocytes to cell death. This effect is also observed in vivo during wound healing after an injury, as shown in dorsal skin of PPARβ/δ+/+ and PPARβ/δ+/− mice.
Keywords: PPARs, inflammation, apoptosis, differentiation, keratinocytes
Peroxisome proliferator-activated receptors (PPARs), which control many cellular and metabolic processes, are members of the superfamily of ligand-inducible transcription factors known as nuclear receptors. Three isotypes called PPARα (NR1C1), PPARβ/δ (NR1C2), and PPARγ (NR1C3) (Nuclear Receptors Nomenclature Committee 1999) have been identified in vertebrates. They display differential tissue distribution, suggesting that each of them fulfills specific functions. PPARα and PPARγ play important roles in lipid homeostasis and in inflammation (Desvergne and Wahli 1999). In contrast, the exact functions of PPARβ remain an enigma. Although fatty acids can bind and activate PPARβ, studies on this isotype have so far been impeded by the lack of information about the nature of its physiological ligands and by its remarkably broad tissue distribution. Recently however, PPARβ was implicated in reverse cholesterol transport (Oliver et al. 2001), in oligodendrocyte maturation and in membrane sheet formation (Saluja et al. 2001).
In addition, PPARs may play an important role in skin development, as PPARα ligands can accelerate fetal rat epidermal development (Hanley et al. 1998). In epidermis, the three PPAR isotypes are expressed during development, but their levels decrease to become undetectable in the interfollicular keratinocytes 5–9 d after birth (Michalik et al. 2001). However, the expression of both PPARα and PPARβ is reactivated upon proliferative stimuli, such as treatment with the phorbol ester TPA or hair plucking and at the wound edges after skin injury. Although PPARβ−/− and PPARβ+/− mice are not affected by apparent skin defects, they display an increased hyperplasic response to TPA treatment (Peters et al. 2000; Michalik et al. 2001). More interestingly, wound closure is delayed in PPARβ+/− mice as compared with wild-type animals (Michalik et al. 2001). Taken together, these observations suggest that PPARβ may play an important role in skin, particularly in stress situations.
The epidermis in which keratinocyte is the predominant cell type is characterized by a lifelong polarized pattern of epithelial growth and cell differentiation in association with normal skin renewal (Spellberg 2000). In pathological conditions, for example, psoriasis, or after epidermal injury, keratinocytes produce and respond to growth factors and cytokines, they become migratory, and synthesize components of the basement membrane. These activated wound keratinocytes express a specific set of keratins that are distinct from those present in normal epidermis. The expression of keratin 1 (K1) and keratin 10 (K10), normally produced in the suprabasal and differentiating keratinocytes, is suppressed and replaced by the expression of keratin 6 (K6) and keratin 16 (K16) (Mansbridge and Knapp 1987). These responses are orchestrated by a number of signaling pathways, including the stress-associated kinase pathway (SAPK/JNK), which function both in sequential and parallel fashions (Dotto 1999).
Interestingly, whereas keratinocyte differentiation occurs during both normal epidermis renewal and wound repair, PPARβ up-regulation is observed only in the latter. Hence, this study was undertaken to unveil the roles of PPARβ during wound healing. The results obtained indicate that the up-regulation of the PPARβ gene is closely associated with necrosis. Moreover, pro-inflammatory cytokines, for example, TNF-α, can both increase PPARβ expression via the stress kinases signaling pathway and trigger the production of ligands for this receptor. Consistent with an important role of PPARβ in mediating inflammation-induced keratinocyte differentiation, keratinocytes derived from PPARβ−/− mice are both severely delayed in inflammatory cytokine-stimulated differentiation and more sensitive to TNF-α-induced apoptosis.
Results
Reactivation of PPARβ is associated with cell death by necrosis
The initial approach to this study was to identify the cascade of events leading to the injury-dependent activation of the PPARβ gene, which is normally silent in adult interfollicular keratinocytes (Michalik et al. 2001). Because the response to tissue injury can be mimicked in mixed leukocyte reactions (MLR), we used this assay to identify potential signals that trigger PPARβ expression (Gallucci et al. 1999). In MLR, immature bone-marrow-derived dendritic cells (DCs) are activated by exposure to necrotic cells. The activated DCs are, in turn, potent stimulators of T-cell proliferation (Banchereau and Steinman 1998). The factors secreted by these DCs/T-cells are representative of the signals produced after injury and comprise pro-inflammatory cytokines. In this study, necrosis was achieved by freeze thawing primary fibroblasts or by mincing skin. As a control, MLR was also performed with DCs exposed to UVB-induced apoptotic fibroblasts. Conditioned medium (CM) collected from the activated DCs/T-cell cocultures was used to culture mouse primary keratinocytes. Its effects on their differentiation and PPARβ expression were analyzed.
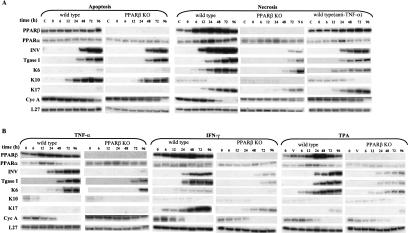
Keratinocytes cultured in control medium — not exposed to DCs/T-cells — did not differentiate over the experimental time period (data not shown), whereas keratinocytes cultured in apoptosis-derived CM started to express keratinocyte differentiation markers, such as involucrin (INV), transglutaminase type I (Tgase I), and keratin 10 (K10) (Fig. 1A, first panel). There was no change in the expression of PPARβ and PPARα. In parallel cultures, primary keratinocytes isolated from PPARβ−/− mice and exposed to apoptosis-derived CM displayed a differentiation pattern similar to that of wild-type cells, showing that apoptosis-induced differentiation is a PPARβ-independent process (Fig. 1A, second panel).
Figure 1.
External signals stimulate PPARβ expression and cell differentiation in primary mouse keratinocyte cultures. (A) Apoptosis-derived CM and necrosis-derived CM induce keratinocyte differentiation. Primary keratinocytes from wild-type (first, third, and fifth panels) or from PPARβ−/− mice (second and fourth panels) were exposed at time 0 (usually after two or three passages) to conditioned medium (CM) from mixed leukocyte reactions (MLR), prepared either with apoptotic fibroblasts (first and second panels) or with necrotic cells (third, fourth, and fifth panels). Keratinocytes were lysed after different times of exposure to CM as indicated, and the expression of PPARβ was evaluated by RPA. The expression of involucrin (INV), transglutaminase I (TgaseI), was used as markers of keratinocyte differentiation. The expression of keratin (K) K6, K10, and K17 reflects different pathways of keratinocyte differentiation. Cyclin A (Cyc A) expression indicated the status of the cell with respect to cell cycle. In the fifth panel, the necrosis-derived CM was precleared of TNF-α. (B) The major pro-inflammatory mediators TNF-α, IFN-γ, and TPA up-regulate PPARβ expression. Primary keratinocytes from wild-type (first, third, and fifth panels) or from PPARβ−/− mice (second, fourth, and sixth panels) were cultured in KSFM and exposed at time 0 to TNF-α 5 ng/ml (first and second panels) IFN-γ 5 ng/ml (third and fourth panels). or TPA 20 ng/ml (fifth and sixth panels). In all three treatments, the expression of all differentiation markers is strongly reduced and delayed in PPARβ−/− cells.
In contrast, exposure of keratinocytes to necrosis-derived CM resulted in a rapid and intense up-regulation of PPARβ (Fig. 1A, third panel). This was accompanied by accelerated keratinocyte differentiation and cell cycle arrest, as indicated by both an early onset of INV and Tgase I and a decrease in cyclin A (Cyc A) expression. The expression of the wound-repair-associated keratins, K6 and K17, was dramatically increased, whereas the weak expression of K10 was suppressed (Fig. 1A, third panel). Strikingly, the differentiation pattern induced by necrosis-derived CM was dramatically delayed in PPARβ−/− keratinocytes (Fig. 1A, fourth panel), as revealed by the expression of the differentiation markers. In these cells, the transient and weak PPARα up-regulation seen in wild-type keratinocyte was unaltered. Together, these observations clearly show that PPARβ is essential for mediating necrosis-associated signals that stimulate keratinocyte differentiation.
Both in vivo and in the MLR model, necrosis triggers an inflammatory response mediated by cytokines (Messmer and Pfeilschifter 2000). Hence, we tested whether pro-inflammatory cytokines can induce PPARβ expression and keratinocyte differentiation similarly to the necrosis-derived CM. We first assessed the effect of TNF-α, which is considered as the main coordinator of the inflammation response. TNF-α alone (5 ng/ml) induced PPARβ, INV, Tgase I, and K6 expression, and inhibited Cyc A expression (Fig. 1B, first panel). In contrast, PPARβ−/− keratinocytes exposed to TNF-α exhibited a weak and delayed expression of the differentiation markers accompanied by a gradual decrease in Cyc A level (Fig. 1B, second panel). This delayed response by these mutant cells is unlikely to be due to defective TNF-α signaling, because PPARβ−/− cells, like their wild-type counterparts, expressed high levels of the TNF-α receptor (p55TNF-R1) (data not shown).
To assess the contribution of TNF-α signaling in the necrosis-induced differentiation, we precleared the necrosis-derived CM of TNF-α by incubation with excess anti-TNF-α antibodies prior to addition to the keratinocytes. This pretreatment significantly reduced the potency of the CM to induce PPARβ, INV, Tgase I, and K6 expression. Thus, TNF-α is a trigger of PPARβ expression and a major player in inflammatory-induced keratinocyte differentiation (Fig. 1A, fifth panel). However, the anti-TNF-α antibody in the CM did not totally abrogate the differentiation process, nor did it alter the expression of K17 (Fig. 1A, fifth panel), indicating that additional factors in necrosis-derived CM participate to PPARβ-induced keratinocyte differentiation. Another important inflammatory cytokine, IFN-γ, also produced a similar differentiation profile as necrosis-derived CM or TNF-α. Interestingly, IFN-γ strongly induced K17 but not K6 expression (Fig. 1B, third and fourth panels). Finally, we also investigated the effect of the phorbol ester TPA, known to induce inflammation in skin (Puignero and Queralt 1997). TPA treatment resulted in a ninefold increase in the expression of PPARβ, associated with increased K6, but not K17 expression (Fig. 1B, fifth and sixth panels). Both IFN-γ and TPA-induced differentiation patterns are PPARβ dependent, as seen from the results of the parallel experiments performed with PPARβ−/− keratinocytes.
These observations show that necrosis-derived differentiation signals, such as produced after injury, lead to elevated PPARβ expression. The peak of PPARβ expression typically coincides with the switch to cell cycle arrest and the onset of differentiation. Western blot analyses, performed as reported by Michalik et al. (2001), verified that corresponding changes occurred in PPARβ protein levels (data not shown). Thus, pro-inflammatory cytokines that are produced in response to necrosis act as primary triggers of elevated PPARβ expression. Upon exposure to necrosis-derived CM or pro-inflammatory cytokines, PPARβ−/− keratinocytes displayed dramatically impaired differentiation, showing that PPARβ plays a critical role in enabling keratinocytes to differentiate in response to inflammation.
Reactivation of PPARβ in keratinocytes is mediated by an AP-1 site in the PPARβ promoter.
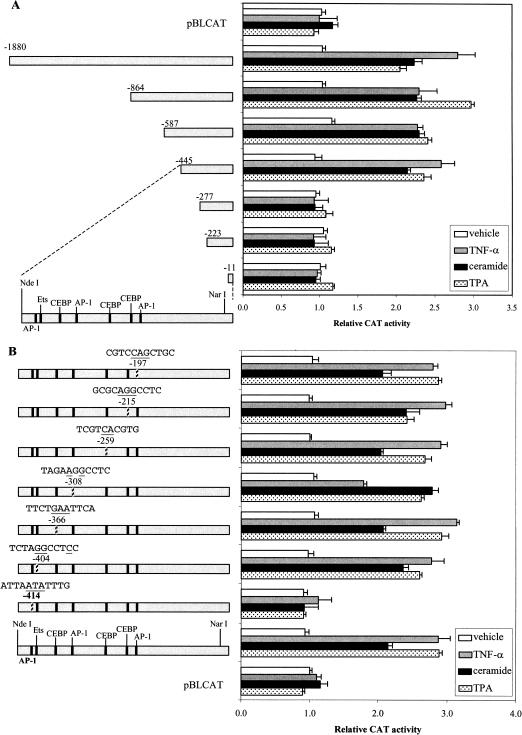
Treatment of cultured primary keratinocytes with TNF-α resulted in an expression profile of PPARβ, which was closest to that observed after injury in an in vivo model (Michalik et al. 2001). Therefore, the molecular events by which TNF-α regulates the promoter of the PPARβ gene were studied next. The mouse PPARβ promoter was isolated (accession no. AF187850) and a 1.9-kb fragment containing the transcription initiation site was subcloned into a CAT reporter gene. The effect of TNF-α on the activity of a series of truncations of this 1.9-kb PPARβ promoter region were analyzed by use of transfection into primary keratinocytes. The effects of ceramide (Cer), a downstream effector of TNF-α (Hannun and Luberto 2000), and TPA were also studied. The addition of each of the three agents increased the promoter activity of pPPARβ(−1880), as well as that of the shorter promoter constructs pPPARβ(−864), pPPARβ(−587), and pPPARβ(−445), by two to threefold (Fig. 2A). In contrast, pPPARβ(−277), pPPARβ(−223), and pPPARβ(−11) were not responsive to TNF-α, TPA, or Cer, suggesting that the response elements are located between −445 and −277.
Figure 2.
Identification of the region in the PPARβ promoter that mediates the TNF-α, TPA, and ceramide responses. (A) PPARβ promoter 5′-deletion mutants. Plasmids are named according to the length of the promoter region they contain upstream from the transcription start site. Keratinocytes in culture were cotransfected with a CAT reporter driven by the various PPARβ promoter constructs and pCMV-β-galactosidase as a control of transfection efficiency. Keratinocyte cultures were treated for 24 h with the inducers indicated. Results were normalized to β-galactosidase activity and are presented as relative CAT activity compared with that of the promoterless vector pBLCAT alone. (B) TNFα-, TPA-, and Cer-signaling target an AP-1(−414) of the PPARβ promoter. The hatched bar depicts site-directed mutations of the transcription factor binding sites. The mutations introduced into the site are indicated as underlined nucleotides above the respective hatched bar. The numbers associated with the sites indicate their position relative to the transcription start site. Data are means of six independent experiments. Vehicles for TPA, TNF-α, and Cer were ethanol, PBS, and DMSO, respectively.
This proximal part of the PPARβ promoter contains sequences related to AP-1, Ets, and C/EBP transcription factor binding sites (Fig. 2A), as identified by using MatInspector (Wingender et al. 2000). Mutations were introduced in each of these sites and their relative importance was assessed via transactivation studies (Fig. 2B). Whereas mutation of the Ets or C/EBP sites did not affect regulation by the three agents, mutation of the AP-1 site, located at −414, completely abolished responsiveness to all three inducers. Hence, the signaling pathways that are activated by TPA, TNF-α, or Cer terminate at the AP-1(−414) site in the PPARβ promoter.
TNF-α-induced expression of PPARβ is mediated by the factor-associated-with-neutral-sphingomyelinase and the stress-associated kinase cascade
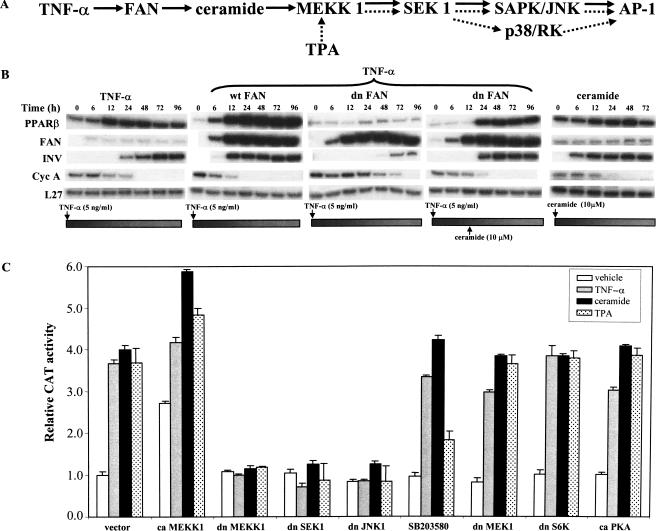
Next, experiments were designed to identify the individual steps relaying the signal initiated from TNF-α to the AP-1(−414) site in the PPARβ promoter. TNFα-induced trimerization of its receptor (p55TNF-R1) leads to the stimulation of several signaling cascades. One of these pathways involves the production of bioactive Cer, which, in turn, acts as second messengers to initiate downstream effects (Kronke 1999). Cer is of particular interest in this study as it is a component of the epidermal stratum corneum. Adam-Klages et al. (1996) reported that FAN (factor associated to neutral sphingomyelinase) couples the p55TNF-R1 to neutral sphingomyelinase (N-Smase), which is the major enzyme responsible for Cer generation via sphingomyelin hydrolysis (Fig 3A). Thus, the involvement of FAN in transducing the effect of TNF-α to the PPARβ promoter was explored. Keratinocytes were transfected with wild-type (wt) or a dominant-negative (dn) FAN and treated with TNF-α. Overexpression of wtFAN potentiated the ability of TNF-α to induce PPARβ expression (Fig. 3B, second panel) and triggered an early onset of keratinocyte differentiation as indicated by the induction of INV expression. In contrast, overexpression of dnFAN abolished the effect of TNF-α on PPARβ expression and resulted in delayed cell differentiation. The repressive effect of dnFAN could be rescued by addition of exogenous Cer (Fig. 3B, third and fourth panels), in accordance with the role of Cer as a downstream effector of FAN. Treatment of untransfected keratinocytes with Cer alone resulted in an elevated PPARβ expression and an early expression of INV.
Figure 3.
Signaling cascades that mediate the effects of TNF-α, TPA, and Cer on the PPARβ gene in keratinocytes. (A) (Overall scheme) The solid arrows represent the major TNFα-activated, FAN-mediated, pathway regulating PPARβ expression. The cascade invoked by TPA is depicted by the dashed arrows. (B) TNF-α induces PPARβ gene expression through FAN and Cer. Keratinocytes were cotransfected with expression vectors for wild-type FAN (wtFAN, second panel) or a dominant-negative form of FAN (dnFAN, third and fourth panels) and pCMV-β-galactosidase as internal control. Transfected keratinocytes were treated with TNF-α and RPA were performed at the indicated times. (fourth panel) Suppression of PPARβ expression by dnFAN was rescued by addition of exogenous ceramide at 12 h post-transfection. (first panel) Control experiment with TNF-α alone; (fifth panel) effect of ceramide in absence of TNF-α treatment. (C) Regulation of PPARβ via the stress-associated kinase pathway. Keratinocytes were transfected with a CAT reporter driven by the pPPARβ(−445) promoter region together with expression vectors encoding dominant negative (dn) or constitutively active (ca) kinases. The empty vector pCDNA 3.1 was used as a control (vector). Transfected keratinocytes were treated with an inducer (TNF-α, ceramide, or TPA) and CAT activity was measured 24 h post-treatment. SB203580 (10 μM) is a specific inhibitor of p38 and was added 1 h prior to treatment with the inducers. Values are means of six independent experiments.
Transmission of the TNF-α signal downstream of Cer involves the mitogen-activated protein (MAP) kinase pathways. These pathways lead to the phosphorylation of transcription factors, such as those belonging to the AP-1 family, and consequently to a modulation of gene expression (Kyriakis and Avruch 1996). TNF-α and Cer are known to activate MEKK1, which is a key player of the MAPK cascade. MEKK1, in turn, can phosphorylate several downstream kinases including SEK1 (Davis 2000; Fig. 3A). To evaluate the role of these kinases in regulating PPARβ expression, the pPPARβ(−445) construct, which harbors the AP-1 site that is responsive to TNF-α, was cotransfected with expression vectors encoding either constitutively active (ca) or dominant-negative forms of several kinases. Alternatively, specific known kinase inhibitors were used. Transfection with dnMEKK1 abolished TNFα-, TPA-, and Cer-dependent promoter activity (Fig. 3C). In contrast, caMEKK1 increased the basal activity by two to threefold and a further twofold upon addition of the cytokines. The cytokine-mediated induction of the PPARβ promoter is also suppressed by dnSEK1.
Downstream of SEK1, both JNK/SAPK and p38 have been shown to modulate AP-1 expression and activity (Davis 2000). The dnJNK/SAPK completely abolished the responsiveness of the PPARβ promoter to the three inducers. Interestingly, SB20358, a specific inhibitor of p38, only partially attenuated the effect of TPA stimulation and had no effect on stimulation by TNF-α or Cer (Fig. 3C). Finally, additional experiments ruled out the possible involvement of S6K, PKA, and MEK1 signaling in regulating PPARβ expression (Fig. 3C). Hence, activation of the PPARβ promoter by TNF-α, TPA, and Cer is mediated through MEKK1, SEK1, and SAPK/JNK (Fig. 3C).
Taken together, these observations show that TNF-α, TPA, and Cer utilize overlapping signal transduction cascades that converge to JNK/SAPK and AP-1. Incidentally, the cascades that are activated by the three agents are stress-associated pathways and are invoked in response to inflammation (Kyriakis and Avruch 1996). Hence, these data attest to an important role for PPARβ in the stress- and injury-associated responses of keratinocytes.
Expression and activation of PPARβ lead to keratinocyte differentiation
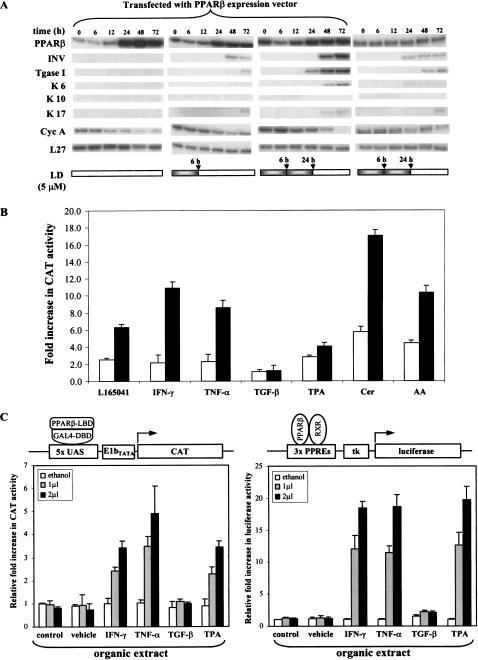
The results presented so far show that expression of PPARβ, induced by inflammatory cytokines, correlates with keratinocyte cell cycle arrest and differentiation. Furthermore, experiments with PPARβ−/− keratinocytes have shown that PPARβ is required for these processes to take place (Fig. 1). Therefore, we hypothesized that PPARβ overexpression would be sufficient to induce keratinocyte differentiation in our primary cell culture assay. To test this possibility, primary keratinocytes overexpressing PPARβ were treated or not with the synthetic PPARβ ligand L165041 (LD). A high level of PPARβ alone was not sufficient to trigger keratinocyte differentiation, whereas in conjunction with a 6-h LD treatment, these cells expressed the differentiation marker, INV (Fig. 4A, first and second panels). Repeated LD exposure of the PPARβ overexpressing cells led both to cell cycle arrest and to keratinocyte differentiation (Fig. 4A, third panel). However, the same treatment applied to untransfected keratinocytes resulted only in a mild expression of INV (Fig. 4A, fourth panel). Thus, the induction of keratinocyte differentiation requires both an elevated expression of PPARβ and the presence of a PPARβ ligand that activates the receptor.
Figure 4.
PPARβ accelerates keratinocyte differentiation. (A) Overexpression and activation of PPARβ stimulates keratinocyte differentiation. Keratinocytes were transfected with a wtPPARβ expression vector and lysed for RPA at various time points post-transfection, as indicated. Overexpression of PPARβ alone is not sufficient to trigger keratinocyte differentiation (first panel), which requires a 6-h exposure to a PPARβ-specific ligand L165041 (LD) (second panel). Better results were obtained with a repeated exposure to LD [6 h, followed by an additional 18 h with fresh medium containing the second dose (third panel)]. This latter dose regime was also able to slightly induce the expression of differentiation markers in absence of ectopical expression of PPARβ (fourth panel). (B) Activation of the PPARβ LBD by cytokines. Keratinocytes were cotransfected with an expression vector for a GAL4 DBD–PPARβ LBD, the Gal4 responsive reporter vector pG5CAT (Clontech), and the pCMV-β-galactosidase vector. CAT and β-galactosidase activities were measured after 24-h exposure to two different concentrations (open and solid bars) of PPARβ selective ligand L165041 (LD; 1, 5 μM), IFN-γ (10, 25 ng/ml), TNF-α (10, 25 ng/ml), TGF-β (10, 25 ng/ml), TPA (10, 20 ng/ml), Ceramide (Cer; 10, 20 μM), or arachidonic acid (AA; 10, 20 μM). Exposure to IFN-γ, TNF-α, TPA, and Cer resulted in a dose-dependent increase of the CAT reporter activity. (C) Total lipid extract of cytokine-activated keratinocytes activates PPARβ. Total lipid extract were prepared from keratinocytes untreated (control), or exposed to vehicle or to various pro-inflammatory mediators, as indicated. (Left) Transfections were performed as in A. Cells were then exposed to 1 or 2 μL of a total lipid extract for 24 h and CAT activity was measured. (Right) Keratinocytes were transfected with the wtPPARβ expression vector, pGL-3xPPRE-tk-luc reporter gene, and pCMV-β-galactosidase. Normalized reporter activity is shown as fold increase as compared with control.
TNF-α, TPA, and ceramide can generate endogenous ligands of PPARβ
Together, our results suggest that signaling by inflammatory molecules result not only in stimulation of the PPARβ gene but also leads to the production of an endogenous ligand. To test this hypothesis, an expression vector encoding a fusion protein comprising the GAL4–DNA-binding domain (DBD) and the PPARβ ligand-binding domain (LBD) was constructed. This fusion protein allows for monitoring the activation of the PPARβ LBD independently of the expression of the endogenous PPARβ gene. Keratinocytes were cotransfected with this fusion construct and a CAT reporter gene controlled by GAL4 response elements. The responsiveness of this system was validated with the synthetic PPARβ ligand L165041, which activated the fusion protein in a dose-dependent manner (Fig. 4B). Treatment of the transfected cells with either IFN-γ, TNF-α, TPA, or Cer that were shown above to up-regulate PPARβ gene expression, also resulted in a dose-dependent increase in the transcriptional activity of the fusion protein. Interestingly, both the bioactive lipid Cer and arachidonic acid (AA) led to a dramatic increase in CAT activity (Fig. 4B). In contrast, TGF-β had no effect on CAT activity.
To verify that the activation of the PPARβ LBD was not due to its phosphorylation but to the production of a ligand, we tested the ability of an organic extract from cytokine-activated keratinocytes to transactivate PPARβ. This experiment was performed in keratinocytes transfected either with the GAL4 DBD/PPARβ LBD fusion protein and its corresponding reporter CAT gene (Fig 4C, left), or with wild-type PPARβ and a PPRE-containing reporter luciferase gene (Fig 4C, right). In both systems, a dose-dependent increase in reporter activity was observed, showing the presence of a ligand in the organic extracts. Thus, in addition to elevating PPARβ gene expression, inflammatory signals also trigger the production of endogenous ligand(s) necessary for PPARβ activation.
PPARβ regulates genes associated with apoptosis
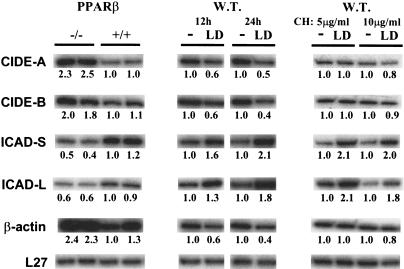
We have established that inflammation stimulates PPARβ expression and produces ligand(s) that activate the receptor, which together induce keratinocyte differentiation. Interestingly, amidst the TNF-α-induced keratinocyte differentiation experiments, we observed that PPARβ−/− keratinocytes, upon a prolonged exposure to TNF-α, died at a higher rate than wild-type cells. This differential response to this cytokine, which is an inducer of apoptosis (Natoli et al. 1998), led us to test whether PPARβ controls genes involved in apoptosis. The expression of such genes was analyzed by RPA in PPARβ−/− versus wild-type keratinocytes. Expression of the proapoptotic genes CIDE-A and CIDE-B (cell death-inducing DFF45-like effector A and B), involved in nuclear condensation and DNA fragmentation (Inohara et al. 1998), was 1.8–2.5-fold higher in the PPARβ−/− cells (Fig. 5). In contrast, the expression of the anti-apoptotic ICAD-S and ICAD-L (inhibitor of caspase-activated deoxyribonuclease short and long forms), which are highly homologous to the human DFF45, were reduced in the PPARβ−/− keratinocytes (Fig. 5). This is consistent with the known role of DFF45 (DNA fragmentation factor 45) in the inhibition of the DNase/apoptotic activity of CIDEs (Inohara et al. 1998). Thus, PPARβ appears to up-regulate antiapoptotic and down-regulate proapoptotic genes in keratinocytes. Whether this regulation is direct or indirect, that is, does not or does require de novo protein synthesis, was analyzed using keratinocytes treated for 12 and 24 h with the PPARβ ligand LD, in the presence or absence of cycloheximide (CH), a protein synthesis inhibitor. Consistent with the results presented above, all the genes tested responded to the PPARβ agonist in the absence of the protein synthesis inhibitor (Fig. 5). However, in the presence of CH, the down-regulation of CIDE-A and CIDE-B was lost, whereas the up-regulation of ICAD-S and ICAD-L was maintained. This result indicates that de novo protein synthesis is required for PPARβ-dependent down-regulation of CIDE-A and CIDE-B, whereas up-regulation of ICAD-S and ICAD-L is a direct effect of PPARβ. A further analysis of the molecular mechanisms by which PPARβ modulates apoptosis is underway. In addition to the genes involved in apoptosis, the expression of β-actin in the PPARβ−/− cells was ∼2.3-fold higher than in wild-type cells, a fact worth mentioning because of the frequent use of β-actin mRNA as a internal control in expression studies. Furthermore, this result is consistent with our previous observation that PPARβ−/− keratinocytes exhibited altered cell morphology (Michalik et al. 2001).
Figure 5.
PPARβ target genes are associated with apoptosis. RPA was performed using total RNA (2.5 μg) isolated from keratinocytes derived from PPARβ+/+ and PPARβ−/− mice (left); or from wild-type keratinocytes not treated (−) or treated for 12 and 24 h with 1μM of the PPARβ specific ligand L165041 (LD; middle); and with 5 or 10 μg/ml of cycloheximide (CH; right). Two representative samples are shown per condition. Numbers below each band represent the fold differences in mRNA expression levels with respect to the wild-type or untreated keratinocytes.
Elevated PPARβ confers increased resistance to TNF-α-induced apoptosis
The above results implicate a role of PPARβ in the repression and stimulation of pro-apoptotic and anti-apoptotic genes, respectively. Regardless of the origin of the apoptotic stimuli, the commitment to apoptosis occurs through the activation of caspases and the fragmentation of nuclear DNA. In accordance with this process, TNF-α receptors can trigger apoptosis via activation of caspase 8, followed by DNA fragmentation. This prompted us to measure this enzymatic activity and to monitor DNA fragmentation in wild-type and PPARβ−/− keratinocytes.
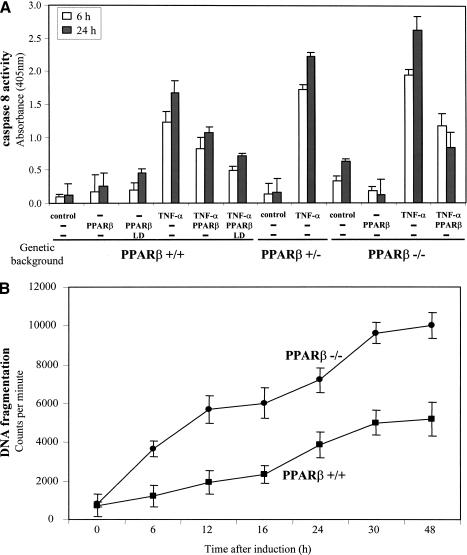
Primary keratinocytes displayed minimal caspase 8 activity (Fig. 6A). Upon treatment with TNF-α, the caspase activity increased by 12- and 14-fold after 6 and 24 h of treatment, respectively. Interestingly, keratinocytes expressing PPARβ ectopically showed a reduced TNFα-induced caspase activity (Fig. 6A). Incubation of these cells with a PPARβ selective ligand LD further reduced the caspase activity by 2.5-fold, when compared with untransfected wild-type keratinocytes. The importance of PPARβ in controlling caspase activity levels was further examined in PPARβ+/− and PPARβ−/− keratinocytes. In unchallenged keratinocytes, only PPARβ−/− cells displayed higher basal levels of caspase 8 activity. However, both mutant keratinocytes exhibited significantly enhanced TNFα-induced caspase activity when compared with wild-type cells (Fig. 6A). Finally, the reduced response to TNF-α seen in wild-type cells could be attributed to PPARβ expression, as the response of PPARβ−/− keratinocytes to TNF-α could be normalized by transfection of a PPARβ-expression vector.
Figure 6.
Inflammation-induced PPARβ expression protects keratinocytes from apoptosis. (A) Elevated PPARβ expression inhibits TNFα-induced apoptosis. Keratinocytes prepared from PPARβ+/+,PPARβ+/−, and PPARβ−/− mice were either transfected with expression vectors harboring the wtPPARβ and/or cultured in the absence or presence of TNF-α and L165041 (LD, 5 μM) as indicated. The level of TNFα-induced apoptosis was monitored by caspase 8 activity. Keratinocytes expressing wtPPARβ were more resistant to apoptosis signals, whereas PPARβ+/− cells showed an increase susceptibility. PPARβ−/− keratinocytes exhibited higher basal caspase 8 activity and were more sensitive to TNFα-induced apoptosis, but could be rescued by transfection with the vector expressing wtPPARβ. Values are means of three independent experiments. (B) PPARβ−/− keratinocytes are more susceptible to TNFα-induced apoptosis. Culture of primary keratinocytes from wild-type and from PPARβ−/− mice were exposed to TNF-α. Apoptosis was measured by radioactive DNA fragmentation assay over indicated periods of time. Values are means of three independent experiments.
The fragmentation of genomic DNA is also a hallmark of apoptosis (Duke 1983) and, hence, was also assessed in PPARβ−/− and PPARβ+/+ keratinocytes upon TNFα-induced apoptosis. As expected from the caspase 8 activity measurements presented above, PPARβ−/− keratinocytes showed higher levels of TNF-α-induced DNA fragmentation than wild-type cells over the time course monitored (Fig. 6B).
The observed higher caspase 8 activity, higher DNA fragmentation rate, and resulting reduced resistance to apoptosis of PPARβ−/− cells, provide evidence for an important role of PPARβ in the control of keratinocyte death during inflammation.
Ablation of a PPARβ allele in vivo results in increased proliferation and apoptosis in mouse epidermis
The relevance of the antiapoptotic role of PPARβ, observed in primary keratinocyte culture, was also examined in vivo during wound repair. We used a model of epidermal repair in PPARβ mutant mice (Michalik et al. 2001). Epidermal repair after a skin wound includes keratinocyte proliferation, migration, and differentiation. For wound closure and efficient healing, the balance between proliferation and cell death has to be tightly controlled. Recently, we have shown that PPARβ expression is up-regulated in the keratinocytes at the wound edges, and that the deletion of one PPARβ allele (PPARβ+/− animals) resulted in delayed wound healing (Michalik et al. 2001).
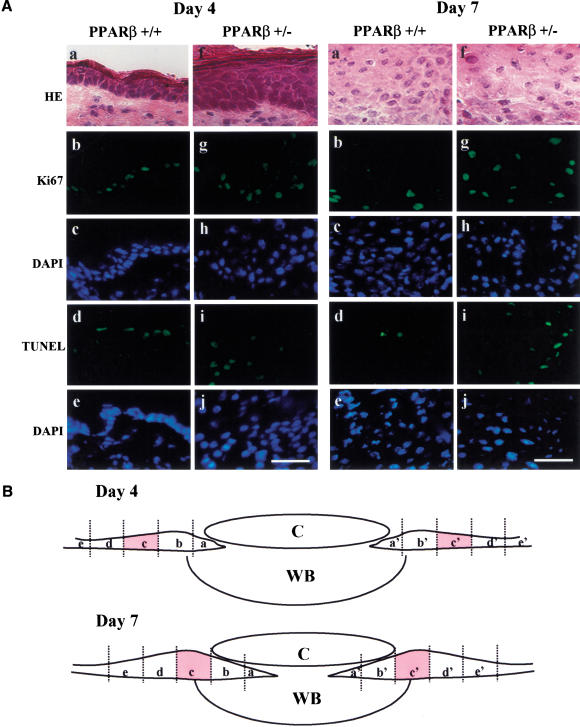
Apoptosis and proliferation were assessed in this model by use of TUNEL and immunofluorescent labeling of the Ki67 nuclear proliferation marker, respectively, on dorsal mouse skin sections from days 4 and 7 after an injury. Figure 7A and Table 1 summarize the observations made at the epidermal wound edges (see Fig. 7B) of PPARβ+/− and wild-type mice. Quantification of the Ki67-positive cells revealed a twofold increase in the number of proliferating keratinocytes in the PPARβ+/− mice at 4 and 7 d post injury. This observation shows that in a physiological situation such as wound healing, PPARβ is involved in the control of epithelial cell proliferation. Furthermore, it is in agreement with results obtained previously by us and others revealing higher keratinocyte proliferation rates in the epidermis of mutant mice after TPA treatment (Peters at al. 2000; Michalik et al. 2001). Even more importantly and consistent with the result obtained here with primary keratinocytes culture, the number of TUNEL-positive keratinocytes in the same region was 10-fold higher in the PPARβ mutant epidermis at day 7 post injury (Fig. 7; Table 1). These data show that in vivo as well as in vitro, PPARβ has an anti-apoptotic role. In the context of skin wound healing, these findings show that PPARβ is necessary for the control of the keratinocyte life span, and, thus, for a well-tuned balance between cell death and proliferation.
Figure 7.
PPARβ controls the balance between apoptosis and proliferation of keratinocytes in vivo. (A) Increased proliferation and elevated apoptosis in the PPARβ+/− mice. The Ki67 and TUNEL-positive keratinocytes were revealed by fluorescent labeling on skin sections obtained at days 4 and 7 after a dorsal skin injury. The figure shows representative fields of the labelings obtained at the wound edges (region c/c' of the epithelium, according to B). PPARβ+/+ (a–e) and PPARβ+/− (f–j) mice skin sections after hematoxylin/eosin staining (HE) (a,f), Ki67 immunostaining (b,g), TUNEL (d,i) and DAPI staining (c,h,e,j). Magnification bar, 50 μm. (B) Regions of a skin wound at days 4 and 7 after a full-thickness biopsy. (C) Clot; (WB) wound bed. The positions of the 10 microscope fields in which apoptosis and cell proliferation were quantified is indicated as regions a–e and a' –e'. The quantification (summarized in Table 1) is a mean of the number of proliferative or apoptotic positive cells counted in regions e to e' of the epithelium. A shows pictures taken in region c and c', as representative fields.
Table 1.
Number of Ki67 or TUNEL positive cells quantified based on respective fluorescent labelings
|
|
Day 4
|
Day 7
|
||
|---|---|---|---|---|
| Ki67
|
TUNEL
|
Ki67
|
TUNEL
|
|
| PPARβ+/+ | 12.3 ± 1.28 | 9.0 ± 2.0 | 5.1 ± 1.4 | 1.3 ± 0.8 |
| PPARβ+/− | 24.1 ± 3.13** | 7.5 ± 1.2 | 10.3 ± 1.7* | 15.1 ± 2.3* |
Values represent the mean cell number counted from 10 standardized microscope fields per section (Fig. 7B, regions a to e and a′ to e′), performed on three different animals for each genotype ± SEM. Asterisks indicate that the difference is statistically significant (*P < 0.05; **P < 0.01).
Discussion
The results presented herein show that, in keratinocytes, PPARβ is an important transcription factor relaying inflammatory signals at the cell surface into specific gene expression patterns, which define appropriate cellular responses in sudden stress situations. Firstly, signals such as TNF-α or IFN-γ activate the stress-associated signaling pathway leading to the stimulation of the PPARβ gene via an AP-1 site in its promoter. Secondly, they trigger the production of PPARβ ligands. Thirdly, the resulting increase in PPARβ transcriptional activity strongly accelerates the differentiation of keratinocytes and increases their resistance to apoptotic signals. Finally, increased proliferation and death of keratinocytes at the edges of epidermal wounds in PPARβ+/− mice are likely to participate in the healing delay observed previously in these animals.
PPARβ promotes terminal differentiation of keratinocytes
As mentioned above, IFN-γ and TNF-α-induced activation of PPARβ accelerates keratinocyte differentiation in addition to the antiapoptotic effect of the receptor. The inflammatory cytokines trigger both elevated expression and ligand-induced activation of PPARβ, which is required to achieve their effects. We were able to recapitulate the cytokine-dependent differentiation simply by both high-ectopic expression of PPARβ in undifferentiated keratinocytes and its activation by a specific synthetic ligand. With respect to keratinocyte differentiation, it is unclear as yet why Peters et al. (2000) did not observe any difference in INV expression upon TPA treatment between PPARβ+/+ and PPARβ−/− mice. Possible explanations are that their expression data were normalized to the β-actin expression level that, as shown here, is increased in PPARβ−/− cells. An increase in INV expression, if paralleled by an elevation of β-actin synthesis would obviously have remained undetected using this normalization procedure. Alternatively, the pleiotropic effects of TPA treatment might activate both PPARβ-dependent and independent pathways of keratinocyte differentiation (Dotto 1999). Finally, topical application of TPA on skin may also have effects on the dermal fibroblasts that might modulate the keratinocyte response (Szabowski et al. 2000).
Whether the PPARβ-dependent cell differentiation noted here is keratinocyte specific or if activation of PPARβ in other tissues can accelerate cell differentiation as well, remains uncertain. In support of the latter, ectopic expression of PPARβ in NIH-3T3 fibroblasts was reported to promote early adipocyte differentiation that was dependent on the addition of PPARβ-selective ligands (Amri et al. 1995; Hansen et al. 2000). Also, PPARβ agonists were shown to stimulate oligodendrocyte differentiation and regulate the size of membrane sheets (Saluja et al. 2001).
PPARβ expression is regulated by pro-inflammatory cytokines
The epidermis is the first line of defense of the organism against aggressions from the environment, and thus must often respond to various types of injuries. As such, epidermal injury is also typically associated with inflammation, particularly during wound repair, which involves modulation of cell growth and differentiation of keratinocytes. Although PPARβ is undetectable by in situ hybridization in adult mouse interfollicular epidermis, its reactivation after a topical application of TPA, during wound repair and the healing delay observed in PPARβ+/− mice (Michalik et al. 2001), suggested a link between this receptor and keratinocyte biology during inflammation. The findings reported herein reveal that pro-inflammatory cytokines, such as TNF-α and IFN-γ produced during skin injury, can increase expression of PPARβ and produce its ligands. The inflammatory response is one of the major differences between normal skin renewal and wound repair (Savill and Fadok 2000). The response of keratinocytes to inflammatory signals was tested by use of necrotic cells in the MLR assay, and also by treating them with the noncytokine inflammatory agent, TPA. The stimulation of the PPARβ gene by the inflammatory molecules, as studied herein using TNF-α, comprises the recruitment of FAN by p55TNF-R1, which, via activation of N-Smase, induces the production of ceramide. In turn, ceramide activates a stress-associated kinase cascade terminating at an AP-1 site in the PPARβ promoter. It is worth noting that FAN-deficient mice, which show an impaired N-Smase activation, have a delayed cutaneous barrier homeostasis and repair phenotype (Kreder et al. 1999). Hence, PPARβ up-regulation and activation is clearly a stress-associated event, such as it occurs after injury.
Inflammation-induced PPARβ expression and increased resistance to apoptosis
Certainly, the up-regulation of PPARβ in any biological system can lead to several outcomes. Here, these effects will be considered in the context of keratinocytes exposed to TNF-α. This cytokine induces oligomerization of the receptor p55TNF-R1, which leads to the formation of a multimolecular signal-transducing complex, from which several pathways originate rapidly. Paradoxically, activation of p55TNF-R1 initiates two different types of signals, the first leading to cell death, and the second protects the cell from it. p55TNF-R1 interacts with TRADD, which recruits the downstream transducers FADD and TRAF2. FADD interacts with apoptotic proteases triggering cell death (Natoli et al. 1998). In contrast, TRAF2 can activate the transcription factors NFκB, c-Jun, ATF, TCF/Elk, or SAPK/JNK, which are necessary, but not sufficient to confer protection against TNFα-induced apoptosis (Natoli et al. 1998). This points to a critical role for additional p55TNF-R1 complex components that are involved in cytoprotection against TNFα-induced apoptosis. We showed herein that such an event is the stimulation of PPARβ expression and its ligand-dependent activation, which confer increased resistance to TNFα-induced apoptosis, as measured by caspase activity and DNA fragmentation. PPARβ up-regulates antiapoptotic and down-regulates proapoptotic genes, in response to inflammatory signals in keratinocytes. Thus, up-regulation of PPARβ by the FAN-mediated pathway, which is independent of the TRAF2-associated signaling (Adam-Klages et al. 1996), is an additional way of directing TNF-α signaling away from apoptosis and toward cell survival and differentiation. This novel pathway should contribute to avoid excessive cell death during inflammation (see below). Further support to this anti-apoptotic role of PPARβ comes from the observation that increased PPARβ levels in colorectal carcinoma confers resistance to NSAID-induced apoptosis (He et al. 1999).
Roles of elevated PPARβ expression during wound repair
Wound repair requires the integration of interdependent processes that involve, among others, soluble mediators, inflammatory cytokines produced by a variety of cell types, and extracellular matrix components. It comprises three successive major phases, that is, inflammation, re-epithelization, and tissue remodeling (Martin 1997). During the initial phase of inflammation, keratinocytes are exposed to a battery of pro-inflammatory cytokines and bioactive lipids. PPARβ expression is activated in the very early phase of the injury and remains high until wound closure, whereas PPARα expression is reactivated only transiently during the inflammation phase (Michalik et al. 2001). In agreement with the in vivo study, we also observed a transient increase in PPARα expression in cultured primary keratinocytes upon exposure to inflammatory signals. This transient PPARα expression, together with the fact that inflammatory eicosanoids, for example, LTB4 and 8S-HETE are PPARα ligands (Devchand et al. 1996), and an alteration in the recruitment of inflammatory cells to the wound bed in PPARα−/− mice (Michalik et al. 2001), reinforces the hypothesis that PPARα participates in the control of the inflammatory response. It is worth noting, with respect to the present work on the role of PPARβ, that the expression of PPARα in response to inflammatory signals is very similar in wild-type and PPARβ−/− keratinocytes, indicating that PPARα does not compensate for the lack of PPARβ. It also indicates that PPARα expression is not controlled by PPARβ.
The injury-triggered release of pro-inflammatory cytokines serves as an instant danger signal. Paradoxically, whereas these factors are important for hemostasis, recruitment of macrophages and removal of infectious agents, they are also apoptotic signals (Hannun and Luberto 2000). A role of PPARβ expression, induced by cytokines and bioactive lipids, would be to confer resistance against apoptotic signals as discussed above, thereby providing a critical window for the action of other factors, such as KGF, which modulate keratinocytes behavior (Werner 1998). A deficiency in PPARβ during this phase would result in an increased apoptosis of keratinocytes, hence, reducing the number of proliferating and migrating cells that are vital for wound closure. The results obtained in this study at wound edges of PPARβ+/− mice are in agreement with this hypothesis. Although we observed higher keratinocyte proliferation rates at the wound edges in mutant mice, the number of apoptotic cells was increased dramatically. Also consistent with data herein, Michalik et al. (2001) reported that the most significant differences in the rate of wound closure between wild-type and PPARβ+/− mice are observed during the first week of wound repair.
As wound repair enters into the re-epithelization phase, migrating keratinocytes have a major important role in rapid wound closure. The signals for the continued PPARβ activation at that stage might be related to T-cell activation that occurs at that time. The accelerated keratinocyte differentiation sustained by elevated and prolonged activation of PPARβ as seen in keratinocyte cultures, is likely to be important during the re-epithelialization phase. Furthermore, the role of PPARβ might then also be related to a different, specific spatio-temporal role. For example, we have reported that PPARβ+/− keratinocytes in culture are defective in substrate adhesion (Michalik et al. 2001). At present, it is unclear which are the PPARβ target genes that contribute to this phenotype.
In conclusion, PPARβ expression and activation might participate in the sequential regulatory events taking place during wound healing or inflammatory challenges. As deviation from this pattern may result in skin disorders, for example, psoriasis (Nickoloff et al. 2000), the exploration of the role of PPARβ in skin disorders might open important therapeutic perspectives and lead to the discovery of additional, so far unknown functions of this PPAR isotype.
Materials and methods
Reagents
All cytokines were purchased from PeproTech, Inc. N-acetyl-D-erythro-sphingosine (ceramide) and SB203580 were from CalBiochem. Anti-mouse-TNF-α antibodies were from R & D Systems. Keratinocyte serum-free culture medium (KSFM), Iscove's Modified Eagles Medium (IMDM), and Dispase were obtained from GIBCO-BRL. Fetal calf serum (FCS, <10 EU/mL) was from Sigma. Sterile nylon wool for enrichment of T-cells was from Robbins Scientifics. The in situ cell death detection kit was from Roche.
Cell cultures
Dendritic cells (DCs) were cultured as described by Gallucci et al. (1999). Splenic T lymphocytes were isolated as described by Peterson et al. (2000). Mouse keratinocytes were obtained from 3-day-old pups from PPARβ+/+, PPARβ+/−, and PPARβ−/− (Michalik et al. 2001) and isolated from epidermis as reported by Hager et al. (1999) with some modifications; epidermis was separated from the dermis following overnight incubation at 4°C in 2.5 U/mL of Dispase. Epidermis was placed in a 50-mL centrifuge tube with 10 mL of KSFM, and the tube was given 50 firm shakes. Keratinocytes were resuspended in KSFM containing 0.05 mM Ca2+ and 0.1 ng/ml epidermal growth factor, and seeded at 5 × 104 cells/cm2. Keratinocytes were used after two to three passages. Fibroblasts were similarly isolated from the dermal pieces. Fibroblasts were plated in IMDM containing 5% FCS and were used after three to five passages.
Mixed leukocyte reaction
Induction of apoptosis in fibroblasts was performed by UV irradiation as reported (Fadok et al. 1998). Apoptotic efficiency was monitored by caspase 8 activity. Necrosis was achieved by repeated freezing at −80°C and thawing at 37°C, or by mincing newborn mouse skin (1g/mL in PBS). Large cellular debris and any remaining viable cells were removed by centrifugation at 2000g. Necrosis was assessed by trypan blue uptake.
MLR were carried out as reported (Gallucci et al. 1999). Briefly, DCs were exposed for 24 h to supernatant of necrotic cells or medium from apoptotic cells in culture, after which the medium was discarded and the activated DCs were cocultured with T-cells for 48 h. Conditioned medium obtained from this MLR (CM) was clarified of cells by centrifugation and desalted via ultrafiltration with 3000 MW cutoff YM membrane (Ambion), prior to addition to keratinocytes. Ca2+ concentration of ultrafiltered medium was below 0.02 mM.
For TNF-α neutralization experiment, necrosis-derived CM was preincubated at 37°C for 1 h with 20 μg/mL of anti-TNF-α antibody. After treatment, the CM was added onto the keratinocytes. The 50% neutralization dose of anti-TNF-α was 0.6μg/mL in the presence of 0.25 ng/mL of TNF-α.
RNase protection assay
The cDNA corresponding to the mouse PPARα and PPARβ A/B domain, as well as gene-specific probes corresponding to Cyc A, INV, TgaseI, K6, K10, K17, and L27 were subcloned into pGEM3Zf(+) (Promega). Gene-specific antisense riboprobes were synthesized by in vitro transcription with either T7 or Sp6 RNA polymerase (Ambion). For all riboprobes, except L27, a ratio of 1:1 of α32P-UTP to cold UTP was used, whereas a ratio 1:20 was used for L27 probe.
RPA was carried out as described by the manufacturer (Ambion) with the following modifications: 1–2 × 106 keratinocytes were lysed in 200 μL of Lysis/Denaturation buffer and clarified by centrifugation through a Qiashredder (QIAGEN). A total of 45 μL of lysate was incubated with 1 ng of gene-specific riboprobes (1 × 109 cpm/μg) and 10 ng of L27 probe (1 × 107 cpm/μg). RPA products were resolved in a 6% electrolyte-gradient denaturing polyacrylamide gel. Gels were dried and exposed to X-ray film or to Phosphor screen.
Transfection of primary keratinocytes
Keratinocytes were trypsinized and replated into 12-well culture plates, and incubated for 24 h to reach 40%–50% confluence. Cells were transfected using Superfect (QIAGEN). At the indicated time, cells were rinsed with phosphate buffer and lysed in Reporter Lysis buffer (Promega) for β-galactosidase and CAT assay or in Lysis/Denaturation Solution for RPA. Activity or mRNA of cotransfected pCMV-β-galactosidase reporter was used to monitor transfection efficiency. The various MKK7 and SEK1 expression vectors were from E. Nishida (Kyoto University, Japan) and the various MEKK1 and SAPK/JNK expression constructs were from R.J. Davis (University of Massachusetts Medical School, Worcester) and D.J. Templeton (University of Virginia Medical School, Charlottesville).
The proximal 1.9-kb mouse PPARβ promoter region was subcloned upstream of the CAT reporter gene in pBLCAT. The 5′ deletion promoter mutants were generated using available restriction enzyme sites. Site-directed mutagenesis of transcription factor binding sites was achieved using the QuikChange Site-Directed mutagenesis kit (Stratagene).
The LBD of PPARβ, corresponding to amino acids 136–440, was PCR amplified, and subcloned into EcoRI/HindIII sites of pMGAL4 (Clontech). The PPRE-TK-Luc reporter was from R. Evans (Salk Institute for Biological Sciences, La Jolla, CA). Lipids of cytokine-induced keratinocytes were extracted as reported (Buck et al. 1991). Briefly, 5 × 106 keratinocytes were treated with indicated agents for 12 h. The organic extract was vacuum dried and the pellet dissolved in 10 μL of ethanol.
Apoptosis assays
TNF-α-induced apoptosis was carried out as reported (Reinartz et al. 1996). The apoptotic keratinocytes were measured using ApoAlert Caspase 8 Colorimetric assays (Clontech). Apoptosis was also monitored using radioactive DNA fragmentation assay (Duke 1983). Briefly, the cells, at a density of 2 × 104 cells/cm2, were prelabeled via incubation with 5 μCi/mL of [3H]thymidine for 24 h. TNFα-induced apoptosis were performed on these labeled cells. As background control, labeled cells were unchallenged. At indicated time points, the cells were lysed in 150 μL of Cell Lysis buffer (Boehringer Mannheim), and subsequently, 100 μL of supernatant were counted.
In vivo proliferation and apoptosis assays
The number of proliferative and apoptotic keratinocytes in the PPARβ+/+ and PPARβ+/− mice epidermis was analyzed on frozen sections of skin wounds, 4 and 7 d after a dorsal full-thickness wound. The detection of the Ki67 proliferation marker was performed as described (Michalik et al. 2001). The apoptotic keratinocytes were detected using the TUNEL assay according to manufacturer's protocol (Roche). Briefly, 7-μm cryosections of dorsal wounds were fixed (4% formaldehyde/PBS, 20 min at room temperature), treated with Proteinase K (20 mg/ml, 10 min at room temperature), and the fragmented DNA was labeled by use of fluorescent nucleotides. The slides were subsequently counterstained (DAPI), washed, and mounted before microscopic observation.
Acknowledgments
We thank E. Nishida, R.J. Davis, D.J. Templeton, and R. Evans for the gift of plasmids; J. Tschopp for critical reading of the manuscript; and N. Deriaz, T. Favez, and M. Narce for their assistance. This work was supported by grants from the SNSF, the Etat de Vaud, and the HFSP to B.D. and W.W. and by the Novartis Foundation to N.N.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL walter.wahli@iba.unil.ch; FAX 41-21-692-4115.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.207501.
References
- Adam-Klages S, Adam D, Wiegmann K, Struve S, Kolanus W, Schneider-Mergener J, Kronke M. FAN, a novel WD-repeat protein, couples the p55 TNF-receptor to neutral sphingomyelinase. Cell. 1996;86:937–947. doi: 10.1016/s0092-8674(00)80169-5. [DOI] [PubMed] [Google Scholar]
- Amri EZ, Bonino F, Ailhaud G, Abumrad NA, Grimaldi PA. Cloning of a protein that mediates transcriptional effects of fatty acids in preadipocytes. Homology to peroxisome proliferator-activated receptors. J Biol Chem. 1995;270:2367–2371. doi: 10.1074/jbc.270.5.2367. [DOI] [PubMed] [Google Scholar]
- Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Buck J, Myc A, Garbe A, Cathomas G. Differences in the action and metabolism between retinol and retinoic acid in B lymphocytes. J Cell Biol. 1991;115:851–859. doi: 10.1083/jcb.115.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- Devchand PR, Keller H, Peters JM, Vazquez M, Gonzalez FJ, Wahli W. The PPARalpha-leukotriene B4 pathway to inflammation control. Nature. 1996;384:39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]
- Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev. 1999;20:649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- Dotto GP. Signal transduction pathways controlling the switch between keratinocyte growth and differentiation. Crit Rev Oral Biol Med. 1999;10:442–457. doi: 10.1177/10454411990100040201. [DOI] [PubMed] [Google Scholar]
- Duke RC. Endogenous endonuclease-induced DNA fragmentation: An early event in cell-mediated cytolysis. Proc Natl Acad Sci. 1983;80:6361–6365. doi: 10.1073/pnas.80.20.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok VA, Bratton DL, Konowal A, Freed PW, Westcott JY, Henson PM. Macrophages that have ingested apoptotic cells in vitro inhibit pro-inflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J Clin Invest. 1998;101:890–898. doi: 10.1172/JCI1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallucci S, Lolkema M, Matzinger P. Natural adjuvants: Endogenous activators of dendritic cells. Nat Med. 1999;5:1249–1255. doi: 10.1038/15200. [DOI] [PubMed] [Google Scholar]
- Hager B, Bickenbach JR, Fleckman P. Long-term culture of murine epidermal keratinocytes. J Invest Dermatol. 1999;112:971–976. doi: 10.1046/j.1523-1747.1999.00605.x. [DOI] [PubMed] [Google Scholar]
- Hanley K, Jiang Y, He SS, Friedman M, Elias PM, Bikle DD, Williams ML, Feingold KR. Keratinocyte differentiation is stimulated by activators of the nuclear hormone receptor PPARalpha. J Invest Dermatol. 1998;110:368–375. doi: 10.1046/j.1523-1747.1998.00139.x. [DOI] [PubMed] [Google Scholar]
- Hannun YA, Luberto C. Ceramide in the eukaryotic stress response. Trends Cell Biol. 2000;10:73–80. doi: 10.1016/s0962-8924(99)01694-3. [DOI] [PubMed] [Google Scholar]
- Hansen JB, Zhang H, Rasmussen TH, Petersen RK, Flindt EN, Kristiansen K. PPARdelta-mediated regulation of preadipocyte proliferation and gene expression is dependent on cAMP signaling. J Biol Chem. 2000;276:3175–3182. doi: 10.1074/jbc.M005567200. [DOI] [PubMed] [Google Scholar]
- He TC, Chan TA, Vogelstein B, Kinzler KW. PPARdelta is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell. 1999;99:335–345. doi: 10.1016/s0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Koseki T, Chen S, Wu X, Nunez G. CIDE, a novel family of cell death activators with homology to the 45kDa subunit of the DNA fragmentation factor. EMBO J. 1998;17:2526–2533. doi: 10.1093/emboj/17.9.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreder D, Krut O, Adam-Klages S, Wiegmann K, Scherer G, Plitz T, Jensen JM, Proksch E, Steinmann J, Pfeffer K, et al. Impaired neutral sphingomyelinase activation and cutaneous barrier repair in FAN-deficient mice. EMBO J. 1999;18:2472–2479. doi: 10.1093/emboj/18.9.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronke M. Involvement of sphingomyelinases in TNF signaling pathways. Chem Phys Lipids. 1999;102:157–166. doi: 10.1016/s0009-3084(99)00084-5. [DOI] [PubMed] [Google Scholar]
- Kyriakis JM, Avruch J. Protein kinase cascades activated by stress and inflammatory cytokines. BioEssays. 1996;18:567–577. doi: 10.1002/bies.950180708. [DOI] [PubMed] [Google Scholar]
- Mansbridge JN, Knapp AM. Changes in keratinocyte maturation during wound healing. J Invest Dermatol. 1987;89:253–263. doi: 10.1111/1523-1747.ep12471216. [DOI] [PubMed] [Google Scholar]
- Martin P. Wound healing–aiming for perfect skin regeneration. Science. 1997;276:75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- Messmer UK, Pfeilschifter J. New insights into the mechanism for clearance of apoptotic cells. BioEssays. 2000;22:878–881. doi: 10.1002/1521-1878(200010)22:10<878::AID-BIES2>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Michalik L, Desvergne B, Tan NS, Basu-Modak S, Escher P, Rieusset J, Peters JM, Kaya G, Gonzalez FJ, Zakany J, et al. Impaired skin wound healing in PPARα and PPARβ mutant mice. J Cell Biol. 2001;154:799–814. doi: 10.1083/jcb.200011148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Costanzo A, Guido F, Moretti F, Levrero M. Apoptotic, non-apoptotic, and anti-apoptotic pathways of tumor necrosis factor signalling. Biochem Pharmacol. 1998;56:915–920. doi: 10.1016/s0006-2952(98)00154-3. [DOI] [PubMed] [Google Scholar]
- Nickoloff BJ, Schroder JM, von den Driesch P, Raychaudhuri SP, Farber EM, Boehncke WH, Morhenn VB, Rosenberg EW, Schon MP, Holick MF. Is psoriasis a T-cell disease? Exp Dermatol. 2000;9:359–375. doi: 10.1034/j.1600-0625.2000.009005359.x. [DOI] [PubMed] [Google Scholar]
- Nuclear Receptors Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- Oliver WR, Jr, Shenk JL, Snaith MR, Russell CS, Plunket KD, Bodkin NL, Lewis MC, Winegar DA, Sznaidman ML, Lambert MH, et al. A selective peroxisome proliferator-activated receptor delta agonist promotes reverse cholesterol transport. Proc Natl Acad Sci. 2001;98:5306–5311. doi: 10.1073/pnas.091021198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Lee SS, Li W, Ward JM, Gavrilova O, Everett C, Reitman ML, Hudson LD, Gonzalez FJ. Growth, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor beta(delta) Mol Cell Biol. 2000;20:5119–5128. doi: 10.1128/mcb.20.14.5119-5128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KE, Iwashiro M, Hasenkrug KJ, Chesebro B. Major histocompatibility complex class I gene controls the generation of gamma interferon-producing CD4(+) and CD8(+) T cells important for recovery from friend retrovirus-induced leukemia. J Virol. 2000;74:5363–5367. doi: 10.1128/jvi.74.11.5363-5367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puignero V, Queralt J. Effect of topically applied cyclosporin A on arachidonic acid (AA)- and tetradecanoylphorbol acetate (TPA)-induced dermal inflammation in mouse ear. Inflammation. 1997;21:357–369. doi: 10.1023/a:1027358102096. [DOI] [PubMed] [Google Scholar]
- Reinartz J, Bechtel MJ, Kramer MD. Tumor necrosis factor-alpha-induced apoptosis in a human keratinocyte cell line (HaCaT) is counteracted by transforming growth factor-alpha. Exp Cell Res. 1996;228:334–340. doi: 10.1006/excr.1996.0333. [DOI] [PubMed] [Google Scholar]
- Saluja I, Granneman JG, Skoff RP. PPARdelta agonists stimulate oligodendrocyte differentiation in tissue culture. Glia. 2001;33:191–204. [PubMed] [Google Scholar]
- Savill J, Fadok V. Corpse clearance defines the meaning of cell death. Nature. 2000;407:784–788. doi: 10.1038/35037722. [DOI] [PubMed] [Google Scholar]
- Spellberg B. The cutaneous citadel: A holistic view of skin and immunity. Life Sci. 2000;67:477–502. doi: 10.1016/s0024-3205(00)00653-6. [DOI] [PubMed] [Google Scholar]
- Szabowski W, Mass-Szabowsk N, Andrecht S, Kolbus A, Schorpp-Kistner M, Fusenig NE, Angel P. c-Jun and JunB antagonistically control cytokine-regulated mesenchymal-epidermal interaction in skin. Cell. 2000;103:745–755. doi: 10.1016/s0092-8674(00)00178-1. [DOI] [PubMed] [Google Scholar]
- Werner S. Keratinocyte growth factor: A unique player in epithelial repair processes. Cytokine Growth Factor Rev. 1998;9:153–165. doi: 10.1016/s1359-6101(98)00010-0. [DOI] [PubMed] [Google Scholar]
- Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Prüß M, Reuter I, Schacherer F. TRANSFAC: An integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]