Abstract
Recent studies have implicated multipotential mesenchymal stem cells (MSCs) as an aid to breast cancer cell proliferation and metastasis, partly as a result of the MSCs secretome. As the tumor gets beyond 2 mm in diameter, the stromal cells could undergo starvation due to the lack of sufficient nutrients in solid tumor microenvironment. In this study, we investigated the survival mechanisms used by stressed stromal cells in breast cancers. We used serum-deprived mesenchymal stem cells (SD-MSCs) and MCF-7 breast cancer cells as model system with a hypothesis that stromal cells in the nutrient-deprived core utilize survival mechanisms for supporting surrounding cells. We tested this hypothesis using in vivo tumor xenografts in immunodeficient mice, which indicated that SD-MSCs supported MCF-7 tumor growth by protection from apoptosis. Histochemical assays showed that SD-MSCs-injected tumors exhibited higher cellularity, decreased apoptosis and decreased differentiation. Beclin-1 staining indicated autophagic areas surrounded by actively proliferating cells. Furthermore, in vitro studies demonstrate that SD-MSCs survive using autophagy and secrete paracrine factors that support tumor cells following nutrient/serum deprivation. Western blot and immunocytochemistry analysis of SD-MSCs demonstrated upregulation and perinuclear relocation of autophagy key regulators such as beclin-1, ATG10, ATG12, MAP-LC3 and lysosomes. Electron microscopic analysis detected a time-dependent increase in autophagosome formation and HDAC6 activity assays indicated the upregulation of autophagy. Taken together, these data suggest that under nutrient-deprived conditions that can occur in solid tumors, stromal cells utilize autophagy for survival and also secrete anti-apoptotic factors that can facilitate solid tumor survival and growth.
Introduction
Breast cancer remains as the leading form of carcinoma affecting women. Current knowledge regarding the biology of breast cancer stroma indicates that mesenchymal stem cells (MSCs) provide the supportive stroma for tumors either by homing to tumor or by surrounding the tumors without infiltration, suggesting that the effect MSCs have on tumor growth kinetics could be a result of stromal factors and paracrine signaling (1). Karnoub et al. (2) proposed that the metastatic traits of breast cancer cells are acquired through exposure of the epithelial cells to mesenchymal cells in the tumor-associated stroma. Both extracellular matrix and growth factors are important regulators of stromal–tumor cell interactions in mammary tumor progression (3–6). Specifically, the role of MSCs in providing survival advantage and drug resistance to various hematological tumors has been described (7–9). However, there is a significant gap in our understanding of the survival mechanisms used by stromal cells under stressful conditions normally observed within solid tumors, such as hypoxia or nutrient deprivation.
Autophagy is a highly conserved catabolic program that has been shown to act as both a pro-survival or pro-death mechanism in different physiological and pathological conditions (10–12). Autophagy is an essential component of growth regulation and maintenance of homeostasis in multicellular organisms in which degradation of organelles and proteins provides macromolecules needed for cell survival (13). Most primary cells resort to this pathway for short-term survival during brief periods of serum/nutrient deprivation followed by full recovery when nutrients are replaced. Prolonged serum/nutrient deprivation, however, induces apoptosis in these primary cells (14–16).
In this study, we report for the first time that the stromal cell property of MSCs in breast cancers is associated with upregulation of autophagy. Xenograft studies using MCF-7 cells and serum-deprived mesenchymal stem cells (SD-MSCs) indicated that tumors with SD-MSCs were less differentiated. Interestingly, in vitro studies replicating the nutrient-deprived conditions of solid tumor showed that MSCs cultured in serum-free medium survive prolonged serum deprivation. Our data suggest that SD-MSCs utilize autophagy to recycle macromolecules and synthesize anti-apoptotic factors for facilitating the survival and growth of surrounding tumor cells.
Methods and materials
In vivo assays
Orthotopic (mammary fat pad) injections were done on 15 8-week-old female Fox Chase SCID Beige mice (CB17.Cg-PrkdcSCIDLystbg/crl strain; Charles River Laboratories, Wilmington, MA), with two injections per mouse (10 injections per group total). We used the MCF-7 breast cancer cell line and two different male donors for MSCs. These cells were mixed and co-injected at a ratio of 2 million MCF-7 cells to 1 million MSCs or SD-MSCs (2:1 ratio) according to previously optimized condition for high tumor implantation rate (17) in 100 μl of Hanks Balanced Salt Solution; Invitrogen, Carlsbad, CA) with 50 μl of growth factor reduced basement membrane Matrigel matrix or Cultrex basement membrane (BD Biosciences, San Jose, CA). Tumors were collected after 15 days for histological studies.
Cell culture
Frozen vials of characterized human MSCs at passages two to four were obtained from the Adult Stem Cell Core of the Louisiana Cancer Research Consortium (http://www.lcrc.info/research/stemCell.html). The serum-deprived MSCs were obtained as described previously (18). Briefly, cells were cultured until they reached 80% confluency, washed three times with phosphate-buffered saline (PBS) and then cultured without fetal bovine serum (FBS). Serum-deprived conditioned media was obtained by collection and 0.22 μm filtration of the supernatant media from serum-deprived MSCs cells. MCF-7 and KHOS cell lines were obtained from ATCC, Manassas, VA. Both cancer cell lines were maintained in culture with Dulbecco’s modified essential medium (Gibco BRL), 10% FBS; Atlanta Biologicals, Lawrenceville, GA), 100 U/ml penicillin, 100 μg/ml streptomycin.
Transwell assay
MSCs were plated in inserts of Transwell Permeable Support 0.4 μm pore size (Corning, Lowell, MA) (105 cells per insert) and MCF-7 breast cancer cells were plated in 12-well Transwell plate (105 cells per well). After 6 h, some inserts were switched to serum-deprived media for 16 h. Then, MCF-7 were placed in serum-free media and incubated 24 h with inserts containing MSCs or SD-MSCs. Total MCF-7 cells DNA content was evaluated using Cyquant assay (Invitrogen, Carlsbad, CA) as described previously (18).
Detection of apoptosis in tissue sections (TUNEL assay)
Paraffin tissue sections were rehydrated and stained with the FragEL™ DNA fragmentation detection kit (EMD Chemicals, Gibbstown, NJ) following the manufacturer protocol. Briefly, sections were permeabilized 20 min with 20 μg/ml of Proteinase K. After equilibration, samples were incubated 1.5 h at 37°C with the terminal deoxynucleotidyl transferase labeling mixture, mounted and visualized by fluorescence microscopy. Fluorescence intensity was evaluated by ImageJ software (W.S.Rasband, ImageJ; US National Institutes of Health, Bethesda, MD, http://rsb.info.nih.gov/ij/, 1997–2008). Results are expressed as ratio of fluorescein isothiocyanate/4′,6-diamidino-2-phenylindole intensity.
Immunohistochemistry
Tumor paraffin sections were deparaffinized, rehydrated and permeabilized 10 min in PBS containing 0.25% Triton X-100. Sections were blocked overnight at 4°C in PBS containing 10% goat serum, 1% bovine serum albumin, 0.05% Tween-20 and then incubated overnight with a mouse monoclonal Beclin-1 antibody (1:50, #AM1818a; Abgent, San Diego, CA) in PBS with 1% bovine serum albumin. The next day, sections were washed and incubated 1 h with a goat anti-mouse AlexaFluor 488 antibody (1:250; Invitrogen). After washes, sections were mounted with Supermount (BioGenex, Fremont, CA) containing 0.5 μg/ml Hoechst 33342 (Invitrogen).
Detection of apoptosis in living cells
KHOS cells were treated with 1 μM staurosporine for 72 h in the presence of 50% SD-MSCs conditional medium or alpha-Minimum Essential Medium as control. The level of apoptosis in living cells was analyzed by Magic Red Caspase detection kit (Immunochemistry Technologies, Bloomington, MN). The cells were visualized by fluorescence microscopy.
Alkaline comet assay
MCF-7 breast carcinoma cells were cultured at 500 cells/cm2 in Dulbecco’s modified essential medium supplemented with 10% FBS. Plates at 75% confluency were treated with 200 μM H2O2 for 18 h in the presence of 50% SD-MSCs conditional medium or alpha-Minimum Essential Medium (control). After 18 h treatment, the medium was changed to Dulbecco’s modified essential medium 10% FBS for 4 h. The alkaline comet assay was performed as described previously (18).
Chick embryo survival assay
Fertilized day 0 eggs were obtained from Charles River Laboratories and left at room temperature for 2 days, which drops survival of embryos to ∼50%. We injected 200 μl of SD-MSCs conditional medium, or alpha-Minimum Essential Medium as control media, to the eggs. The embryos were harvested at 9 days for viability and classified as fully developed or undeveloped.
Electron microscopy
Cells grown in 25 cm2 flasks were fixed in a solution of 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer pH 7.4 for 1 h at room temperature. Then, samples were washed three times for 5 min in 0.1 M sodium cacodylate buffer and sent to the Electron Microscopy Core Facility of the Department of Cell Biology at the Yale University School of Medicine (New Haven, CT).
Western blotting
Proteins were extracted and fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis as described previously (19). Dilution and provider of primary antibodies used were as follows: ATG12 (1:100, #AM1816a; Abgent), ATG10 (1:100, #AP1815a; Abgent), RGS19 (1:100, #AP1820a; Abgent), MAP-LC3 (1:100, #sc28266; Santa Cruz Biotechnology, Santa Cruz, CA) and β-actin (1:1000, #ab20272; Abcam, Cambridge, MA).
Autophagy inhibitors assay
MSCs were placed in serum deprived condition and treated 24 h with 100 μM chloroquine or with 100 nM bafilomycin A1 (Sigma–Aldrich, Saint Louis, MO). Cells were both trypsinized for counting and lysed to evaluate DNA content by Cyquant assay (Invitrogen).
Beclin-1 shRNA transfection
Human MSCs (5 × 104 cells) were transfected using 2 μg of pLKO.1 lentivirus vector containing the hairpin sequence or with the vector alone in 100 μl of buffer provided by the Human MSC Nucleofector kit and using Nucleofector® II device (Lonza, Basel, Switzerland). After 48 h, transfected cells were selected with puromycin (0.5 μg/ml; Sigma–Aldrich) during 8 days. Selected cells were subcultured and grown until 80% confluency before starting serum deprivation. After 5 days of starvation, cells were harvested and lysed and analyzed for total protein per dish using BCA assay (Thermo Fisher Scientific, Rockford, IL). Among five different hairpin sequences tested, we selected the clone ID TRCN0000033552 (Open Biosystems, Huntsville, AL), which give the strongest beclin-1 downregulation.
Immunocytochemistry
MSCs and SD-MSCs were grown on coverslips at densities between 200–500 cells/cm2 for a period of 20 days. Briefly, cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton in PBS for 10 min. Slides were then incubated for 1 h in Tris-buffered saline (0.05% Tween-20 in PBS) with 1% bovine serum albumin as a blocking solution, 2 h with primary antibodies, followed by incubation for 1 h at room temperature with fluorescein isothiocyanate secondary antibodies (1:500) or Alexa Fluor-conjugated secondary antibodies (1:200; Invitrogen), unless otherwise noted. Coverslips were mounted in Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (Vector laboratories, Burlingame, CA) and observed by a fluorescence microscope. Dilution and origin of primary antibodies used in this study were as follows: rabbit monoclonal anti-beclin-1 (1:100, #ab51031; Abcam); mouse monoclonal anti-acetylated α-tubulin (1:100, # ab24610; Abcam); rabbit polyclonal anti-HDAC6 (1:250, #H2287; Sigma–Aldrich) and rabbit polyclonal anti-ATG12 (1:200, #AP1816a; Abgent).
Motility assay
MSCs and SD-MSCs were incubated with 5 μM CellTracker™ Green CMFDA (5-chloromethylfluorescein diacetate Celltracker (Invitrogen) for 45 min, washed and replated in a 24-well HTS FluoroBlok plate insert system, 8.0 μm pore size (BD Biosciences), in serum-free media in the presence or absence of 20 μM Tubacin. Cell motility was measured as described previously (20).
Results
Stromal support of MSCs in large MCF-7 tumors is associated with upregulation of autophagy
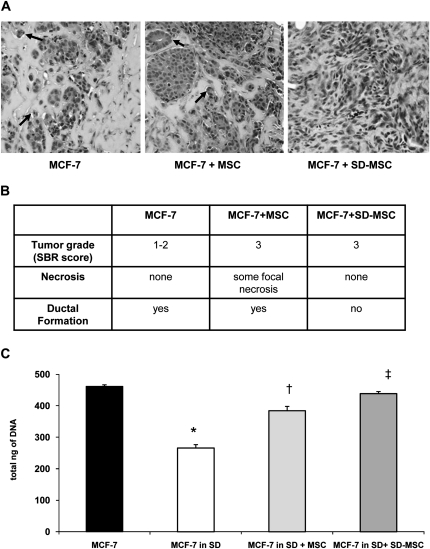
Recent studies have established that MSCs provide sufficient stromal support for tumor cells (21). To determine whether MSC-secreted growth factors would enhance the proliferation and survival of breast cancer cells, we injected MCF-7 cells alone or with MSCs in Matrigel into mammary fat pads of immunodeficient SCID-beige mice. Serum-deprived MSCs were included as surrogate model cells to test the hypothesis that MSCs survive the transient nutrient deprivation associated with large tumors and secrete growth factors. Previous experiments to optimize the MCF-7:MSC tumor model indicated that a ratio of 2:1 had close to 90% tumor implantation rate (17). Therefore, we chose to use an established model to test the concept of role of autophagy in stromal support. Hematoxylin–eosin sections of the tumors indicated higher cellularity in both MSCs and SD-MSC co-injected tumors (compare with MCF-7 control group) but with no significant difference between MSCs and SD-MSCs groups (Figure 1A). However, the tumors generated from SD-MSCs co-injected group had decreased ductal formation compared with the MSC co-injected or control groups (Figure 1A and B).
Fig. 1.
SD-MSCs enhance MCF-7 breast carcinoma proliferation (A) hematoxylin–eosin staining of a representative tumors from three groups, black arrows indicate ductal formation (magnification ×200). (B) Pathological analysis of the tumors described in above using Scarff-Bloom-Richardson (SBR) score, necrosis and ductal formation (C) Transwell assay showing that SD-MSC supports the survival of MCF-7 placed in serum deprivation (SD) condition (*P < 0.001 compared with MCF-7, n = 3, †P< 0.001 compared with MCF-7 in SD, ‡P < 0.01 compared with MCF-7 in SD + MSC).
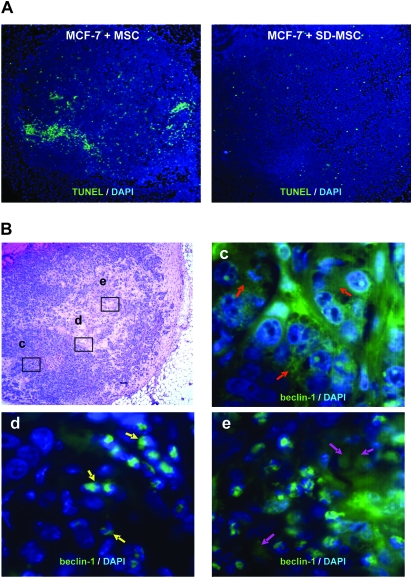
MCF-7 cells placed in serum-deprived conditions show a significant decrease in cell viability (by 60 ± 1%, P < 0.001, n = 3), but in vitro co-culture assays demonstrated that MSC or SD-MSCs helped to maintain survival of MCF-7 cells (by 83 ± 3% and 95 ± 1%, respectively) under serum-deprived conditions. These data support our in vivo observations suggesting that SD-MSCs have better stromal supportive properties than MSCs (Figure 1C), supporting our hypothesis that MSCs retain the stromal supportive properties under nutrient-deprived conditions. In addition, previously published data on secretion of stromal supportive factors during hypoxia further support our hypothesis (20,22). Next, we sought to identify the mechanism underlying the survival of these MSCs in the presumably nutrient-deprived core of the tumor. TUNEL staining showed a significant decrease of apoptotic nuclei in tumors from the group of MCF-7 cells with SD-MSCs compared with tumors from MCF-7 with MSCs (1.8 ± 0.6 versus 5.7 ± 0.7, P < 0.001, n = 6) (Figure 2A). However, immunohistochemical detection assays for autophagic cells showed that similar sized tumors from both SD-MSC and MSC co-injections demonstrated characteristic expression of the autophagy marker, beclin-1, bordering the proliferating cells on one side and necrotic cells on the other side (Figure 2B). These data confirm previously published observations regarding the presence of autophagic cells in breast tumors (23,24). Fluorescent in situ hybridization assays for detection of the Y chromosome were performed 2 weeks later confirmed the continued presence of human MSCs in the tumors (supplementary Figure S1 is available at Carcinogenesis Online).
Fig. 2.
Autophagic SD-MSCs prevent apoptosis in tumor (A) MCF-7 cells were suspended in Matrigel and either injected alone or co-injected with MSCs or SD-MSCs in the mammary fat pad of SCID beige mice. After 2 weeks, tumor sections were stained for apoptotic cells using a TUNEL assay (magnification ×40). (B) Autophagy beclin-1 marker is gradually relocalized to the nucleus in the inner MCF-7 tumor. MCF-7 and SD-MSCs were co-injected in mammary fat pad of SCID beige mice. After 2 weeks, tumors were collected, sectioned and stained. Top left: hematoxylin–eosin staining of a representative section from a fully developed tumor. Top right (background set high): immunohistochemistry of beclin-1 revealed a diffuse staining in growing parts of the tumor (red arrows). Bottom left: at the frontier of the necrotic part of the tumor, beclin-1 staining is perinuclear in some cells (yellow arrows). Bottom right: in the necrotic part, most of the cells showed a relocation of beclin-1 around the nucleus (purple arrows), in addition to expressing condensed nuclei compared with the other part of the tumor. Magnification: top left panel: ×40; c, d and e: ×1000.
SD-MSCs release anti-apoptotic or pro-survival factors during serum deprivation
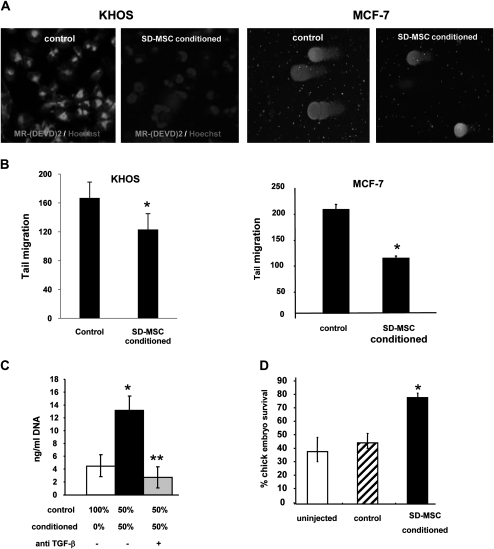
It was important to determine whether the novel stromal supportive properties that we had observed were limited only to MSCs as a physiological adaptation with in vivo consequences. Previously, we had demonstrated, using cytokine blots, that conditioned medium from SD-MSCs expressed secreted growth factors and anti-apoptotic factors (19). Notable pleiotropic factors involved in growth, apoptosis and tumor growth identified in the conditioned medium were insulin-like growth factor 1, insulin-like growth factor 2, transforming growth factor (TGF) β and IGFBP-2 (19). Therefore, we sought to test the role of these secreted factors in both in vitro and in vivo models. We tested for the anti-apoptotic factors secreted by SD-MSCs using two different cell lines, an epithelial cancer (MCF-7) and a mesenchymal cancer (KHOS). First, the KHOS osteosarcoma cell line was treated with apoptosis inducer, staurosporin, either in the presence or absence of conditioned media from SD-MSCs. As shown in Figure 3A, SD-MSCs-conditioned media decreased caspase activation and apoptosis in the KHOS cells (left panel). A second approach to test for anti-apoptotic factors released by SD-MSCs used MCF-7 breast cancer cells and KHOS. Cells were treated with hydrogen peroxide (H2O2) to induce DNA damage either in the presence of conditioned media from SD-MSCs or control medium for 24 h. DNA damage was detected using the COMET assay (25) and the tail length (Figure 3A, right panel) was quantified in 70 comets for each group. As indicated in Figure 3B, the presence of conditioned medium from SD-MSCs resulted in significant decrease in DNA damage. To determine whether TGFβ secreted by SD-MSCs played a role in the anti-apoptotic properties of SD-MSCs-conditioned media, we performed a similar apoptosis assay using conditioned media that had been immunodepleted for TGFβ. The data show that depletion of TGFβ decreased the protective effect of conditioned media on apoptosis (Figure 3C).
Fig. 3.
SD-MSCs secrete anti-apoptotic and survival factors during serum deprivation (A) (Two left panels) dual staining of osteosarcoma KHOS cells with Hoechst 33342 and MR-(DEVD)2 following 24 h 1 μg/ml staurosporin treatment either in the presence of serum-deprived conditioned medium or with control medium. Apoptotic cells bearing red lysosomal bodies with less intense blue nuclei are detected in control. SD-MSCs show non-apoptotic cells bearing bright blue nuclei and absent or reduced lysosomal staining (magnification ×600). (Two right panels) show representative images of Comet assay performed on MCF-7 cells obtained with and without SD-MSC-conditioned media. (B) Quantification of tail migration in a Comet assay of KHOS (left panel) MCF-7 (right panel) cells pre-treated with H2O2 either in the presence of SD-MSC-conditioned or -control media. (*P < 0.01). (C) KHOS osteosarcoma cells were treated as in (3A) either in the presence of control medium, serum-deprived conditioned medium or serum-deprived conditioned medium immunodepleted for TGF-β. The surviving cells were quantified by a Cyquant DNA quantification method (*P < 0.05 compare with control 100%; **P < 0.01 compare with conditioned 50% without anti-TGFβ, n = 3) (D) Day 2 chick embryos were either injected with both control medium or conditioned medium from SD-MSCs and embryo survival was determined at day 9 (*P < 0.01 compare with control).
The preceding in vitro models demonstrated that conditioned medium from SD-MSCs protected against apoptotic insults in an unrelated cell type. Next, we tested whether conditioned medium could protect against apoptosis in an in vivo model using a developing chick embryo. Delaying the start of incubation by incubating fertilized eggs at (the non-permissive) room temperature can reduce the survival of embryos by ∼50%. However, injecting these eggs with 200 μl of SD-MSC-conditioned medium increased their survival rates from 50 to 80% at the non-permissive temperature (Figure 3D).
Autophagosomes are formed during serum deprivation of MSCs
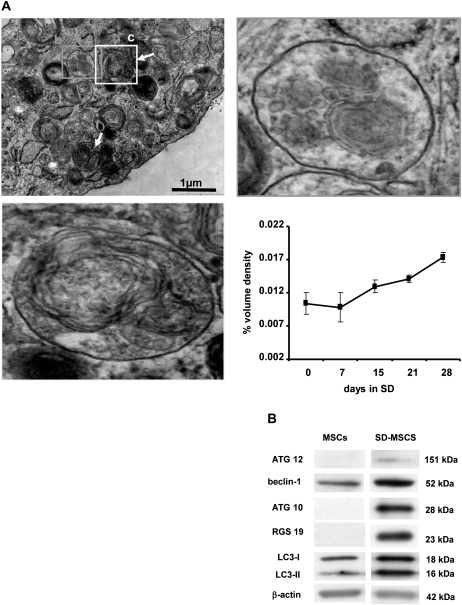
Previous studies from our laboratory have uncovered a unique survival property of MSCs in response to serum deprivation (18). Identifying the molecular pathways that regulate the survival mechanisms will be critical in understanding the stromal properties of these cells. As a first step, we performed gene expression and 2D proteomic assays on SD-MSCs compared with MSCs that were cultured in complete serum for 48 h to understand the transcriptome and proteome of MSCs during serum deprivation (supplementary Figure S2 is available at Carcinogenesis Online). In the gene expression microarrays, hierarchical clustering of the genes by their patterns of expression indicated an upregulation of autophagy-related genes (supplementary Table S1 and Figure S2 are available at Carcinogenesis Online). Time course electron microscopic analysis to determine autophagosome formation by MSCs during serum deprivation showed the presence of characteristic double membrane organelles (Figure 4A). To measure autophagosomes formation, the ratio of autophagosome volume to cellular volume was determined and compared with normal MSCs (day 0). As shown in the graph, in the lower right panel of Figure 4A, the number of autophagosomes increased with time of incubation in serum-deprived medium.
Fig. 4.
Autophagosomes are formed during serum deprivation of MSCs. (A) Top left: electron micrograph showing a small portion of SD-MSCs on day 15 of serum deprivation. In addition to autophagosomes (orange arrows and magnified area at top right) and autolysosomes (yellow arrows and magnified area at bottom left). Bottom right: quantification of autophagosomes by densitometry analysis of organelles. Data are represented as the volume ratio of autophagosomes to cellular volume. (B) Western blot showing upregulation of indicated autophagy proteins in SD-MSCs after 28 days of serum deprivation.
Both protein and messenger RNA expression profiles detected upregulated proteins involved in various steps of autophagolysosome formation. We examined the expression of Beclin-1, ATG10, ATG12 and MAP-LC3 by both immunohistochemistry and western blotting. As shown in Figure 4B, autophagy markers are upregulated in SD-MSCs compared with control MSCs. In addition, we observed that the autophagy-related markers were relocalized to perinuclear areas (supplementary Figure S3 is available at Carcinogenesis Online).
Autophagy is essential for MSC survival during serum deprivation
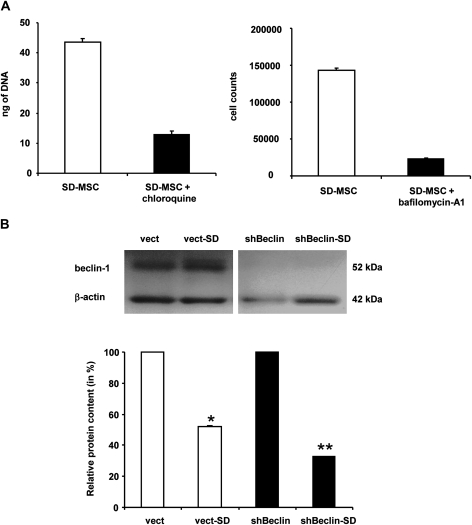
Next, logical step to address the role of autophagy in the stromal support of MSCs is to test the effect of autophagy knock down on the survival of SD-MSCs. Exposure of SD-MSCs to chloroquine (100 mM) and bafilomycin-A1 (100 nM), known pharmacological autophagy inhibitors (26,27), led to 4-fold decrease in cell survival (Figure 5A). To further confirm the role of autophagy in SD-MSC survival, MSCs were transducted with lentiviral shRNA to beclin-1. Western blot assays showed decreased beclin-1 levels in MSCs following shRNA transduction as compared with control vector (Figure 5B). The stable transfectants were cultured in serum-free medium showed a significant decrease in survival of MSCs that expressed shRNA to beclin-1 (66% versus 48% in control cells, P < 0.001) (Figure 5C).
Fig. 5.
Autophagy is essential for SD-MSC survival. (A) Effect of autophagy inhibitors on serum-deprived cell: MSCs were serum-deprived (SD-MSC) and treated for 24 h with 100 μM of chloroquine or 100 nM of bafilomycin-A1. (B) shBeclin-1 transfection induced SD-MSC cell death. Top: co-detection of beclin-1 and β-actin expression in MSCs transfected by control vector or shBeclin vector. Bottom: total protein expression in MSCs after 4 days in serum deprived conditions (*P < 0.001 compare with control vector, **P < 0.001 compare with vector-SD, n = 3).
Serum deprivation induces HDAC6 activity and increases motility in MSCs
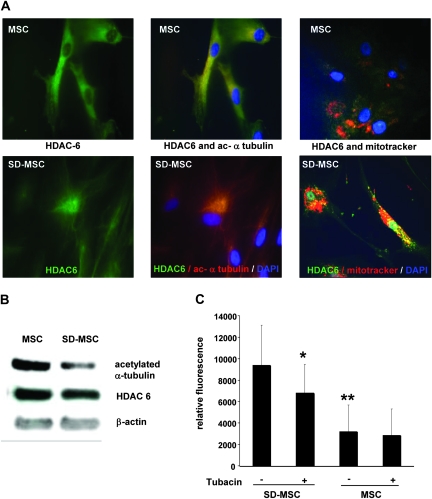
Unlike the other histone deacetylases, HDAC6 activity can be detected in the cytoplasm and results in deacetylation of α-tubulin necessary for dynein motor complex migration. Recent evidence shows that HDAC6, a ubiquitin-binding deacetylase, is a central player in autophagy, targeting protein aggregates and damaged mitochondria by controlling the fusion of autophagosomes to lysosomes (28). HDAC6 recruits actin-remodeling machinery that stimulates autophagosome–lysosome fusion and substrate degradation (29). A decrease in acetylated α -tubulin was observed in proteomic assays of SD-MSCs (supplementary Figure S2 is available at Carcinogenesis Online). This was further confirmed by immunocytochemical assays that demonstrated a perinuclear colocalization of HDAC6 with acetylated α-tubulin coupled to a decrease in acetylated α-tubulin expression (Figure 6A and B). Furthermore, immunocytochemistry showed perinuclear colocalization of HDAC6 and mitochondria in SD-MSCs. Thus, unlike in MSCs, where mitochondria are not colocalized with HDAC6 and acetylated α-tubulin is expressed evenly on the cell membrane (Figure 6A), the data presented here suggest that the components of autophagy are relocalized to perinuclear locations in SD-MSCs.
Fig. 6.
Increased HDAC6 activity and motility in SD-MSCs. (A) Immunocytochemistry of MSCs and SD-MSCs at day 20 indicate the change in the localization of HDAC6 (in green) and acetylated α-tubulin proteins (in red). Magnification ×1000 (B) western blot analysis indicating decreased acetylated α-tubulin, with minimal change in HDAC6 expression. (C) Tubacin, a specific HDAC6 inhibitor, decreased SD-MSCs migration but has no effect on MSCs (**P < 0.05 between Tubacin treated SD-MSCs and untreated, **P < 0.005 between untreated MSCs and untreated SD-MSCs).
Decrease in acetylated α-tubulin is also associated with increased cell motility (30–32). To demonstrate the functional activation of HDAC6 in response to autophagy induction in SD-MSCs, we performed migration assays for SD-MSCs and MSCs in the presence of specific HDAC6 inhibitor, Tubacin. First, we observed that SD-MSCs showed a 3-fold increase in the migration capacity compared with MSCs (Figure 6C). Second, addition of Tubacin significantly decreased the migration ability of SD-MSCs but not MSCs suggesting a role for HDAC6 activity on SD-MSCs mobility (Figure 6C). Next, we determined whether upregulation of HDAC6 activity was associated with increased cell mobility that coupled with increased matrix metalloproteinase (MMP) activity in MSCs. We tested the MMP expression and activity in SD-MSCs as compared with MSCs; both microarray assays, MMP activity assays showed a decrease in the MMP activity in SD-MSC suggesting that the HDAC6 activity is linked to the autophagosome formation rather than increased cell motility (supplementary Figure S4 is available at Carcinogenesis Online).
Taken together, these data suggest that the stromal cells of breast cancers secrete survival factors to support the tumor progression. In addition, these cells survive in the nutrient-deprived core by utilizing autophagic recycling mechanisms. These data further suggest that the pro- survival properties of SD-MSCs in response to serum deprivation were not just limited to the MSCs but that the release of survival factors could protect adjacent unrelated cells from apoptosis as demonstrated by the chicken embryo experiment and the preceding in vitro cell culture experiments.
Discussion
The present study combined with our previous publications (20,22) has implications for poorly vascularized epithelial solid tumors that harbor regions in which both the transformed epithelial cells and surrounding stromal cells are nutrient deprived. A large body of evidence exists suggesting that epithelial cells in breast, prostate and ovarian tumors resort to autophagy or self-catabolism to survive during periods of serum/nutrient deprivation and to prevent necrosis (33–35). This is the first study to address the role of autophagy in the stromal component of a tumor. Interestingly, autophagy in stromal cells occurs concomitantly with secretion of survival and supportive factors that inhibit apoptosis in the cancer cells (34,36).
Our laboratory had previously shown that subjecting early passage MSCs to serum deprivation for 2–4 weeks selects for a distinct subpopulation of cells; the SD-MSCs are remarkable in that they survive complete serum deprivation for prolonged periods of time, have longer telomeres and enhanced expression of genes that are expressed primarily in early progenitor cells (18). The results described herein demonstrate that SD-MSCs exhibit stromal cell properties including (i) survival under adverse conditions of nutrient deprivation in the core of the solid tumor (ii) secretion of survival/wound healing factors for self-support and for the support of surrounding cells and (iii) reduced MMP activity to decrease transmigration of stromal cells.
SD-MSCs expressed several proteins that were not expressed by control MSCs cultured in complete medium containing serum. This observation is particularly intriguing in light of the fact that these proteins were synthesized de novo during serum deprivation.
To our knowledge, this is the first attempt to decipher molecular mechanisms behind the stromal properties of MSCs under serum-deprived conditions. The data obtained here corroborates previously published observations both from our laboratory and others in hypoxia model, another important consideration for stromal cell survival (20–22). The in vivo tumor data from Figures 1 and 2 indicate that SD-MSCs have a general effect on supporting the survival of surrounding cells. SD-MSCs are a subpopulation of the MSCs that survive serum deprivation therefore the data corroborates our hypothesis that the SD-MSCs represent the cell population that provides stromal supportive functions. Apparent differences in cellularity of the tumor and neo-angiogenesis explain the role of autophagy in theserum/nutrient-deprived stromal compartment of the tumors. Autophagy has been shown to play a role in solid tumors both as tumor promoting and inhibiting. Though these data indicate higher levels of autophagy in tumors co-injected with SD-MSCs, the possibility of tumor cells undergoing autophagy cannot be ruled out. Recent evidence indicates that, at least in cells with intact apoptotic machinery, autophagy is primarily a pro-survival and not a pro-death process (37). Presumably, autophagic recycling of macromolecules is linked to the capacity of SD-MSCs to sustain life during prolonged starvation.
The data presented here are the first demonstration of autophagosome formation in MSCs under conditions that mimic serum/nutrient deprivation during disease processes such as ischemia, infection and the central serum/nutrient- and oxygen-deprived regions of large solid tumors. Our published (18) and current data have uncovered a potentially novel and unique survival mechanism for MSCs during prolonged serum deprivation: autophagic metabolism to provide needed nutrients for long-term survival and for directed secretion of survival factors that aid in both survival of MSCs and the cells around them. The data obtained using serum-deprived media is specific to the MSC phenotype without interference from the serum in normal growth media. Taken together, these studies are an important advancement in understanding the survival properties of MSCs, autophagy as a survival mechanism for MSCs and as a stromal support for cancer cells.
Supplementary material
Supplementary Methods and Figures S1–S4 can be found at http://carcin.oxfordjournals.org/
Funding
Some of the materials employed in this work were provided by the Tulane Center for Gene Therapy through a grant from NCRR of the National Institutes of Health (grant # P40RR017447) and grants National Institutes of Health (AR 47796, AR 48323), the Oberkotter Foundation, the HCA the Health Care Company and the Louisiana Gene Therapy Research Consortium to Dr. Darwin J.Prockop. Some of the materials for comet assays were provided by Tulane Cancer Center through a grant from NCRR of the National Institutes of Health (grant # P20RR020152).
Supplementary Material
Acknowledgments
We would like to thank Darwin J.Prockop for funding support.We also thank Kenneth Williams and Terence Wu and Mark Pypaert, Yale University-CT, for their assistance in Mass spectrophotometry and Electron microscopy. We would like to thank Stuart L.Schreiber, Chemistry and Chemical Biology, Harvard University, Broad Institute of Harvard and MIT for the generous gift of Tubacin.
C.S. and P.P.—conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing; A.Z.O., A.G.B., D.Z.C. and S.D.—collection and/or assembly of data; A.K., D.S.K., B.G.R.—provision of study material, manuscript writing and R.R.P.—conception and design, data analysis and interpretation, manuscript writing, financial support, final approval of manuscript.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- FBS
fetal bovine serum
- MMP
matrix metalloproteinase
- MSC
mesenchymal stem cell
- PBS
phosphate-buffered saline
- SD-MSC
serum-deprived mesenchymal stem cell
- TGF
transforming growth factor
References
- 1.Stagg J. Mesenchymal stem cells in cancer. Stem. Cell Rev. 2008;4:119–124. doi: 10.1007/s12015-008-9030-4. [DOI] [PubMed] [Google Scholar]
- 2.Karnoub AE, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 3.Cullen KJ, et al. Regulation of human breast cancer by secreted growth factors 9. Acta Oncol. 1989;28:835–839. doi: 10.3109/02841868909092318. [DOI] [PubMed] [Google Scholar]
- 4.Hirtenlehner K, et al. Influences of stroma-derived growth factors on the cytokine expression pattern of human breast cancer cell lines 2. Arch. Gynecol. Obstet. 2002;266:108–113. doi: 10.1007/s00404-001-0278-z. [DOI] [PubMed] [Google Scholar]
- 5.Seeger H, et al. Influence of stroma-derived growth factors on the estradiol-stimulated proliferation of human breast cancer cells 1. Eur. J. Gynaecol. Oncol. 2004;25:175–177. [PubMed] [Google Scholar]
- 6.Yee D, et al. The insulin-like growth factors, their receptors, and their binding proteins in human breast cancer 8. Cancer. Treat. Res. 1991;53:93–106. doi: 10.1007/978-1-4615-3940-7_5. [DOI] [PubMed] [Google Scholar]
- 7.Konopleva M, et al. Stromal cells prevent apoptosis of AML cells by up-regulation of anti-apoptotic proteins. Leukemia. 2002;16:1713–1724. doi: 10.1038/sj.leu.2402608. [DOI] [PubMed] [Google Scholar]
- 8.Lwin T, et al. Bone marrow stromal cells prevent apoptosis of lymphoma cells by upregulation of anti-apoptotic proteins associated with activation of NF-kappaB (RelB/p52) in non-hodgkin's lymphoma cells. Leukemia. 2007;21:1521–1531. doi: 10.1038/sj.leu.2404723. [DOI] [PubMed] [Google Scholar]
- 9.Nefedova Y, et al. Bone marrow stromal-derived soluble factors and direct cell contact contribute to de novo drug resistance of myeloma cells by distinct mechanisms. Leukemia. 2003;17:1175–1182. doi: 10.1038/sj.leu.2402924. [DOI] [PubMed] [Google Scholar]
- 10.Adhami F, et al. Cerebral ischemia-hypoxia induces intravascular coagulation and autophagy. Am. J. Pathol. 2006;169:566–583. doi: 10.2353/ajpath.2006.051066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castino R, et al. Autophagy-dependent cell survival and cell death in an autosomal dominant familial neurohypophyseal diabetes insipidus in vitro model. FASEB J. 2005;19:1024–1026. doi: 10.1096/fj.04-3163fje. [DOI] [PubMed] [Google Scholar]
- 12.Kang C, et al. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev. 2007;21:2161–2171. doi: 10.1101/gad.1573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelekar A. Autophagy. Ann. N. Y. Acad. Sci. 2005;1066:259–271. doi: 10.1196/annals.1363.015. [DOI] [PubMed] [Google Scholar]
- 14.Fuertes G, et al. Role of proteasomes in the degradation of short-lived proteins in human fibroblasts under various growth conditions. Int. J. Biochem. Cell Biol. 2003;35:651–664. doi: 10.1016/s1357-2725(02)00382-5. [DOI] [PubMed] [Google Scholar]
- 15.Ollinger K, et al. Nutrient deprivation of cultured rat hepatocytes increases the desferrioxamine-available iron pool and augments the sensitivity to hydrogen peroxide. J. Biol. Chem. 1997;272:23707–23711. doi: 10.1074/jbc.272.38.23707. [DOI] [PubMed] [Google Scholar]
- 16.Uchiyama Y. Autophagic cell death and its execution by lysosomal cathepsins. Arch. Histol. Cytol. 2001;64:233–246. doi: 10.1679/aohc.64.233. [DOI] [PubMed] [Google Scholar]
- 17.Rhodes LV, et al. Adult human mesenchymal stem cells enhance breast tumorigenesis and promote hormone independence. Breast Cancer. Res.Treat. 2010;121:293–300. doi: 10.1007/s10549-009-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pochampally RR, et al. Serum deprivation of human marrow stromal cells (hMSCs) selects for a subpopulation of early progenitor cells with enhanced expression of OCT-4 and other embryonic genes. Blood. 2004;103:1647–1652. doi: 10.1182/blood-2003-06-1967. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez C, et al. Epigenetic reprogramming of IGF-1 and leptin genes by serum deprivation in multipotential mesenchymal stromal cells. Stem Cells. 2008;27:375–382. doi: 10.1634/stemcells.2008-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung SC, et al. Short-term exposure of multipotent stromal cells to low oxygen increases their expression of CX3CR1 and CXCR4 and their engraftment in vivo. PLoS One. 2007;2:e416. doi: 10.1371/journal.pone.0000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ame-Thomas P, et al. Human mesenchymal stem cells isolated from bone marrow and lymphoid organs support tumor B-cell growth: role of stromal cells in follicular lymphoma pathogenesis. Blood. 2007;109:693–702. doi: 10.1182/blood-2006-05-020800. [DOI] [PubMed] [Google Scholar]
- 22.Hung SC, et al. Angiogenic effects of human multipotent stromal cell conditioned medium activate the PI3K-Akt pathway in hypoxic endothelial cells to inhibit apoptosis, increase survival, and stimulate angiogenesis. Stem Cells. 2007;25:2363–2370. doi: 10.1634/stemcells.2006-0686. [DOI] [PubMed] [Google Scholar]
- 23.John S, et al. Regulation of estrogenic effects by beclin 1 in breast cancer cells. Cancer Res. 2008;68:7855–7863. doi: 10.1158/0008-5472.CAN-07-5875. [DOI] [PubMed] [Google Scholar]
- 24.Schoenlein PV, et al. Autophagy facilitates the progression of ERalpha-positive breast cancer cells to antiestrogen resistance. Autophagy. 2009;5:400–403. doi: 10.4161/auto.5.3.7784. [DOI] [PubMed] [Google Scholar]
- 25.Choucroun P, et al. Comet assay and early apoptosis. Mutat. Res. 2001;478:89–96. doi: 10.1016/s0027-5107(01)00123-3. [DOI] [PubMed] [Google Scholar]
- 26.Bellodi C, et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J. Clin. Invest. 2009;119:1109–1123. doi: 10.1172/JCI35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solomon VR, et al. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur. J. Pharmacol. 2009;625:220–233. doi: 10.1016/j.ejphar.2009.06.063. [DOI] [PubMed] [Google Scholar]
- 28.Lee JY, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthias P, et al. HDAC6 a new cellular stress surveillance factor. Cell Cycle. 2008;7:7–10. doi: 10.4161/cc.7.1.5186. [DOI] [PubMed] [Google Scholar]
- 30.Hubbert C, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 31.Tran AD, et al. HDAC6 deacetylation of tubulin modulates dynamics of cellular adhesions. J. Cell Sci. 2007;120:1469–1479. doi: 10.1242/jcs.03431. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol. Cell. 2007;27:197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J, et al. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- 34.Jones RG, et al. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maiuri MC, et al. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ. 2009;16:87–93. doi: 10.1038/cdd.2008.131. [DOI] [PubMed] [Google Scholar]
- 36.Bursch W, et al. Cell death and autophagy: cytokines, drugs, and nutritional factors. Toxicology. 2008;254:147–157. doi: 10.1016/j.tox.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 37.Dalby KN, et al. Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy. 2010;6:322–329. doi: 10.4161/auto.6.3.11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.