Abstract
The magnitude of benefit is variable for advanced non-small cell lung cancer (NSCLC) patients receiving platinum-based chemotherapy. The purpose of this study is to determine whether genetic variations in the transforming growth factor-beta (TGF-β) pathway are associated with clinical outcomes in NSCLC patients receiving first-line platinum-based chemotherapy. Five hundred and ninety-eight advanced-stage NSCLC patients who received first-line platinum-based chemotherapy with or without radiotherapy were recruited at the MD Anderson Cancer Center between 1995 and 2007. DNA from blood was genotyped for 227 single nucleotide polymorphisms (SNPs) in 23 TGF-β pathway-related genes to evaluate their associations with overall survival. In individual SNP analysis, 22 variants were significantly associated with overall survival, of which the strongest associations were found for BMP2:rs235756 [hazard ratio (HR) = 1.45; 95% confidence interval (CI), 1.11–1.90] and SMAD3:rs4776342 (HR = 1.25; 95% CI, 1.06–1.47). Fifteen and 18 genetic loci displayed treatment-specific associations for chemotherapy and chemoradiation, respectively, identifying a majority of the cases who would be predicted to respond favorably to a specific treatment regimen. BMP2:rs235753 and a haplotype in SMAD3 were associated with overall survival for both treatment modalities. Cumulative effect analysis showed that multiple risk genotypes had a significant dose-dependent effect on overall survival (Ptrend = 2.44 x 10−15). Survival tree analysis identified subgroups of patients with dramatically different median survival times of 45.39 versus 13.55 months and 18.02 versus 5.89 months for high- and low- risk populations when treated with chemoradiation and chemotherapy, respectively. These results suggest that genetic variations in the TGF-β pathway are potential predictors of overall survival in NSCLC patients treated with platinum-based chemotherapy with or without radiation.
Introduction
Lung cancer is the leading cause of cancer-related death worldwide and non-small cell lung cancer (NSCLC) accounts for ∼85% of all lung cancer. Approximately 65–75% of patients with NSCLC have unresectable advanced or metastatic disease at the time of diagnosis (1). Currently, the standard therapy for patients with advanced-stage disease is platinum-based chemotherapy, and patients with locally advanced disease are generally treated with a combination radiation therapy plus platinum-based chemotherapy (2). However, the overall response rate for platinum-based therapy is only in the range of 17–32% (3), and the median survival continues to be dismal at 7–9 months (4). Some patients derive no or little apparent benefit, some others have modest prolongation of survival, whereas still others will survive ≥2 years. It would be helpful to be able to predict which patients are most and least probable to benefit, so that those who are unlikely to benefit could be spared toxicity of the platinum chemotherapy.
The transforming growth factor-beta (TGF-β) pathway has important roles in cellular proliferation, angiogenesis, differentiation, migration, apoptosis and epithelial–mesenchymal transition via interactions with SMAD (5) and via cross talk with other signaling pathways such as extracellular signal-regulated kinase, c-jun N-terminal kinase, PI3K/AKT and Rho-like guanosine triphosphatases (6). Despite the prominent role, TGF-β pathway appears to play in regulating multiple cellular processes, there have not been many studies published regarding genetic polymorphisms in this pathway. It has been reported that the TGF-β receptor 1 variant rs11466445 (TGFBR1*6A) was associated with an increased risk for several cancers (7–9), but not for lung cancer (10). The published study of TGF-β pathway polymorphisms on cancer outcome indicted that rs868 located in the 3′-untranslated region of TGFBR1 was an independent predictor of bladder cancer mortality (11). To our knowledge, no genetic polymorphisms in the TGF-β pathway have been reported to be associated with response to chemotherapy. However, previous studies have demonstrated that TGF-β enhances the lethal effects of DNA-damaging agents in human lung cancer lines (12). Moreover, non-SMAD pathways, such as PI3K/AKT, play a key role in the development of resistance to platinum (13), and activation of PI3K/AKT is able to inhibit TGF-β mediated apoptosis (14). The possible mechanism underlying these observations is that a wide range of cytotoxic drugs, including cisplatin and carboplatin, regardless of their distinct mechanisms of action, directly and indirectly kill susceptible cells by inducing apoptosis (15). Therefore, it stands to reason that decreased ability to activate apoptotic machinery might play a role in the development of resistance to chemotherapeutic agents (16). Given the role of the TGF-β pathway in such essential cellular processes, it is conceivable that genetic variations within TGF-β pathway genes could emerge as potential predictors of chemotherapy efficacy.
The purpose of our study was to determine whether genetic variations in the TGF-β pathway are associated with survival in NSCLC patients, who have undergone platinum-based chemotherapy or chemoradiation and to identify subgroups who would be more probably to have favorable clinical outcome from specific treatment regimens.
Materials and methods
Study population and data collection
The study was based on 598 newly diagnosed, histologically confirmed NSCLC patients at the University of Texas MD Anderson Cancer Center. None of the patients had undergone chemotherapy or radiotherapy prior to study enrollment. To minimize heterogeneity in the study population, we limited our analysis to patients who had unresectable stage IIIA–IV disease and who had received first-line platinum-based chemotherapy at MD Anderson. All study participants provided signed informed consent and completed a 45 min inperson interview by trained M.D. Anderson staff interviewers. This interview collected epidemiological data, which included demographics, smoking history, alcohol consumption, family history of cancer, medical history and occupational exposures. After each interview, a 40 ml peripheral blood sample was drawn into coded and heparinized tubes for subsequent analysis. Clinical and follow-up information, such as vital status, treatment regimens, stage and pretreatment performance status (PS), was obtained from the patients’ medical records. Study approval was obtained from the MD Anderson Institutional Review Board.
Polymorphism selection and genotyping
In this study, a total of 227 SNPs in 23 candidate genes were selected based on the following criteria (supplementary Table 1 is available at Carcinogenesis Online). Briefly, we utilized Gene Oncology (http://www.geneontology.org) and National Center for Biotechnology Information (NCBI) PubMed (http://www.ncbi.nlm.nih.gov) to identify a list of TGF-β pathway-related genes. For each gene, we selected haplotype-tagging SNPs located within 10 kb upstream of transcriptional start site or 10 kb downstream of transcriptional stop site based on data from the International HapMap Project (http://www.hapmap.org). Using the linkage disequilibrium (LD) select program (http://droog.gs.washington.edu/ldSelect.html) and the UCSC Golden Path Gene Sorter program (http://genome.ucsc.edu), we further divided identified SNPs into bins based on an r2 threshold of 0.8 and minor allele frequency >0.05 in Caucasians to select tagging SNPs. We also included SNPs in the coding (synonymous SNPs and non-synonymous SNPs) and regulatory regions (promoter, splicing site, 5′-untranslated region and 3′-untranslated regions). In addition, functional SNPs and SNPs previously reported to be associated with cancer were also included, such as rs868 (11), rs334354 (Int7G24A) (17), rs928180 (LD with TGFBR1*6A, D′ = 0.98, r2 = 0.91) (11), rs4522809 (18), rs2228048 (19) and rs3087465 (20). The complete set of selected SNPs was submitted to Illumina technical support for Infinium chemistry designability, beadtype analyses and iSelect Infinium Beadchip synthesis. The genes (number of SNPs) were: TGFB1 (7), BMP1 (18), BMP2 (14), BMP4 (5), GDF1 (3), INHA (4), INHBC (5), NODAL (6), TGFBR1 (3), TGFBR2 (3), ACVR1B (3), ACVR1C (3), ACVR2A (3), ACVR2B (3), AMHR2 (2), SMAD1 (8), SMAD2 (8), SMAD3 (50), SMAD4 (4), SMAD5 (8), SMAD6 (29), SMAD7 (21) and SMAD8 (17).
Genomic DNA was extracted from peripheral blood lymphocytes and stored at −80°C until use. Among 227 SNPs, 221SNPs were genotyped using Illumina’s iSelect custom genotyping platform according to manufacturer’s protocol (Illumina, San Diego, CA). Genotypes were called using the Illumina BeadStudio software. Any single nucleotide polymorphisms (SNPs) with a call rate <95% was excluded from further analysis. The mean call rate for this study was 99.9%. Six SNPs (rs868, rs334354, rs928180, rs4522809, rs2228048 and rs3087465) were genotyped using TaqMan assay (21).
Statistical analysis
The endpoint of the study was overall survival, which was calculated from pathologic diagnosis to death regardless of the cause. Statistic analyses were performed using STATA software (STATA Corporation, College Station, TX). The χ2 test or Fisher’s exact test was used to compare clinical characteristics with vital status. Cox’s proportional hazards model was used to estimate hazard ratios (HRs) and their 95% confidence intervals (CIs) for the multivariate survival analyses, adjusting for age, sex, ethnicity, smoking status, clinical stage, pretreatment PS and therapy modality. The genotype affect on patients’ overall survival was analyzed using Kaplan–Meier curves and log-rank tests. The Haploview program was applied to estimate pairwise LD between markers and to partition haplotype blocks (22). Haplotype frequencies were inferred using the expectation-maximization algorithm implemented in the HelixTree program (Golden Helix, Bozeman, MT). The combined effects of unfavorable genotypes analysis included those SNPs showing statistical significance in the main analysis (P < 0.05). Patients were then categorized into low-risk, medium-risk and high-risk groups based on the tertile distribution of the number of unfavorable genotypes. Higher order gene–gene interactions were evaluated using survival tree analysis implemented in the STREE program (http://masal.med.yale.edu/stree/), which uses recursive partitioning to identify subgroups of individuals with similar risk. Bootstrap resampling was performed 100 times to internally validate the results from our analyses (23). All statistical analyses were two sided and P < 0.05 was considered statistically significant.
Results
Patient characteristics
As shown in Table I, 598 patients were enrolled in this study, including Caucasian (78.6%), African-American (15.9%) and other ethnicities (5.5%). At the time of analysis, 142 (23.7%) were still alive and 456 (76.3%) had died. There were no significant differences for those patients who were alive compared with those who had died with respect to age (P = 0.88), ethnicity (P = 0.94), smoking status (P = 0.86) and weight loss at the time of diagnosis (P = 0.39). Using American Joint Committee on TNM-staging system, 82 (13.7%) were stage IIIA, 142 (23.7%) were stage IIIB without pleural effusion (dry), 39 (6.5%) were stage IIIB with pleural effusion (wet) and 335 (56.1%) were stage IV. Of these 598 patients, 243 (40.6%) received concurrent radiation with curative intent. All chemotherapeutic regimens were platinum based. Those alive at last follow-up were less probably to be stage IV (43.0 versus 60.1% for dead versus alive at last follow-up, P = 0.0035), to have a PS of 2–4 (5.9 versus 15.9%, P = 0.002) and to be male (43.0 versus 57.5%, P = 0.002). Consistent with the literature (24,25), the comparisons of clinical variables based on gender, stage and PS were statistically significant, but we controlled for these variables in the subsequent multivariate analyses. Overall, the median survival time (MST) in this study was 12.9 months and median follow-up time was 11.8 months.
Table I.
Selected characteristics of NSCLC patient population (N = 598)
| Characteristic | Alive | Dead | P-value |
| Age, mean (SD), years | 59.6 (10.02) | 59.7 (10.63) | 0.88 |
| Gender, n (%) | |||
| Male | 61 (43.0) | 262 (57.5) | 0.002 |
| Female | 81 (57.0) | 194 (42.5) | |
| Ethnicity, n (%) | |||
| Caucasian | 112 (78.9) | 358 (78.5) | 0.94 |
| African-American | 23 (16.2) | 72 (15.8) | |
| Others | 7 (4.9) | 26 (5.7) | |
| Smoking status, n (%) | |||
| Never | 28 (19.7) | 84 (18.4) | 0.86 |
| Former | 59 (41.5) | 184 (40.4) | |
| Current | 55 (38.7) | 188 (41.2) | |
| Stage (TNM), n (%) | |||
| IIIA | 25 (17.6) | 57 (12.5) | 0.004 |
| IIIB (dry) | 42 (29.6) | 100 (21.9) | |
| IIIB (wet) | 14 (9.9) | 25 (5.5) | |
| IV | 61 (43.0) | 274 (60.1) | |
| PS, n (%) | |||
| 0 | 47 (34.8) | 96 (23.1) | 0.002 |
| 1 | 80 (59.3) | 254 (61.1) | |
| 2–4 | 8 (5.9) | 66 (15.9) | |
| Weight loss, n (%) | |||
| Weight gain or stable | 77 (61.1) | 218 (54.5) | 0.39 |
| 1–5% | 14 (11.1) | 66 (16.5) | |
| 5–10% | 22 (17.5) | 66 (16.5) | |
| >10% | 13 (10.3) | 50 (12.5) | |
| Histology grade, n (%) | |||
| Well differentiated | 4 (4.7) | 23 (7.6) | 0.48 |
| Moderate differentiated | 16 (18.8) | 45 (14.9) | |
| Poorly differentiated | 65 (76.5) | 234 (77.5) | |
Individual SNPs and overall survival
The association of all SNPs with overall survival is summarized in supplementary Table 1, available at Carcinogenesis Online. We identified 22 SNPs with a P < 0.05 and 2 SNPs with a P < 0.01. The association for 15 SNPs was further demonstrated by bootstrap resampling with strong statistical support (55–100% of 100 bootstrap resamplings at P < 0.05, Table II).
Table II.
Significant SNPs associated with overall survival (N = 598)
| Gene | SNP | Modela | HRb | P | BootstrapcP < 0.05 |
| BMP2 | rs235756 | rec | 1.45 (1.11–1.90) | 0.006 | 100 |
| SMAD3 | rs4776342 | add | 1.25 (1.06–1.47) | 0.009 | 96 |
| SMAD3 | rs6494633 | rec | 0.73 (0.56–0.93) | 0.01 | 99 |
| SMAD3 | rs12102171 | dom | 1.32 (1.07–1.63) | 0.01 | 95 |
| SMAD6 | rs12913975 | dom | 1.29 (1.05–1.59) | 0.01 | 93 |
| SMAD7 | rs7227023 | dom | 0.70 (0.53–0.92) | 0.01 | 100 |
| SMAD9 | rs7333607 | rec | 0.52 (0.31–0.87) | 0.01 | 96 |
| TGFB1 | rs4803455 | dom | 1.31 (1.04–1.65) | 0.02 | 87 |
| ACVR2A | rs1424954 | rec | 0.63 (0.43–0.93) | 0.02 | 81 |
| SMAD4 | rs948588 | dom | 0.70 (0.52–0.95) | 0.02 | 61 |
| SMAD6 | rs12906898 | add | 1.19 (1.03–1.38) | 0.02 | 33 |
| BMP1 | rs3857979 | rec | 0.77 (0.60–0.97) | 0.03 | 54 |
| INHBC | rs4760259 | rec | 0.68 (0.48–0.96) | 0.03 | 85 |
| SMAD1 | rs11939979 | dom | 0.77 (0.61–0.97) | 0.03 | 55 |
| SMAD1 | rs11724777 | rec | 1.37 (1.04–1.81) | 0.03 | 85 |
| SMAD4 | rs12456284 | dom | 1.25 (1.02–1.54) | 0.03 | 61 |
| BMP1 | rs7838961 | dom | 1.28 (1.01–1.62) | 0.04 | 26 |
| BMP4 | rs8014071 | add | 1.17 (1.00–1.36) | 0.04 | 22 |
| BMP4 | rs17563 | add | 1.15 (1.01–1.32) | 0.04 | 11 |
| SMAD3 | rs11632964 | add | 0.86 (0.75–0.99) | 0.04 | 40 |
| SMAD3 | rs750766 | rec | 1.29 (1.01–1.64) | 0.04 | 41 |
| SMAD8 | rs511674 | dom | 0.64 (0.43–0.97) | 0.04 | 16 |
Note. a Genetic model of inheritance: dom, dominant model; rec, recessive model; add, additive model.
Adjusted for age, sex, ethnicity, smoking status, clinical stage, PS, chemotherapy regimen and therapy modality.
Bootstrap analysis was performed using 100 replicates to determine statistical support.
BMP genetic variations
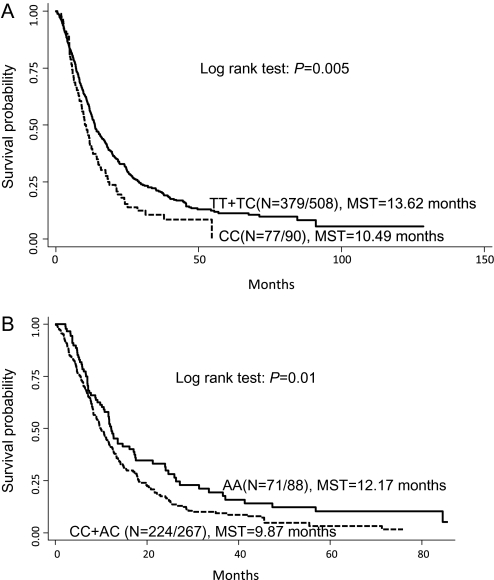
The most significant association in this study was BMP2:rs235756 in a recessive model, which was associated with an increased risk of dying among patients carrying the homozygous variant genotype (HR = 1.45; 95% CI 1.11–1.90). These patients had a significantly shorter MST of 10.49 months compared with 13.62 months for those with the wild-type or heterozygous genotype (Log rank P = 0.005, Figure 1a). BMP4:rs17563 was the only non-synonymous SNP (Val52Ala) that reached statistical significance in this study. Patients carrying variant allele had a significantly increased risk of dying in a dose-dependent manner (HR = 1.15; 95% CI: 1.01–1.32), but the association was not strongly supported by bootstrap analysis. This increase in risk resulted in a decreased overall survival by nearly 3 months from 15.03 months for those with the wild-type genotype to 11.84 months for patients carrying the variant-containing genotypes (Log rank P = 0.016).
Fig. 1.
Kaplan–Meier survival curves for NSCLC patients according to TGF-β-signaling pathway polymorphisms. (a) BMP2:rs235756 in the total population; (b) TGFB1:rs4803455 in patients treated with chemotherapy; MST in months.
Genetic variations of TGF-β/activin/inhibin signaling
A significantly increased risk was shown for patients with at least one variant TGFB1:rs4803455 allele (HR = 1.31; 95% CI 1.04–1.65), whereas two SNPs exhibited significantly decreased risk of death: INHBC: rs4760259 (HR = 0.68; 95% CI, 0.48–0.96) and ACVR2A: rs1424954 (HR = 0.63; 95% CI 0.43–0.93).
SMAD genetic variations
Among the eight SMAD genes analyzed, 14 SNPs reached statistical significance (P < 0.05) for association with overall survival. Under the additive model, SMAD3:rs4776342 was significantly associated with poorer overall survival with an HR of 1.25 (95% CI 1.06–1.47, P < 0.01). A significantly improved overall survival was shown for patients carrying variant-containing genotypes of SMAD7:rs7227023 (HR = 0.70; 95% CI 0.53–0.92). These individuals had a nearly 6 months survival advantage with an MST of 18.19 compared with 12.37 months for those with the wild-type genotype (Log rank P = 0.001).
Combined effects of unfavorable genotypes on overall survival
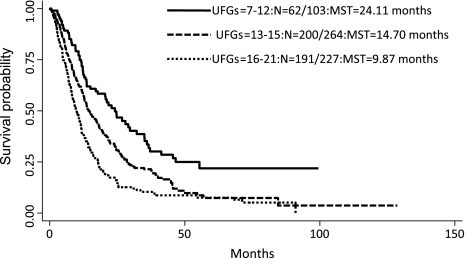
Because abnormal TGF-β signaling can result in the activation of multiple downstream genes and 22 SNPs reached statistical significance in the main effect analysis, we therefore used combined analysis to evaluate whether multiple unfavorable genotypes in TGF-β pathway would have an additive effect on NSCLC survival. There was a significant dose–response trend of increased risk of dying and reduced overall survival time with increasing number of unfavorable genotypes (Figure 2). Compared with the low-risk group consisting of subjects with 0–12 unfavorable genotypes, the medium-risk group with 13–15 unfavorable genotypes had a 2.07-fold (95% CI 1.51–2.85) increased risk, whereas the high-risk group with 16–21 unfavorable genotypes was at a 3.45-fold (95% CI 2.49–4.77) increased risk (Ptrend = 2.44 x 10−15; Figure 2b). The MSTs were 24.11, 14.70 and 9.87 months for patients in the low-, medium- and high-risk groups, respectively (log rank P = 5.65 x 10−10).
Fig. 2.
Kaplan–Meier survival curves for NSCLC patients grouped by the number of unfavorable genotypes (UFGs) in the TGF-β-signaling pathway MST in months.
Effect of TGF-β genetic variation by treatment regimen
We also performed an exploratory analysis stratified by treatment modality (Table III) in an attempt to identify SNPs that may have differential effects on survival in patients receiving chemotherapy only or chemoradiation. This analysis may categorize SNPs into four groups: group 1 SNPs confer poor survival in patients receiving chemotherapy only but good survival in patients receiving chemoradiation; group 2 SNPs has opposite effects on survival compared with group 1 SNPs; group 3 SNPs have beneficial effect on survival in either treatment groups and group 4 SNPs confer poor survival in either treatment groups. One interesting SNP, BMP2:rs235753, belonged to group 1. The wild-type genotype was associated with an increased risk of death in patients receiving chemotherapy only (HR = 1.20, 95% CI 1.01–1.45) but displayed a reduced risk of death in patients treated with chemoradiation (HR = 0.54, 95% CI 0.33–0.88), suggesting that thoracic radiation may improve the survival of patients with the wild-type genotype of this SNP.
Table III.
Effect of wild-type genotype by treatment regimen
| Gene | SNP | Chemoradiation (N = 243) |
Chemotherapy (N = 355) |
Group | ||||
| Effect on survival | HRa | P | Effect on survival | HRa | P | |||
| BMP2 | rs235753 | Favorable | 0.54 (0.33–0.88) | 0.01 | Unfavorable | 1.20(1.01-1.45) | 0.038 | Group 1 |
| TGFBR1 | rs868 | Favorable | 0.63 (0.43–0.92) | 0.02 | Unfavorable | 1.10(0.66–1.85) | 0.85 | |
| ACVR1B | rs877869 | Favorable | 0.69 (0.53–0.91) | 0.008 | Unfavorable | 1.12(0.87–1.43) | 0.61 | |
| SMAD3 | rs11071933 | Favorable | 0.68 (0.47–1.00) | 0.05 | Unfavorable | 1.03(0.79–1.35) | 0.79 | |
| SMAD4 | rs948588 | Favorable | 0.96 (0.56–1.64) | 0.88 | Unfavorable | 1.47(1.01–2.13) | 0.04 | |
| SMAD6 | rs3934908 | Favorable | 0.90 (0.60–1.33) | 0.59 | Unfavorable | 1.35(1.05–1.72) | 0.02 | |
| SMAD6 | rs4776318 | Favorable | 0.73 (0.56–0.93) | 0.01 | Unfavorable | 1.10(0.81–1.52) | 0.57 | |
| SMAD6 | rs4075546 | Favorable | 0.68 (0.53–0.88) | 0.003 | Unfavorable | 1.11(0.76–1.61) | 0.60 | |
| TGFB1 | rs4803455 | Unfavorable | 1.35 (0.90–2.04) | 0.14 | Favorable | 0.67(0.51–0.89) | 0.006 | Group 2 |
| BMP2 | rs235757 | Unfavorable | 1.47 (1.10–1.96) | 0.01 | Favorable | 0.83(0.58–1.19) | 0.40 | |
| TGFBR1 | rs928180 | Unfavorable | 1.72 (1.05–2.78) | 0.03 | Favorable | 0.95(0.69–1.32) | 0.78 | |
| BMP2 | rs173107 | Unfavorable | 1.49 (1.11–2.00) | 0.007 | Favorable | 0.92(0.63–1.33) | 0.81 | |
| SMAD3 | rs9972423 | Unfavorable | 1.23 (0.94–1.61) | 0.13 | Favorable | 0.81(0.67–0.98) | 0.03 | |
| SMAD3 | rs12915039 | Unfavorable | 2.50 (1.01–6.25) | 0.05 | Favorable | 0.80(0.47–1.35) | 0.40 | |
| SMAD4 | rs1787111 | Unfavorable | 1.39 (0.63–3.03) | 0.41 | Favorable | 0.51(0.32–0.81) | 0.006 | |
| SMAD8 | rs9531986 | Unfavorable | 2.00 (0.83–2.27) | 0.21 | Favorable | 0.76(0.58–0.99) | 0.04 | |
| TGFB1 | rs11466345 | Favorable | 0.61 (0.38–0.99) | 0.04 | Favorable | 0.85(0.62–1.16) | 0.31 | Group 3 |
| BMP1 | rs4076873 | Favorable | 0.74 (0.55–0.98) | 0.04 | Favorable | 0.88(0.69–1.14) | 0.66 | |
| BMP2 | rs235756 | Favorable | 0.64 (0.37–1.10) | 0.11 | Favorable | 0.72(0.53–0.99) | 0.04 | |
| BMP4 | rs8014071 | Favorable | 0.64 (0.44–0.93) | 0.04 | Favorable | 0.76(0.52–1.14) | 0.46 | |
| SMAD3 | rs4776342 | Favorable | 0.71 (0.54–0.94) | 0.02 | Favorable | 0.84(0.68–1.03) | 0.09 | |
| SMAD3 | rs1210217 | Favorable | 0.64 (0.47–0.86) | 0.03 | Favorable | 0.81(0.62–1.05) | 0.12 | |
| SMAD4 | rs7244227 | Favorable | 0.66 (0.46–0.96) | 0.03 | Favorable | 0.80(0.56–1.14) | 0.91 | |
| SMAD4 | rs12456284 | Favorable | 0.86 (0.59–1.25) | 0.44 | Favorable | 0.77(0.6–0.99) | 0.04 | |
| SMAD6 | rs12913975 | Favorable | 0.61 (0.42–0.90) | 0.01 | Favorable | 0.76(0.56–1.02) | 0.29 | |
| BMP4 | rs8014363 | Unfavorable | 1.61 (1.03–2.56) | 0.04 | Unfavorable | 1.14(0.87–1.49) | 0.60 | Group 4 |
| ACVR2A | rs1424954 | Unfavorable | 1.82 (0.93–3.45) | 0.08 | Unfavorable | 1.72(1.03–2.86) | 0.04 | |
| SMAD1 | rs11939979 | Unfavorable | 1.33 (0.87–2.04) | 0.89 | Unfavorable | 1.39(1.05–1.82) | 0.02 | |
| SMAD1 | rs2118438 | Unfavorable | 1.47 (0.53–4.17) | 0.95 | Unfavorable | 1.33(1.03–1.75) | 0.03 | |
| SMAD3 | rs6494633 | Unfavorable | 1.43 (0.91–2.27) | 0.12 | Unfavorable | 1.20(1.01–1.43) | 0.04 | |
| SMAD3 | rs11632964 | Unfavorable | 1.37 (0.94–1.96) | 0.92 | Unfavorable | 1.52(1.05–2.17) | 0.02 | |
| SMAD6 | rs4776831 | Unfavorable | 1.16 (0.76–1.75) | 0.95 | Unfavorable | 1.37(1.04–1.79) | 0.02 | |
Adjusted for age, sex, ethnicity, smoking status, clinical stage, PS and chemotherapy regimen. Results in bold are significant at P < 0.05.
Association of TGF-β genetic variation and stage
Given 341 (96.1%) patients with stage IIIB (wet) + IV were treated with chemotherapy alone and 210 (86.4%) patients with stage IIIA + IIIB (dry) received chemoradiation in this study, we further examined the correlations of SNPs with stage to assist in differentiating prognostic factors from predictive factors. The frequencies of genotypes significantly different between the stage IIIA + IIIB (dry) and stage IIIB (wet) + IV were listed in supplementary Table 2, available at Carcinogenesis Online. None of the above-mentioned four groups of significant SNPs showed correlation with stage.
Haplotype analysis and treatment outcome
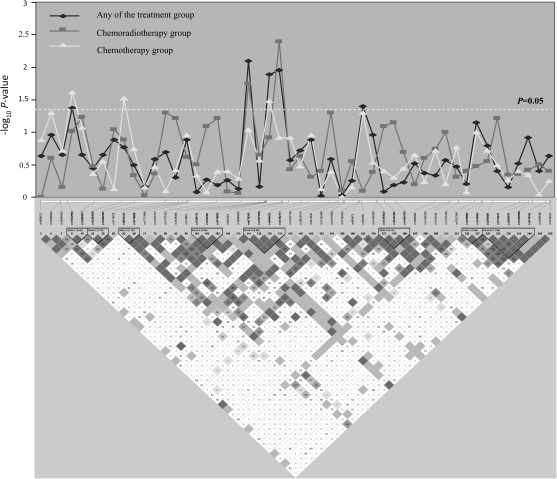
Eight SMAD3 variant loci exhibited association with overall survival either in the total population or when stratified by treatment modality. Therefore, we addressed whether there were any specific haplotypes within SMAD3 associated with treatment outcome. All 49 SNPs in SMAD3 genotyped in this study were examined to identify eight haplotype blocks within the gene. Significant associations between haplotypes and overall survival were observed for haplotype block 5 of SMAD3, which contained rs4776343, rs11071938, rs694633 and rs12102171 (Figure 3). Compared with the most common H1 haplotype (W-W-M-W; W: wild-type allele, M: variant allele), the H2 (W-M-W-W) and H3 (M-W-W-M) haplotypes exhibited significant associations with treatment outcome for chemotherapy with HRs of 1.42 (95% CI 1.13–1.79) and 1.37 (95% CI 1.04–1.81), respectively. The H3 haplotype also reached a significant association with treatment outcome for chemoradiation with an HR of 1.45 (95% CI 1.05--2.01; supplementary Table 3 is available at Carcinogenesis Online).
Fig. 3.
Graphical representation of the P-value obtained from individual SNP analysis and LD structure of SMAD3.
Higher order gene–gene interactions and treatment outcome
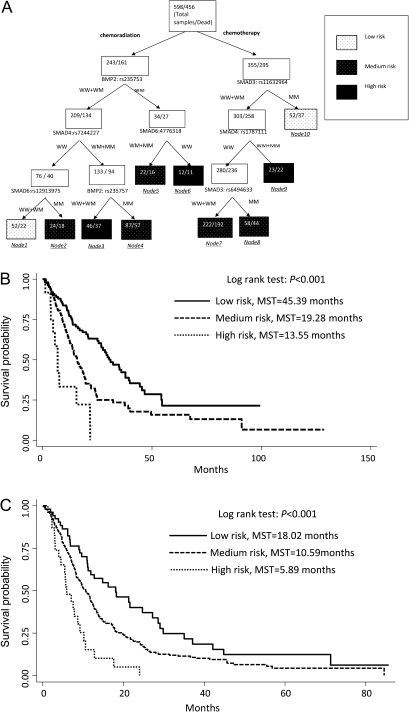
Given there were a large number of individual genetic loci associated with survival for patients who received either chemoradiotherapy or chemotherapy, we explored higher order gene–gene interactions to clarify whether complex interactions among these SNPs could determine the functional outcome (Figure 4a). For patients treated with chemoradiation, survival tree analysis identified several higher order gene–gene interactions among BMP2:rs235753, SMAD4:rs7244227, SMAD6:rs4776318, SMAD6:rs12913975 and BMP2:rs235757. BMP2:rs235753 was identified as the initial split, suggesting its potential predictive role for chemoradiation outcome. The final tree structure identified six terminal nodes with significantly different overall survival times. The low-risk genetic profile group had an MST of 30.39 months (nodes 1). In comparison, the high-risk genetic profile group (HR = 4.45; 95% CI 2.51–7.89) had an MST of 6.81 months (nodes 3 and 6) (Figure 4b). For patients treated with chemotherapy, higher order gene–gene interactions were found among SMAD3:rs11632964, SMAD4:rs1787111 and SMAD3:rs6494633. SMAD3:rs11632964 were the primary split in tree structure. The low-risk genetic profile group had an MST of 18.02 months (node 10), whereas the high-risk group (HR = 4.03; 95% CI, 2.30–7.05) had an MST of 5.89 months and a (node 9) (Figure 4c).
Fig. 4.
Potential higher order gene–gene interactions among TGF-β pathway polymorphisms. (a) Tree structure identifying subgroups of patients with different genetic backgrounds; (b) Kaplan–Meier survival curves for patients based on survival tree analysis in chemoradiation group (c) and chemotherapy group. W, wild-type allele; M, variant allele.
Discussion
In this study, we systematically evaluated genetic variations in TGF-β pathway as predictors of survival in advanced NSCLC. Previous studies have suggested that candidate SNPs and haplotypes in selected genes of TGF-β pathway were associated with the risk of developing lung cancer (26,27) and acute lung injury in mice (28). This is the first study to link TGF-β pathway SNPs with lung cancer survival.
In the main effect analysis, BMP2:rs235756 and SMAD3:rs4776342 showed strong association with NSCLC outcomes. The oncogenic activities of BMP signaling have been shown to be involved in the development of several cancers and associated with poor prognosis (29,30). The mature active form of BMP2 was overexpressed in 98% of NSCLC samples and may be involved in promoting tumor growth, progression and angiogenesis (31). BMP2:rs235756 is located in the flanking region of the BMP2 gene and has already been shown to alter normal BMP function. One study suggests that this variant can alter serum ferritin levels in hemochromatosis (32). Other SNPs in BMP genes have also been shown to result in higher production of the gene product. One example is the non-synonymous BMP4 SNP rs17563, which was also associated with NSCLC survival in our results. This SNP has been related to higher plasma concentrations of BMP4 and increased transcriptional activity, which were shown to predispose individuals to the development malignant melanoma (33). Thus, it is plausible that BMP2:rs235756 and BMP4:rs17563 may increase the production of BMP protein and favor a tumor promotion role of BMP signaling in cancer progression. Our study also suggests that SMAD3:rs4776342, or a functional SNP tagged by this locus, may inactivate SMAD3 resulting in restrained TGF-β signals, which is consistent with the finding that ∼85% of lung cancer cell lines become resistant to the growth inhibitory effect of TGF-β (34).
A single SNP often provides a modest or undetectable effect, whereas the amplified effects of combined SNPs in the same pathway may enhance predictive power. Because multiple deleterious variants with the TGF-β pathway may have a similar effect on overall survival, we analyzed the association with clinical outcomes in patients with increasing number of adverse genotypes. A clear and significant trend was evident for worsening outcomes with increasing number of unfavorable genotypes. These results suggest a cumulative influence by multiple genetic variants within the TGF-β-signaling pathway were able to further enhance the separation of patients based on clinical outcome.
In this study, multiple SMAD3 genotypes were associated with overall survival, including in treatment-specific analyses. Therefore, to understand the combinatory effect of multiple SMAD3 alleles functioning within a haplotype, we determined the LD structure of SMAD3. Haplotype 3 (H3) of SMAD3 in block 5 had consistent effect on variation in two clinical outcomes and also showed an adverse prognosis. These results suggest that haplotype-based analysis may be more informative in prediction of prognosis for NSCLC patients compared with single SNP analysis. Resequencing of DNA samples from individuals carrying the high-risk and low-risk survival haplotypes may be able to discriminate various prognostic subpopulations.
Epistasis is a ubiquitous component of the genetic architecture of common diseases. Complex interactions could determine the functional outcome over the independent main effects of any one susceptibility gene (35). In order to evaluate the effects of TGF-β signaling variation on overall survival for patients receiving chemotherapy or chemoradiotherapy more clearly, we explored higher order gene–gene interactions using survival tree analysis. The discriminative ability of the analysis identified subgroups of patients with different risk levels based on combinations of genotypes. BMP2:rs235753 had the strongest influence on clinical outcome for the chemoradiation groups in this population as this locus was the primary split in survival tree structure. This observation seems to be consistent with our findings in stratified analysis (Table III). SMAD3:rs11632964 was the initial split on the survival tree for the chemotherapy group. It is known that TGF-β signals are transduced through SMAD3 and in vivo studies have shown that blocking TGF-β signaling can result in resistance to DNA-damaging agents including cisplatin (36,37). Therefore, the effect of SMAD3:rs11632964 in our study is in agreement with SMAD3–SMAD4 controlled upregulation of Bim playing an important role in TGF-β -induced apoptosis (38). It is important to point out that the statistical modeling of an interaction does not amount to a true biological interaction and caution should be paid when interpreting these results. It has been generally accepted that activated SMAD3 forms heterogenic complexes with SMAD4 through complementary MH2 domains (39) and a recent study has already demonstrated that BMP2 activates SMAD6 gene transcription through the bone-specific transcription factor Runx2 (40). The biological plausibility of these results is intriguing, especially considering that the terminal nodes with different survival times, if further validated by the analysis for untreated control group, will assist in the identification of subgroups who will receive benefit from specific treatment modalities.
To gain better power and greater insight, we took a comprehensive approach to assess individual and collective effects of genetic variants in the TGF-β pathway on NSCLC clinical outcomes. The major strength of our study is the large population size from a single institution to minimize treatment heterogeneity. Furthermore, we have available to us comprehensive epidemiologic and clinical data for each of these patients. Although function is not known for all genotyped loci, our results have biological plausibility and warrant further study. Further research, including validation and functional studies, is needed to move this information into the clinic.
In conclusion, these results suggest that genetic variations in the TGF-β pathway modulate clinical outcomes in advanced NSCLC patients. We were able to identify subgroups of patients with differences in predicted survival when treated with chemotherapy with or without radiation.
Supplementary material
Supplementary Tables 1–3 can be found at http://carcin.oxfordjournals.org/
Funding
National Cancer Institute (R01 CA111646, P50 CA070907, R01 CA127219, R01 CA55769 and R03 CA128079).
Supplementary Material
Acknowledgments
Conception and design: M.L., D.J.S., M.A.T.H., X.W.; provision of study materials or patients: M.R.S., C.L., S.M.L., X.W.; collection and assembly of data: M.L., J.L., X.W.; data analysis and interpretation: M.L., M.H., M.A.T.H., J.L., X.W. and manuscript writing: M.L., M.A.T.H., J.G., X.W.
Conflict of Interest Statement: None declared.
Abbreviations
Abbreviations
- CI
confidence interval
- HR
hazard ratio
- MST
median survival time
- NSCLC
non-small cell lung cancer
- LD
linkage disequilibrium
- PS
performance status
- TGF-β
transforming growth factor-beta
- SNP
single nucleotide polymorphism
References
- 1.Esteban E, et al. Pemetrexed in first-line treatment of non-small cell lung cancer. Cancer Treat. Rev. 2009;35:364–373. doi: 10.1016/j.ctrv.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Spiro SG, et al. The treatment of advanced non-small cell lung cancer. Curr. Opin. Pulm. Med. 2005;11:287–291. doi: 10.1097/01.mcp.0000166590.03042.56. [DOI] [PubMed] [Google Scholar]
- 3.Gebbia V. Does an optimal therapeutic sequence exist in advanced non-small cell lung cancer? Expert Opin. Pharmacother. 2008;9:1321–1337. doi: 10.1517/14656566.9.8.1321. [DOI] [PubMed] [Google Scholar]
- 4.Schiller JH, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N. Engl. J. Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 5.Pardali K, et al. Actions of TGF-beta as tumor suppressor and pro-metastatic factor in human cancer. Biochim. Biophys. Acta. 2007;1775:21–62. doi: 10.1016/j.bbcan.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasche B, et al. Somatic acquisition and signaling of TGFBR1*6A in cancer. JAMA. 2005;294:1634–1646. doi: 10.1001/jama.294.13.1634. [DOI] [PubMed] [Google Scholar]
- 8.Bian Y, et al. TGFBR1*6A may contribute to hereditary colorectal cancer. J. Clin. Oncol. 2005;23:3074–3078. doi: 10.1200/JCO.2005.00.281. [DOI] [PubMed] [Google Scholar]
- 9.Kaklamani VG, et al. Combined genetic assessment of transforming growth factor-beta signaling pathway variants may predict breast cancer risk. Cancer Res. 2005;65:3454–3461. doi: 10.1158/0008-5472.CAN-04-2961. [DOI] [PubMed] [Google Scholar]
- 10.You W, et al. No association between TGFBR1*6A and lung cancer. J. Thorac. Oncol. 2007;2:657–659. doi: 10.1097/JTO.0b013e318070ccd7. [DOI] [PubMed] [Google Scholar]
- 11.Castillejo A, et al. TGFB1 and TGFBR1 polymorphic variants in relationship to bladder cancer risk and prognosis. Int. J. Cancer. 2009;124:608–613. doi: 10.1002/ijc.24013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raynal S, et al. Transforming growth factor-beta1 enhances the lethal effects of DNA-damaging agents in a human lung-cancer cell line. Int. J. Cancer. 1997;72:356–361. doi: 10.1002/(sici)1097-0215(19970717)72:2<356::aid-ijc26>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 13.Chang F, et al. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia. 2003;17:590–603. doi: 10.1038/sj.leu.2402824. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, et al. Transforming growth factor beta induces apoptosis through repressing the phosphoinositide 3-kinase/AKT/survivin pathway in colon cancer cells. Cancer Res. 2008;68:3152–3160. doi: 10.1158/0008-5472.CAN-07-5348. [DOI] [PubMed] [Google Scholar]
- 15.Kaufmann SH, et al. Induction of apoptosis by cancer chemotherapy. Exp. Cell Res. 2000;256:42–49. doi: 10.1006/excr.2000.4838. [DOI] [PubMed] [Google Scholar]
- 16.Segal-Bendirdjian E, et al. Alteration in p53 pathway and defect in apoptosis contribute independently to cisplatin-resistance. Cell Death Differ. 1998;5:390–400. doi: 10.1038/sj.cdd.4400357. [DOI] [PubMed] [Google Scholar]
- 17.Chen T, et al. Int7G24A variant of transforming growth factor-beta receptor type I is associated with invasive breast cancer. Clin. Cancer Res. 2006;12:392–397. doi: 10.1158/1078-0432.CCR-05-1518. [DOI] [PubMed] [Google Scholar]
- 18.Biros E, et al. A genetic polymorphism in transforming growth factor beta receptor-2 is associated with serum osteopontin. Int. J. Immunogenet. 2009;36:241–244. doi: 10.1111/j.1744-313X.2009.00855.x. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda H. Mutational analysis of transforming growth factor-beta receptor type II and Smad3 tumor suppressor genes in prolactinomas. Brain Tumor Pathol. 2006;23:7–12. doi: 10.1007/s10014-006-0196-7. [DOI] [PubMed] [Google Scholar]
- 20.Jin G, et al. TGFB1 and TGFBR2 functional polymorphisms and risk of esophageal squamous cell carcinoma: a case-control analysis in a Chinese population. J. Cancer Res. Clin. Oncol. 2008;134:345–351. doi: 10.1007/s00432-007-0290-1. [DOI] [PubMed] [Google Scholar]
- 21.Hildebrandt MA, et al. Genetic variations in the PI3K/PTEN/AKT/mTOR pathway are associated with clinical outcomes in esophageal cancer patients treated with chemoradiotherapy. J. Clin. Oncol. 2009;27:857–871. doi: 10.1200/JCO.2008.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee HJ, et al. Single-nucleotide polymorphisms and haplotype LD analysis of the 29-kb IGF2 region on chromosome 11p15.5 in the Korean population. Hum. Hered. 2005;60:73–80. doi: 10.1159/000088269. [DOI] [PubMed] [Google Scholar]
- 23.Sauerbrei W, et al. A bootstrap resampling procedure for model building: application to the Cox regression model. Stat. Med. 1992;11:2093–2109. doi: 10.1002/sim.4780111607. [DOI] [PubMed] [Google Scholar]
- 24.Hanagiri T, et al. Gender difference as a prognostic factor in patients undergoing resection of non-small cell lung cancer. Surg. Today. 2007;37:546–551. doi: 10.1007/s00595-006-3453-9. [DOI] [PubMed] [Google Scholar]
- 25.Chansky K, et al. The International Association for the Study of Lung Cancer Staging Project: prognostic factors and pathologic TNM stage in surgically managed non-small cell lung cancer. J. Thorac. Oncol. 2009;4:792–801. doi: 10.1097/JTO.0b013e3181a7716e. [DOI] [PubMed] [Google Scholar]
- 26.Park KH, et al. Single nucleotide polymorphisms of the TGFB1 gene and lung cancer risk in a Korean population. Cancer Genet. Cytogenet. 2006;169:39–44. doi: 10.1016/j.cancergencyto.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 27.Lei Z, et al. TGFBR1 haplotypes and risk of non-small-cell lung cancer. Cancer Res. 2009;69:7046–7052. doi: 10.1158/0008-5472.CAN-08-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leikauf GD, et al. Haplotype association mapping of acute lung injury in mice implicates activin A receptor, type 1. Am. J. Respir. Crit. Care Med. 2011 doi: 10.1164/rccm.201006-0912OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Page C, et al. BMP-2 signaling in ovarian cancer and its association with poor prognosis. J. Ovarian Res. 2009;2:4. doi: 10.1186/1757-2215-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothhammer T, et al. Bone morphogenic proteins are overexpressed in malignant melanoma and promote cell invasion and migration. Cancer Res. 2005;65:448–456. [PubMed] [Google Scholar]
- 31.Langenfeld EM, et al. Expression of bone morphogenetic proteins in human lung carcinomas. Ann. Thorac. Surg. 2005;80:1028–1032. doi: 10.1016/j.athoracsur.2005.03.094. [DOI] [PubMed] [Google Scholar]
- 32.Milet J, et al. Common variants in the BMP2, BMP4, and HJV genes of the hepcidin regulation pathway modulate HFE hemochromatosis penetrance. Am. J. Hum. Genet. 2007;81:799–807. doi: 10.1086/520001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Capasso M, et al. A predicted functional single-nucleotide polymorphism of bone morphogenetic protein-4 gene affects mRNA expression and shows a significant association with cutaneous melanoma in Southern Italian population. J. Cancer Res. Clin. Oncol. 2009;135:1799–1807. doi: 10.1007/s00432-009-0628-y. [DOI] [PubMed] [Google Scholar]
- 34.Levy L, et al. Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17:41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum. Hered. 2003;56:73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- 36.Stoika R, et al. Potential role of transforming growth factor beta1 in drug resistance of tumor cells. Acta Biochim. Pol. 2003;50:497–508. [PubMed] [Google Scholar]
- 37.Kano MR, et al. Improvement of cancer-targeting therapy, using nanocarriers for intractable solid tumors by inhibition of TGF-beta signaling. Proc. Natl Acad. Sci. USA. 2007;104:3460–3465. doi: 10.1073/pnas.0611660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu J, et al. Identification of the gene transcription and apoptosis mediated by TGF-beta-Smad2/3-Smad4 signaling. J. Cell. Physiol. 2008;215:422–433. doi: 10.1002/jcp.21325. [DOI] [PubMed] [Google Scholar]
- 39.Tian M, et al. The TGF-beta paradox in human cancer: an update. Future Oncol. 2009;5:259–271. doi: 10.2217/14796694.5.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q, et al. Bone morphogenetic protein 2 activates Smad6 gene transcription through bone-specific transcription factor Runx2. J. Biol. Chem. 2007;282:10742–10748. doi: 10.1074/jbc.M610997200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.