Abstract
We evaluated a previously described quantitative real-time PCR (qPCR) for quantifying and differentiating Ureaplasma parvum and U. urealyticum. Because of nonspecific reactions with Staphylococcus aureus DNA in the U. parvum PCR, we developed a modified qPCR and designed new primers. These oligonucleotides eradicated cross-reactions, indicating higher specificity. The detection limits of the qPCR were determined at 1 and 3 colony-forming units/ml for U. parvum and U. urealyticum, respectively. The quantification limits of the assay for both Ureaplasma species ranged from 2.106 to 2.101 copy numbers per PCR. A total of 300 patient samples obtained from the lower genital tract were tested with this newly designed qPCR assay and compared with culture results. Of the samples, 132 (44.0%) were culture positive, whereas 151 (50.3%) tested positive using qPCR. The U. parvum and U. urealyticum species were present in 79.5% and 12.6% of the qPCR-positive samples, respectively. Both species were found in 7.9% of those samples. Quantification of U. parvum and U. urealyticum in the samples ranged from less than 2.5 × 103 to 7.4 × 107 copies per specimen. In conclusion, the modified qPCR is a suitable method for rapid detection, differentiation, and quantification of U. parvum and U. urealyticum.
The genus Ureaplasma belongs to the Mycoplasmataceae family. Ureaplasmas are small prokaryotic cells that lack a cell wall. Previously, there was only one known species found in humans (namely, Ureaplasma urealyticum) comprising 14 serotypes. In 2002, Robertson et al1 proposed subdividing this species into U. parvum, comprising serotypes 1, 3, 6, and 14; and U. urealyticum, comprising serotypes 2, 4, 5, and 7 through 13. These subtypes cannot be distinguished from each other with routine microbiological methods and are, therefore, usually referred to as Ureaplasma species. They are found in the lower genital tract of nearly 50% of pregnant women as part of the normal vaginal flora. However, in some cases, Ureaplasma species have interfered with normal fetal development by causing an ascending infection.2–7 The reason for this infection is not fully understood but may be associated with the virulence of the microorganism, the host immune system, or local factors present in the lower genital tract. Species differentiation might be important because previous studies2,8,9 suggest that nongonococcal urethritis and an adverse pregnancy outcome with respect to birth weight, gestational age, and preterm delivery are implicated with the presence of U. urealyticum and not with U. parvum.
Because strains can only be differentiated with labor-intensive serotyping, an easy-to-perform sensitive method for differentiating the two species is necessary on initiation of pathogenicity studies. Quantitative real-time PCR (qPCR) presents an interesting option because the strains can be differentiated together, with evaluation of the microbial burden in the lower genital tract. Moreover, qPCR can be performed within two hours because of the elimination of postamplification handling.
In a recent study,10 qPCR was used to detect and quantify U. parvum and U. urealyticum in specimens from the lower genital tract. This test was evaluated in our laboratory and did allow the differentiation and quantification of U. urealyticum and U. parvum in two separate reaction mixtures. However, cross-reactions with Staphylococcus aureus DNA were encountered in the U. parvum mixture.
In this study, we investigated the nature of these cross-reactions by sequence analysis of the nonspecific amplification product, and we compared this sequence with the National Center for Biotechnology Information database. We designed new primers targeting the urease gene of U. parvum to improve the specificity of the qPCR. Furthermore, the method was adapted to be more practical for a routine laboratory setting and the phocine herpesvirus 1 (PhHV1) was included as an internal control to monitor for the presence of inhibitory factors in the clinical samples.
Materials and Methods
Investigating the Issue of Specificity in the Original Method
Reaction mixtures and cycling conditions were performed as previously described.10 Primer and probe sequences were blasted with the GenBank database to confirm specificity (http://www.ncbi.nlm.nih.gov, accession date April 25, 2008). Assay specificity was assessed with the 14 reference strains of U. parvum (serotypes 1, 3, 6, and 14) and U. urealyticum (serotypes 2, 4, 5, and 7 through 13). In addition to the 14 reference strains of Ureaplasma species, assay specificity was also determined with 11 human Mycoplasma species, 29 bacterial species, 1 yeast, and 1 viral pathogen: Acinetobacter baumannii, Bacteroides fragilis, Bordetella pertussis, Burkholderia cepacia, Citrobacter freundii, Chlamydia trachomatis, Enterobacter aerogenes, Escherichia coli, Gardnerella vaginalis, Herpes simplex virus 1/2, Listeria monocytogenes, Micrococcus species, Mobiluncus mulieris, Moraxella catarrhalis, Mycoplasma hominis, Mycoplasma amphiforme, Mycoplasma fermentans, Mycoplasma genitalium, Mycoplasma orale, Mycoplasma penetrans, Mycoplasma pirum, Mycoplasma primatum, Mycoplasma salivarium, Mycoplasma spermatophilum, Mycoplasma pneumoniae, Pseudomonas aeruginosa, Serratia marcescens, Stenotrophomonas maltophilia, Streptococcus agalactiae (group B), Streptococcus pneumoniae, Streptococcus pyogenes, Bordetella parapertussis, Candida albicans, Hemophilus influenzae, Klebsiella oxytoca, Klebsiella pneumoniae, Lactobacillus species, Morganella morganii, Proteus mirabilis, Staphylococcus aureus oxacillin R, Staphylococcus aureus oxacillin S, and Staphylococcus epidermidis. The last 11 organisms are urease positive, which is important in specificity testing because the urease gene is the target gene for amplification.
Nonspecific reactions were repeatedly tested (10-fold) with reference strains (to confirm the presence of cross-reactions) and with two more clinical strains. Nonspecific amplification products were analyzed by cloning and sequence analysis (ABI3730, Sanger sequencing, Bigdye v3.1; Applied Biosystems, Halle, Belgium) to retrieve the nature of the cross-reactions (BaseClear, Leiden, the Netherlands).
Modified Method
Primer Selection
To amplify U. parvum DNA, a new primer pair was designed, flanking the probe that was used for U. parvum detection in the originally described method. The forward primer, 5′-CATTGATGTTGCACAAGGAG-3′, and the reverse primer, 5′-CGTGATTTTAATGTATCGGCTTTC-3′, sequences were blasted with GenBank sequences in the National Center for Biotechnology Information database; for primer specificity (http://www.ncbi.nlm.nih.gov, accession date October 1, 2008). Primers were chosen within the ureD subunit of the urease gene of U. parvum and U. urealyticum. One copy of this gene is present per cell. The amplicon product sizes were 147 and 146 bp for U. parvum and U. urealyticum, respectively.
PCR Internal Control
PCR inhibition and extraction were monitored by amplifying the PhHV1. The use of this virus as a control for extraction and the presence of amplification inhibitors were originally described by Niesters.11 The virus strain was provided by the Department of Virology, University Hospital Rotterdam, Rotterdam, the Netherlands.
Modified qPCR Protocol
To detect both Ureaplasma species, real-time PCR was performed in two separate 25-μL reaction mixtures. The PhHV1 viral DNA was detected in multiplex PCR with the two reaction mixtures for U. parvum and U. urealyticum DNA. The reaction mixtures contained a ready-to-use 2x real-time PCR reaction mix containing all necessary components without primers and probes (IQ Multiplex Powermix, Bio-Rad, Nazareth-Eke, Belgium), 0.35 μmol/L of each primer, and 0.2 μmol/L of each probe (forward and reverse primer and probe of the target organism and of the internal control). DNA template, 2 μL, was added to the 23-μL reaction mixtures. Probes (Taqman Minor Groove Binder) for U. parvum (5′-FAM-TTGACCACCCTTACGAG-MGB-3′) and U. urealyticum (5′-FAM-TTGTCCGCCTTTACGAG-MGB-3′) were manufactured by Applied Biosystems (Warrington, UK). The Taqman probe for the internal control (Texas 5′-Red-TTTTTATGTGTCCGCCACCATCTGGATC-BHQ1-3′) was manufactured by Eurogentec (Luik, Belgium). Cycling conditions were as follows: 95°C for 3 minutes, followed by 45 cycles of 95°C for 15 seconds and 58°C for 30 seconds. Amplification, detection, and analysis were performed with a commercially available system (iCycler IQ real-time detection system; Bio-Rad). Data collection was performed during the annealing step. The relative fluorescence measurements were plotted against the cycle number for data analysis. Background fluorescence was corrected by manually setting the baseline cycles from 5 to 16. The log-linear portions of the amplification plots were used to determine a fractional cycle number for threshold fluorescence. In every run, a no-template control (sterile water) and a positive control (a 10–3 dilution of Ureaplasma reference strains, serotypes 3 and 10, that were grown to their logarithmic phase in bromothymol blue broth) were included.
Specificity
Assay specificity was assessed with reference strains of all U. urealyticum and U. parvum serotypes and with the 42 microorganisms previously described. To evaluate cross-reactivity with S. aureus in more detail, the assay was performed in 10-fold with the reference strains of S. aureus and in duplicate on two freshly isolated clinical S. aureus strains.
Limit of Detection
For determining the detection limit of the assay, a 10-fold serial dilution was made from U. parvum serotype 3 and U. urealyticum serotype 10 reference strains grown in bromothymol blue broth. Each dilution, 100 μL, was cultured on differential agar A7; and colony-forming units were counted at a ×40 magnification.12,13 The qPCR sensitivity was determined by analyzing the extracted DNA of the serial dilutions.
External Standard Curve for Quantification
Amplification products from U. parvum, serotype 3, and U. urealyticum, serotype 10, reference strains, obtained with real-time PCR, were ligated into a vector (pGEM-T Easy; Promega Benelux, Leiden, the Netherlands). The plasmids were transformed into E. coli DH5α cells (Life Technologies, Carlsbad, CA), which were grown; and the plasmid was purified. Plasmid concentration was determined using a kit (Quant-iT dsDNA Assay; Invitrogen, Breda, the Netherlands). Cloning, sequencing, and plasmid purification were performed by BaseClear, an independent and accredited service laboratory for DNA-based research (Leiden, the Netherlands). A 10-fold serial dilution series composed of 6 standards (2.106 to 2.101 copies per PCR) was used to construct an external standard curve for quantification. To evaluate the standard curve, each external standard was tested in triplicate; this was repeated twice. Because the ureD subunit that was used to prepare the standard curves was identical in all serotypes within the same species, the linearity of one serotype reflects the linearity of all serotypes within this species; only one standard curve per species needs to be used for quantification.
Reproducibility
The intra-assay and interassay reproducibility of the U. parvum and U. urealyticum PCR was tested with two different dilutions of U. parvum and U. urealyticum cultures. High and low concentrations of Ureaplasma species reference strains serotypes 3 and 10 (10−3 and 10−4 dilutions, respectively) that were grown to their logarithmic phase in bromothymol blue broth were used to determine reproducibility of the assay. The experiments were repeated 10 times after extraction (easyMAG; Biomérieux, Boxtel, the Netherlands), and the CV was calculated.
Evaluation of the qPCR with Clinical Samples
Samples
Three-hundred samples from the lower genital tract of healthy pregnant women were obtained at their first prenatal consultation. Two types of genital swabs containing a different type of transport medium were in use in our hospital for sampling the lower genital tract and the preservation of microorganisms: eSwab (Copan Italia, Brescia, Italy) and Universal Transport Medium (UTM, Copan Italia, Brescia, Italy). We aimed to evaluate the transport medium most appropriate for culture and/or PCR detection. One-hundred one samples were transported in eSwab, containing 1 ml of liquid medium; and 199 samples were transported in Universal Transport Medium (UTM), containing 3 ml of liquid medium.
Culture
Liquid transport medium was mixed, and approximately 0.2 ml of medium was inoculated on differential agar medium A7 and in bromothymol blue broth. Cultures were performed as previously described.6 Positive culture results were semiquantitatively reported (ie, when ureaplasmas were detected on the primary agar plate, the number of colonies was evaluated and classified into four groups [×40 magnification]: <1, 1 to 5, 6 to 12, and >12 colonies per field).
DNA Extraction
Before nucleic acid extraction, all clinical samples were spiked with the dilution of PhHV1 that resulted in a Ct value of approximately 30. Sample DNA was extracted using a platform (easyMAG automated extraction platform; Biomérieux, Brussel, Belgium), according to the manufacturer's instructions. Liquid transport medium, 100 μL, was used for DNA isolation.
Quantitative PCR
The qPCR was performed as previously described. Samples with a Ct of less than 40 were considered positive. Standard curves from both species were tested in every clinical sample run. For positive samples, the copy number per specimen was calculated, starting from the copy number per PCR reaction. Samples with copy numbers outside the linear range of the external standard curve were not quantified. Samples with results discordant between culture and qPCR were retested with qPCR using the remaining liquid transport medium, which was re-extracted.
Statistical Analysis
The χ2 test was used to analyze statistical differences in the quality of the eSwab and the UTM for detecting U. parvum and U. urealyticum with culture and qPCR.
Results
Investigating the Issue of Specificity in the Original qPCR Method
The specificity of the original described method was assessed by testing all U. parvum and U. urealyticum reference strains with the two reaction mixtures that were prepared as described by Cao et al.10 No cross-reactivity was seen between the serotypes of the U. parvum species and the serotypes of the U. urealyticum species. Among the 42 other strains that were tested for specificity of the assay, both S. aureus strains (S. aureus oxacillin S and S. aureus oxacillin R) showed a positive result in the mixture for U. parvum detection. Further analysis of this nonspecific reactivity revealed a DNA fragment of approximately 150 bp after gel electrophoresis. A blast search of the sequence of this amplified DNA showed 100% homology with the urease accessory protein ureG from S. aureus.
Evaluating the Modified qPCR Method
Specificity
The specificity tests with the primers designed in our laboratory showed no cross-reactions among U. parvum, U. urealyticum, S. aureus, or any of the other 41 microorganisms. No cross-reactivity was observed with the reference strains of S. aureus and with the isolated clinical S. aureus strains.
Limit of Detection
Sensitivity was determined by analyzing serial dilutions of extracted DNA from U. urealyticum serotype 10 and U. parvum serotype 3. By testing the 10-fold serial dilutions, the lowest levels of detection were determined to be 1 and 3 colony-forming units/ml for U. parvum and U. urealyticum, respectively.
External Standard Curve for Quantification
Figure 1 and Figure 2 show the linear range of the standard curves of both species tested in triplicate. The standard curve for U. urealyticum and U. parvum was linear over a 6-log-range. The upper and lower quantification limits were 2.101 and 2.106 copies per PCR reaction, respectively.
Figure 1.
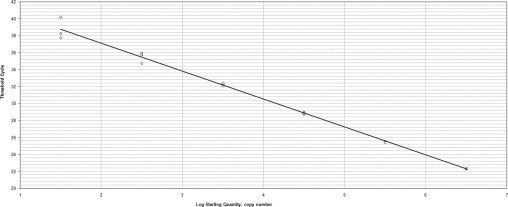
Standard curve (in triplicate) from plasmids containing the U. parvum serotype 3 DNA fragment. Correlation coefficient, 0.996; slope, −3.305; intercept, 43.318; PCR efficiency, 100.7%.
Figure 2.
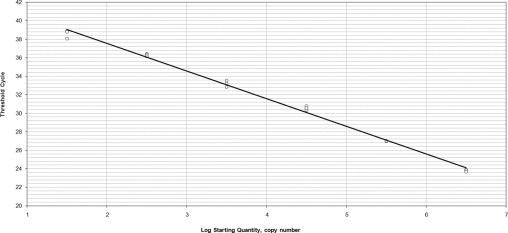
Standard curve in triplicate from plasmids containing the U. urealyticum serotype 10 DNA fragment. Correlation coefficient, 0.995; slope, −3.108; intercept, 43.601; PCR efficiency, 109.8%.
Reproducibility of the qPCR
The intra-run and interrun variations for U. urealyticum and U. parvum, tested at two concentrations (high and low), are summarized in Table 1.
Table 1.
Interrun and Intrarun Variation of the Modified qPCR for U. parvum and U. urealyticum
| Dilution | CV, % |
Mean Ct values | |
|---|---|---|---|
| Intra-run variation | Interrun variation | ||
| U. parvum | |||
| 10−3 (high concentration) | 9.8 | 5.2 | 33.4 |
| 10−4 (low concentration) | 11.5 | 10.8 | 36.8 |
| U. urealyticum | |||
| 10−3 (high concentration) | 12.3 | 7.5 | 32.5 |
| 10−4 (low concentration) | 13.4 | 9.3 | 36.0 |
Ct, cycle threshold.
Evaluating the Modified qPCR with Clinical Samples
Culture and qPCR results of the 300 clinical samples, according to the type of transport medium used, are summarized in Table 2. Of the 300 specimens, 151 (50.3%) were positive by qPCR (49.5% and 50.8% for eSwab and UTM, respectively; P = 0.77); 132 samples (44.0%) were positive by culture (38.6% and 46.7% for eSwab and UTM, respectively; P = 0.155). The positive predictive values of the qPCR for samples transported in eSwab and UTM were 78% and 92%, respectively, using the culture method as a reference method. The negative predictive value of the qPCR for samples transported in eSwab and UTM was 100%.
Table 2.
Culture and qPCR Results of 300 Clinical Specimens
| eSwab |
UTM |
|||
|---|---|---|---|---|
| Variable | Culture-positive results | Culture-negative results | Culture-positive results | Culture-negative results |
| qPCR-positive samples | 39 | 11 | 93 | 8 |
| U. parvum | 33 | 6 | 76 | 5 |
| U. urealyticum | 4 | 4 | 8 | 3 |
| Mixed | 2 | 1 | 9 | 0 |
| qPCR-negative samples | 0 | 51 | 0 | 98 |
| Total, no. (%) | 39 (13.0) | 62 (20.7) | 93 (31.0) | 106 (35.3) |
Discordant results between qPCR and culture were obtained in 19 samples: all those samples were culture negative and qPCR positive. The discordant samples were retested with qPCR using a newly extracted sample from the remaining transport medium, and the results were confirmed. Negative culture results with a positive qPCR result were more frequently found in samples transported in eSwab (11/101) than in samples transported in UTM (8/199). For the 19 culture-negative and qPCR-positive samples, copy numbers in the qPCR ranged from less than 2.5 × 103 to 2.0 × 104 copies per specimen.
Of the 151 qPCR-positive samples, 19 (12.6%) were positive for U. urealyticum, 120 (79.5%) were positive for U. parvum, and 12 (7.9%) were positive for both species. There were no differences observed with the type of transport medium used (Table 2).
The quantitative results of the qPCR, expressed as copy number per specimen, are summarized in Table 3. Fifteen positive samples had results less than the lowest quantification standard of the standard curve, thus not allowing quantification. None of the samples had results higher than the highest standard of the standard curve, and 136 samples had copy numbers within the linear range of the standard curve.
Table 3.
Ranges of qPCR Results per Species and per Type of Transport Medium
| qPCR results, copies per specimen |
||
|---|---|---|
| Variable | eSwab transport medium | UTM |
| U. urealyticum | <2.5 × 103−9.1 × 106 | <7.5 × 103−2.3 × 107 |
| U. parvum | <2.5 × 103−7.4 × 107 | <7.5 × 103−7.4 × 107 |
| In mixed infections | ||
| U. urealyticum | <2.5 × 103−6.0 × 104 | <7.5 × 103−2.2 × 106 |
| U. parvum | <2.5 × 103−7.7 × 105 | <7.5 × 103−3.2 × 107 |
A correlation of the quantitative results of the qPCR and the semiquantitative results of the culture is summarized in Figure 3. Because there were no significant differences in the ranges for mixed infections, they are not presented separately in this figure.
Figure 3.
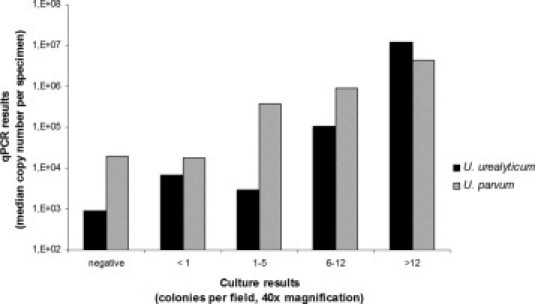
Comparison of the semiquantitative culture results (xaxis) (133 culture-positive samples) and the quantitative PCR results (y axis). The qPCR results are split into two groups: quantification of U. urealyticum and U. parvum.
No PCR inhibition was observed in any of the 300 clinical samples.
Discussion
Assay specificity is of the utmost importance when attempting to detect a specific organism because it yields true-positive results. As defined by the Clinical Laboratory Standards Institute, a panel of microorganisms closely related to the target organism should be used to monitor cross-reactivity. In addition, organisms present in the normal flora of the specimen should be tested.14
In our evaluation of the primer and probe sequences from the previously described study, cross-reactions were observed with S. aureus.10 Cloning and sequencing the amplification product of the nonspecific reaction confirmed the presence of S. aureus DNA and, more specifically, a 150-bp fragment from the urease accessory protein ureG. None of the nine other organisms containing the urease gene (ie, the target gene for the Ureaplasma qPCR) demonstrated a cross-reaction. Because S. aureus is frequently found on mucosal surfaces and the skin of healthy individuals, the primers described for U. parvum were assumed to be inappropriate for testing clinical specimens, requiring another PCR protocol.15,16
Although several PCR methods have already been described for Ureaplasma, no method met our requirements for pathogenicity studies.10,17–20 Determining the microbial burden may also be important for pathogenicity; thus, a PCR protocol enabling species differentiation, together with quantification, is needed.21 Moreover, the protocol should be validated on clinical samples and must be highly specific. Thus, a qPCR protocol using a new primer pair for U. parvum was designed and compared with culture results. With these new primers, no cross-reactivity was seen with staphylococcal DNA, making the protocol more specific. The new qPCR protocol was developed with the inclusion of viral DNA detection as an internal control. Although no inhibition was detected in any of the 300 clinical samples, the inclusion of control DNA for the detection of inhibition is mandatory when testing clinical samples because such inhibitors could generate false-negative results.14,22
The modifications in the protocol did not interfere with the sensitivity: our external standard curve was only linear over a 6-log range, whereas it was a 7-log range in the method originally described.
Most clinical samples belonged to the U. parvum biovar. In this study, the percentages of U. urealyticum, U. parvum, and samples containing both species correlate well with those found in previous studies.10,23–25 This qPCR method, amplifying U. parvum and U. urealyticum in separate reactions, probably represents a more adequate method for the detection of mixed infections.
The positivity rates of U. parvumandU. urealyticum were higher in qPCR than in culture (50.3% versus 44.3%).
Although the difference observed for culture between the UTM and eSwab does not reach significance (46.7% versus 38.6%; P = 0.115), the UTM appears to be more appropriate for Ureaplasma culture. Indeed, the instruction manual of the UTM recommends its use for the culture of mycoplasmas and ureaplasmas, Chlamydia, and viruses, whereas the eSwab instruction manual describes that it can be used for transportation of all kinds of bacteria in general. The medium used in the UTM is probably more suited for the culture of more fastidious organisms. The positivity rates in qPCR for samples transported in UTM and eSwab were comparable (50.3% versus 49.3%). Because we calculated the positive predictive values of the qPCR based on the culture results (gold standard), the positive predictive value of 78% for samples transported in eSwab is probably falsely low and should be further investigated.
Quantification results showed that U. parvum was present in higher concentrations in the clinical specimens compared with U. urealyticum. The difference observed between eSwab and UTM for U. parvum and U. urealyticum quantification in mixed infections (>1 log) is probably because of the few samples containing mixed infections (three transported in eSwab and nine transported in UTM). There was a good correlation between the semiquantitative culture method and the qPCR method for U. parvum and U. urealyticum.
In conclusion, a new qPCR protocol was evaluated and validated as an adequate test for detecting, differentiating, and quantifying U. parvum and U. urealyticum. This assay can be applied in further epidemiological and pathogenicity studies in our laboratory.
Acknowledgments
We thank Jørgen Skov Jensen (Statens Serum Institut, Copenhagen, Denmark) for providing the human Mycoplasma lysates for specificity evaluation.
Footnotes
Supported by a grant from Steunfonds Marguerite-Marie Delacroix.
References
- 1.Robertson J.A., Stemke G.W., Davis J.W., Jr, Harasawa R., Thirkell D., Kong F., Shepard M.C., Ford D.K. Proposal of Ureaplasma parvum sp. nov. and emended description of Ureaplasma urealyticum (Shepard et al. 1974) Robertson et al. 2001. Int J Syst Evol Microbiol. 2002;52:587–597. doi: 10.1099/00207713-52-2-587. [DOI] [PubMed] [Google Scholar]
- 2.Abele-Horn M., Peters J., Genzel-Boroviczeny O., Wolff C., Zimmermann A., Gottschling W. Vaginal Ureaplasma urealyticum colonization: influence on pregnancy outcome and neonatal morbidity. Infection. 1997;25:286–291. doi: 10.1007/BF01720398. [DOI] [PubMed] [Google Scholar]
- 3.Breugelmans M., Vancutsem E., Naessens A., Laubach M., Foulon W. Association of abnormal vaginal flora and Ureaplasma species as risk factors for preterm birth: a cohort study. Acta Obstet Gynecol Scand. 2010;89:256–260. doi: 10.3109/00016340903418769. [DOI] [PubMed] [Google Scholar]
- 4.Foulon W., Naessens A., Dewaele M., Lauwers S., Amy J.J. Chronic Ureaplasma urealyticum amnionitis associated with abruptio placentae. Obstet Gynecol. 1986;68:280–282. [PubMed] [Google Scholar]
- 5.Lamont R.F., Taylor-Robinson D., Wigglesworth J.S., Furr P.M., Evans R.T., Elder M.G. The role of mycoplasmas, ureaplasmas and chlamydiae in the genital tract of women presenting in spontaneous early preterm labour. J Med Microbiol. 1987;24:253–257. doi: 10.1099/00222615-24-3-253. [DOI] [PubMed] [Google Scholar]
- 6.Naessens A., Foulon W., Cammu H., Goossens A., Lauwers S. Epidemiology and pathogenesis of Ureaplasma urealyticum in spontaneous abortion and early preterm labor. Acta Obstet Gynecol Scand. 1987;66:513–516. doi: 10.3109/00016348709015726. [DOI] [PubMed] [Google Scholar]
- 7.Yoon B.H., Romero R., Park J.S., Chang J.W., Kim Y.A., Kim J.C., Kim K.S. Microbial invasion of the amniotic cavity with Ureaplasma urealyticum is associated with a robust host response in fetal, amniotic, and maternal compartments. Am J Obstet Gynecol. 1998;179:1254–1260. doi: 10.1016/s0002-9378(98)70142-5. [DOI] [PubMed] [Google Scholar]
- 8.Maeda S., Deguchi T., Ishiko H., Matsumoto T., Naito S., Kumon H., Tsukamoto T., Onodera S., Kamidono S. Detection of Mycoplasma genitalium: Mycoplasma hominis, Ureaplasma parvum (biovar 1) and Ureaplasma urealyticum (biovar 2) in patients with non-gonococcal urethritis using polymerase chain reaction-microtiter plate hybridization. Int J Urol. 2004;11:750–754. doi: 10.1111/j.1442-2042.2004.00887.x. [DOI] [PubMed] [Google Scholar]
- 9.Yokoi S., Maeda S., Kubota Y., Tamaki M., Mizutani K., Yasuda M., Ito S., Nakano M., Ehara H., Deguchi T. The role of Mycoplasma genitalium and Ureaplasma urealyticum biovar 2 in postgonococcal urethritis. Clin Infect Dis. 2007;45:866–871. doi: 10.1086/521266. [DOI] [PubMed] [Google Scholar]
- 10.Cao X., Wang Y., Hu X., Qing H., Wang H. Real-time TaqMan polymerase chain reaction assays for quantitative detection and differentiation of Ureaplasma urealyticum and Ureaplasma parvum. Diagn Microbiol Infect Dis. 2007;57:373–378. doi: 10.1016/j.diagmicrobio.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Niesters H.G. Quantitation of viral load using real-time amplification techniques. Methods. 2001;25:419–429. doi: 10.1006/meth.2001.1264. [DOI] [PubMed] [Google Scholar]
- 12.Shepard M.C., Lunceford C.D. Differential agar medium (A7) for identification of Ureaplasma urealyticum (human T mycoplasmas) in primary cultures of clinical material. J Clin Microbiol. 1976;3:613–625. doi: 10.1128/jcm.3.6.613-625.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson J.A. Bromothymol blue broth: improved medium for detection of Ureaplasma urealyticum (T-strain mycoplasma) J Clin Microbiol. 1978;7:127–132. doi: 10.1128/jcm.7.2.127-132.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute . Clinical and Laboratory Standards Institute; Pennsylvania: 2009. Molecular Diagnostic Methods for Infectious Diseases (MM3-P2) p. 26. [Google Scholar]
- 15.Chiller K., Selkin B.A., Murakawa G.J. Skin microflora and bacterial infections of the skin. J Investig Dermatol Symp Proc. 2001;6:170–174. doi: 10.1046/j.0022-202x.2001.00043.x. [DOI] [PubMed] [Google Scholar]
- 16.VandenBergh M.F., Verbrugh H.A. Carriage of Staphylococcus aureus: epidemiology and clinical relevance. J Lab Clin Med. 1999;133:525–534. doi: 10.1016/s0022-2143(99)90181-6. [DOI] [PubMed] [Google Scholar]
- 17.Aaltone R., Jalava J., Laurikainen E., Karkkainen U., Alanen A. Cervical Ureaplasma urealyticum colonization: comparison of PCR and culture for its detection and association with preterm birth. Scand J Infect Dis. 2002;34:35–40. doi: 10.1080/00365540110077074. [DOI] [PubMed] [Google Scholar]
- 18.Mallard K., Schopfer K., Bodmer T. Development of real-time PCR for the differential detection and quantification of Ureaplasma urealyticum and Ureaplasma parvum. J Microbiol Methods. 2005;60:13–19. doi: 10.1016/j.mimet.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Yi J., Yoon B.H., Kim E.C. Detection and biovar discrimination of Ureaplasma urealyticum by real-time PCR. Mol Cell Probes. 2005;19:255–260. doi: 10.1016/j.mcp.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Yoon B.H., Romero R., Lim J.H., Shim S.S., Hong J.S., Shim J.Y., Jun J.K. The clinical significance of detecting Ureaplasma urealyticum by the polymerase chain reaction in the amniotic fluid of patients with preterm labor. Am J Obstet Gynecol. 2003;189:919–924. doi: 10.1067/s0002-9378(03)00839-1. [DOI] [PubMed] [Google Scholar]
- 21.Abele-Horn M., Scholz M., Wolff C., Kolben M. High-density vaginal Ureaplasma urealyticum colonization as a risk factor for chorioamnionitis and preterm delivery. Acta Obstet Gynecol Scand. 2000;79:973–978. [PubMed] [Google Scholar]
- 22.Muska A., Peck E., Palmer S. Caiser Academic Press; Norfolk, UK: 2007. Standards and controls: concepts for preparation and use in real-time PCR applications; pp. 101–131. MacKay I. [Google Scholar]
- 23.Kong F., Ma Z., James G., Gordon S., Gilbert G.L. Species identification and subtyping of Ureaplasma parvum and Ureaplasma urealyticum using PCR-based assays. J Clin Microbiol. 2000;38:1175–1179. doi: 10.1128/jcm.38.3.1175-1179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Echahidi F., Van Geel K., Lauwers S., Naessens A. Comparison of two methods for serotyping Ureaplasma urealyticum clinical isolates. J Microbiol Methods. 2002;49:157–161. doi: 10.1016/s0167-7012(01)00359-1. [DOI] [PubMed] [Google Scholar]
- 25.Naessens A., Foulon W., Breynaert J., Lauwers S. Serotypes of Ureaplasma urealyticum isolated from normal pregnant women and patients with pregnancy complications. J Clin Microbiol. 1988;26:319–322. doi: 10.1128/jcm.26.2.319-322.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]


