INTRODUCTION
This review will provide a historical perspective of the evolution of our understanding of the protein transport machinery. One of the interesting findings of the last 2 decades or so is that molecular chaperones are as vital to the process of protein export as they are for the folding of cytosolic proteins. This article will describe the discovery of the protein export machinery and, in particular, will focus on protein transport in Escherichia coli and the involvement of the molecular chaperone SecB.
DISCOVERY OF THE PROTEIN EXPORT MACHINERY
The process of protein secretion is particularly remarkable in that water-soluble secretory proteins overcome a hydrophobic membrane barrier and protein transport occurs with high fidelity and efficiency. Following synthesis, as much as 50% of all cellular protein is exported to an extracytoplasmic compartment. These proteins are synthesized as precursors with additional amino-terminal sequences, called signal sequences, that are required for efficient export. These signal sequences are eventually cleaved by signal peptidase, releasing the mature protein in its final location. The presence of a signal sequence in secretory proteins led Günter Blobel to propose the signal recognition hypothesis. In his hypothesis, Blobel proposed that translation of the signal sequence from a messenger RNA transcript results in a unique hydrophobic sequence on the amino-terminal end of the nascent chain. The emergence of this signal sequence from the large ribosomal subunit initiates entry into the secretory pathway via attachment of the ribosomal-nascent chain complex to the membrane. This provides the topological conditions for the transfer of the nascent chain across the membrane (Blobel and Dobberstein 1975). Although signal sequences are not homologous in primary sequence, they do have some common structural features: (1) a net positive charge in the N-terminus, (2) a hydrophobic core region, and (3) a polar cleavage region. In membrane-mimetic systems such as detergent micelles or lipid vesicles, signal peptides have a high tendency to form α-helices. The ability to adopt a helical conformation appears to be required for the function of a signal sequence, since some peptides that correspond to export-defective signal sequences display reduced α-helical potential. In contrast, functional signal sequences share a high capacity to adopt α-helices (Jones et al 1990; Izard et al 1995). The low degree of sequence conservation and the ability of synthetic signal sequences to interact with lipids led many investigators to initially suggest the direct transfer model of protein secretion. This model suggests that the signal sequence of a nascent polypeptide binds in an α-helical conformation directly to the fatty acyl interior of a membrane. The subsequent docking of the ribosomal complex to its binding site on the membrane would have sufficient energy such that even strongly hydrophilic residues on the emerging polypeptide would partition into the bilayer interior. Quantitative assessment of the energetics of this process suggested that this mechanism could be used both to secrete proteins and to insert transmembrane proteins (von Heijne and Blomberg 1979; Inouye and Halegoua 1980; Engelman and Steitz 1981; De Vrije et al 1989; Jones et al 1990).
However, evidence has accumulated showing that some protein components, such as the signal recognition particle (SRP), are directly involved in the recognition of signal sequences, and this recognition process is crucial for efficient protein export in all systems tested (Phillips and Silhavy 1992; Walter and Johnson 1994; Valent et al 1995). The SRP delivers nascent precursors attached to ribosomes to a membrane-bound SRP receptor (Connolly and Gilmore 1989), with the eventual release of the precursors to the proteinaceous channel complex. Consistent with a prediction that the SRP should have a hydrophobic binding pocket for a signal sequence, recent crystal structures of the signal sequence binding subunit of SRP from Thermus aquaticus and the mammalian counterpart, SRP54, reveal a hydrophobic groove that is likely to contribute to the interaction with signal sequences (Keenan et al 1998; Clemons et al 1999). Furthermore, the channel-forming ability of proteinaceous complexes through which polypeptides can be translocated, called translocons, have been demonstrated in prokaryotic and eukaryotic cells (Hanein et al 1996; Meyer et al 1999). Yeast translocons (Sec61p complexes) in detergent form cylindrical oligomers, with a diameter of about 85 Å and a central pore of about 20 Å. Each oligomer contains 3–4 heterotrimers. Similar ring structures are seen in reconstituted proteoliposomes and native membranes. Similarly, the SecY/E translocon of Bacillus subtilis forms ring structures in detergent micelles and in intact lipid bilayers, often with a quasi-pentagonal appearance in projection. When plasma membrane vesicles and protoplasts of E coli are fused to planar lipid bilayers and subsequently studied using electrophysiological techniques, large transmembrane aqueous channels are opened by addition of 0.2 nM LamB signal peptide to the cytoplasmic side of the membrane. These aqueous pores are similar in conductance to those observed in mammalian endoplasmic reticulum when puromycin is used to release and thus unplug nascent translocating chains (Simon and Blobel 1991, 1992). These results demonstrate that signal peptides are sufficient for opening the protein-conducting channels and suggest that signal sequences are the physiological ligands that open protein-conducting channels at the initiation of protein translocation across prokaryotic plasma membranes and the mammalian endoplasmic reticulum.
Studies of prokaryotic protein translocation have been preceded by those of eukaryotic protein translocation, not only because the signal hypothesis is based on the eukaryotic model system but also because it is relatively easy to handle and use microsomes to study mechanisms of protein transport in vitro. In addition, the study of yeast genetics has rapidly lead to the discovery of several genes involved in the efficiency of transport processes. Understanding the basic principles of protein transport in the eukaryotic system has spurred research in prokaryotes and subcellular compartments, such as mitochondria and chloroplasts. It is now apparent that all export systems share some highly conserved components, typically including an SRP, SRP receptor, and the subunits of the translocation channel, although there are some differences among them. Interestingly, bacterial components such as SecA and SecY are found in thylakoid membranes of chloroplasts. The principles of protein transport in bacteria, therefore, can be extended to several systems.
E COLI TRANSLOCON: THE PROTEIN-CONDUCTING CHANNEL
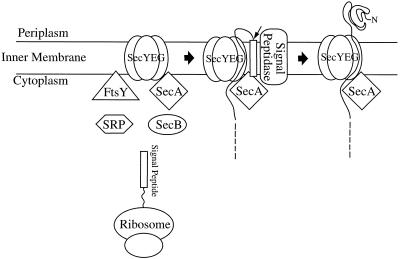
SecA (100 kDa) was the first translocation component to be purified from the inner membrane of E coli in a functional form (Fig 1). The removal of SecA from soluble components prepared from a wild type strain by passage through an anti-SecA antibody column abolishes protein translocation. Translocation activity is completely restored by addition of purified SecA protein, indicating that the observed defects are solely related to the loss of SecA function (Cabelli et al 1988). Salt or urea treatment renders inner membrane vesicles dependent on cytosolic factors for functional binding and translocation of proOmpA. The relevant soluble protein is SecA. It is functional as a dimer (Cunningham et al 1989; Driessen 1993) and is responsible for the hydrolysis of adenosine triphosphate (ATP) that is required for both cotranslational and posttranslational protein translocation (Chen and Tai 1987; Lill et al 1989). SecA alone has a low level of adenosine triphosphatase (ATPase) activity, which is stimulated by anionic lipids and stabilized by interaction with a precursor protein (Lill et al 1990). The fully activated state of SecA ATPase activity, the translocation ATPase, requires a lipid environment, some membrane proteins, and a precursor protein.
Fig 1.
Schematic representation of protein transport across the inner membrane of E coli. The nascent polypeptide is recognized by cytoplasmic proteins such as SRP or SecB. It is delivered to a protein translocation channel, SecYEG, in the membrane by a high-affinity SRP-FtsY or SecB-SecA interaction. Subsequently, the signal peptide is cleaved by signal peptidase (leader peptidase), and translocation through the channel is finally completed. Translocation of the polypeptide is powered by multiple rounds of ATP hydrolysis by SecA
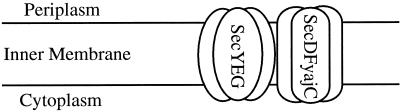
A trimeric complex was isolated from detergent extracts of inner membranes (Brundage et al 1990). Individual fractions obtained during the purification were assayed by forming proteoliposomes through detergent removal and measuring their capacity to support, in the presence of proOmpA, the translocation ATPase activity of SecA. The 3 members of this complex were identified in a sodium dodecyl sulfate–polyacrylamide gel as band 1, SecY, and SecE (Brundage et al 1990). SecY spans the membrane 10 times and SecE spans the membrane 3 times (Akiyama and Ito 1989; Schatz et al 1989). During analysis of the structure-function relationship of SecE, Nishiyama et al (1993, 1994) found that a trichloroacetic acid–soluble fraction obtained from detergent-solubilized E coli cytoplasmic membranes stimulates the translocation activity of reconstituted proteoliposomes more than 20-fold. SecE, SecY, SecA, and ATP are essential for the factor-dependent stimulation of activity. A 12-kDa protein, which has stimulating activity, was purified to homogeneity from the trichloroacetic acid–soluble fraction. The partial amino acid sequence of the protein coincides with that deduced from the DNA sequence of the upstream region of the leuU gene. When the gene encoding p12, secG, is deleted from the chromosome, the deletion strain exhibits cold-sensitive growth. Pulse-chase experiments reveal that precursors of outer membrane protein A (OmpA), maltose-binding protein (MBP), and β-lactamase (Bla) accumulate at 20 °C, but not at 37 °C. Finally, the SecG protein and the band 1 subunit of E coli preprotein translocase were identified as the same protein (Nishiyama et al 1993, 1994; Douville et al 1994). Although SecY, SecE, and SecG can be isolated separately and functionally recombined during proteoliposome formation (Hanada et al 1994), they are physiologically associated as a trimer both in vivo (Joly et al 1994) and in vitro (Brundage et al 1992). The complex of SecA bound stoichiometrically to the membrane-embedded SecYEG (Hartl et al 1990; Douville et al 1995) constitutes the core of the preprotein translocase. The pure enzyme complex supports ATP and ΔH+-dependent preprotein translocation at rates and to extents that are comparable with those seen with the starting membrane vesicles (Bassilana and Wickner 1993). Recent studies have shown that SecYEG is part of a larger complex that also contains SecD, SecF, and YajC (Fig 2). Although sufficient to provide sites for high-affinity binding and membrane insertion of SecA and for its activation as a preprotein-dependent ATPase, SecYE has only a very low capacity to support translocation. The proteins encoded by secD operon-SecD, SecF, and yajC-also form an integral membrane heterotrimeric complex (SecDFyajC). Physical and functional studies show that these 2 trimeric complexes are associated to form SecYEGDFyajC, the hexameric integral membrane domain of the preprotein translocase holoenzyme. Either SecG or SecDFyajC can support the translocation activity of SecYE by facilitating the ATP-driven cycle of SecA membrane insertion and deinsertion at different stages of the translocation reaction (Duong and Wickner 1997). So far, at least 2 different protein transport pathways have been found to use this translocon in E coli: the SRP-dependent and SecB-dependent pathways.
Fig 2.
Schematic representation of the SecYEGDFyajC heterohexameric complex. A heterotrimeric complex of SecY, SecE, and SecG forms a heterohexameric complex with another heterotrimer composed of SecD, SecF, and yajC
THE SRP-DEPENDENT PATHWAY
E coli 4.5S RNA and P48 (also called fifty-four homologue [Ffh]) are analogous to the eukaryotic 7S RNA and SRP54, respectively (Poritz et al 1988; Bernstein et al 1989). These components together suffice to constitute a functional SRP. The phylogenetic description of the secondary structure of the various SRP RNAs reveals a unique and highly conserved structural domain. This domain is also found in the 4.5S RNA of E coli and in the small cytoplasmic RNA of B subtilis (Poritz et al 1988). The human SRP RNA can be divided into 4 structural domains (domains I, II, III, and IV). Domain IV is particularly interesting in that it consists of only 50 nucleotides, which form a rigid well-defined structure. It is composed of 3 short helices that are flanked by internal loops. The sequences of these loops are highly conserved among all SRP RNAs. The 4.5S RNA of the E coli SRP is much smaller than its eukaryotic and archaeal counterparts and that of most other bacteria. Nevertheless, it consists of a domain identical to domain IV found in other SRP RNAs of E coli (Poritz et al 1990; Larsen and Zwieb 1991). It has been postulated that domain IV might confer on SRP the specific ribosome-binding properties required for its signal recognition function. Expression of human SRP 7S RNA in E coli can suppress the lethality caused by depletion of 4.5S RNA. In E coli, both RNAs are associated with Ffh. In vitro, both E coli Ffh and SRP54 specifically bind to 4.5S RNA. These results suggest that 4.5S RNA can functionally be replaced by 7S RNA from human SRP (Ribes et al 1990). When E coli cells are depleted of 4.5S RNA, the translocation of Bla is strongly impaired but that of MBP, ribose-binding protein (RBP), and OmpA are unaffected (Poritz et al 1990; Ribes et al 1990).
Ffh is necessary for viability and efficient protein export, since E coli cells that lack a functional Ffh fail to grow (Phillips and Silhavy 1992). Depletion of Ffh results in an elongated cell phenotype. The translocation of alkaline phosphatase, Bla, and RBP in Ffh-depleted cells is defective, whereas that of MBP, LamB, outer membrane protein F (OmpF), and OmpA is less affected. The sequence of Ffh suggests 3 domains: an amino-terminal N domain of unknown function, a central guanosine triphosphate (GTPase) G domain, and a methionine-rich M domain that binds both SRP RNA and signal peptides (Römisch et al 1990; Zorf et al 1993). Sequence conservation suggests that structurally similar N and G domains are present in FtsY, an E coli SRP receptor. The M domain of Ffh has been suggested to be involved in RNA binding, whereas the N and G domains bind signal peptides (Zheng and Gierasch 1997). The crystal structure of the protease-resistant N and G domains (NG domain) from Ffh of T aquaticus suggests that the N domain senses or controls the nucleotide occupancy of the GTPase domain (Freymann et al 1997).
FtsY, an E coli SRP receptor, is located in the cytosol and the membrane, although it does not have any predicted transmembrane segment (Luirink et al 1994), unlike SRα, the membrane-associated mammalian counterpart (Connolly and Gilmore 1989). The Ffh/4.5S ribonucleoprotein binds tightly to FtsY in a GTP-dependent manner. This interaction results in the stimulation of GTP hydrolysis, which can be inhibited by signal peptides. These properties mimic those of mammalian SRP and its receptor, suggesting that the E coli Ffh/4.5S ribonucleoprotein and FtsY have functions in protein targeting that are similar to those of their mammalian counterparts (Miller et al 1994). A GTPase domain (NG domain) in FtsY is preceded by a highly charged domain (A domain). The structure of the NG domain could be divided into 3 regions: the N domain, a GTPase domain, and an α-β-α insertion (I box) that is unique to the SRP-type GTPases (Bourne et al 1991; Montoya et al 1997).
Using a protein cross-linking approach, Valent et al (1998) have demonstrated that the SRP pathway delivers nascent inner membrane proteins to the membrane. The SRP receptor FtsY, GTP, and inner membranes are required for release of the nascent proteins from the SRP. On release of the SRP at the membrane, the targeted nascent proteins insert into a translocon that contains at least SecA, SecY, and SecG.
THE SecB-DEPENDENT PATHWAY
Kumamoto and Beckwith (1983) isolated mutants with pleiotropic defects in protein secretion. The lesions, which cause secretion defects, have been mapped to a locus called secB. These mutants are defective in the localization of at least MBP, LamB, OmpA, and OmpF (Kumamoto 1990). It is believed that SecB associates with the precursor forms of exported proteins in the cytoplasm and dissociates before or during translocation of precursors across the cell membrane (Kumamoto 1989). DNA sequence analysis of the secB gene revealed that the SecB protein is a 16.6-kDa acidic polypeptide, and centrifugation analysis has revealed it forms a tetramer (Kumamoto and Nault 1989; Watanabe and Blobel 1989). Randall and Hardy (1986) found that before export, precursor MBP (pre-MBP) is found in the cytoplasm in a loosely folded conformation that is sensitive to protease. Furthermore, when export of MBP is inhibited by the presence of uncoupler, the wild type precursor remains in the cytoplasm in a proteolysis-sensitive state. In contrast, under the same conditions, a mutant form (MalE14–2) that folds rapidly into a stable conformation is resistant to proteolysis. A kinetic analysis was carried out in the absence of uncoupler so that the MBPs would be exported. Although the export of the wild type precursor is rapid and complete, export of the mutant precursor is slow, and only 40% of the protein reaches the periplasm. It was found that the kinetics of folding closely parallel the loss of export competence, and once the population of intracellular precursor has folded completely, no further export occurs. This finding suggested that tightly folded pre-MBP molecules are unable to translocate. Therefore, pre-MBP molecules that are destined for export must be in a loosely folded conformation; components of the E coli export apparatus are involved in maintaining this translocation-appropriate form.
Although the precursor protein proOmpA can translocate across purified E coli inner membrane vesicles in the absence of any other soluble proteins, the efficiency of translocation is significantly enhanced if cytoplasmic factors are also present (Crooke et al 1988). In fact, after dilution, proOmpA rapidly loses its ability to become membrane integrated. This suggests that the primary role of soluble factors is to maintain exported proteins in a translocation-competent state. These factors could enhance precursor export by promoting or stabilizing a specific nonnative conformation or simply by obstructing the folding of the protein into its final form. The best characterized example of a soluble factor that serves this purpose is SecB. In the absence of SecB, tightly folded cytoplasmic precursor MBP is observed in vivo (Kumamoto and Gannon 1988), consistent with the notion that one of the functions of SecB is to prevent precursor folding. In addition, purified SecB influences the conformation of precursor proteins in vitro. It retards folding, prevents formation of protease-resistant pre-MBP, promotes membrane translocation of MPB in vitro (Collier et al 1988; Weiss et al 1988), and maintains the translocation competence of proOmpA and prePhoE in vitro (Lecker et al 1989; Kusters et al 1989). In addition, SecB has been shown to associate physically with precursors as predicted from its effects on export competence (Kumamoto 1989; Lecker et al 1989; Randall et al 1990).
A model for the interaction of SecB and ligands, based on studies of peptide binding (Randall 1992), proposes that the tetramer of SecB has multiple sites that bind flexible, positively charged stretches of polypeptide approximately 15 aminoacyl residues in length. Saturation of these binding sites induces a conformational change in SecB, and a hydrophobic site is exposed, which is proposed to interact with the hydrophobic regions of the nonnative polypeptide ligand. However, an effort to define the characteristics of SecB-binding sites on the physiological ligand MBP did not show a consensus sequence involved in binding. It did, however, suggest that the binding frame within a physiological ligand for SecB comprises contiguous sites positioned around the center of the polypeptide chain (Topping and Randall 1994).
Why do some precursor proteins have an absolute requirement for SecB, whereas others do not? Competition experiments show that SecB binds selectively to nonnative proteins with high affinity and with specificity for stretches of a few aromatic and basic residues (Hardy and Randall 1991; Knoblauch et al 1999). The occurrence and affinity of binding regions are similar in SecB-dependent, SecB-independent, and cytosolic proteins. Furthermore, SecB lacks specificity toward signal sequences. Therefore, SecB cannot differentiate between secretory and nonsecretory proteins via its binding specificity. It is suggested that selectivity in binding is due to a kinetic partitioning of polypeptides between folding and association with SecB. By exposing a hydrophobic patch on SecB, Diamond and Randall (1997) have detected a complex between SecB and MBP under conditions in which rapid folding of the polypeptide otherwise precludes formation of a kinetically stable complex. These studies explain how SecB specifically recognizes and binds to its target. However, questions about why and how SecB-independent secretory proteins are not affected by SecB remain unanswered. Several investigators have tried to determine what specifies SecB dependence using fusion proteins of various portions of SecB-dependent and SecB-independent proteins. The amino-terminal 74 aminoacyl residues are responsible for the SecB dependence of MBP, whereas the 60 residues that are C terminal to residue 320 of the mature protein are responsible for the SecB dependence of LamB (Gannon and Kumamoto 1989; Altman et al 1990). For OmpF, the first 11 amino acids of the mature protein seem to influence SecB dependence (Watanabe et al 1988). Yet, it is not clearly understood why these various portions result in SecB dependence of these different proteins.
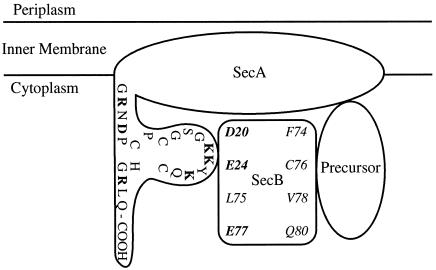
The phenotype of secB null mutants suggests that stabilization of a loosely folded conformation is not the only role of SecB in export (Kumamoto 1991). In the absence of SecB, 75% of the cytoplasmic pre-MBP remains in an unfolded, export-competent form (Kumamoto and Gannon 1988). If the sole function of SecB is to prevent folding, this population of pre-MBP should still be exported even in secB null strains. However, a significant defect is observed in the efficiency with which this pre-MBP is exported. These data suggest that, in addition to its role in maintaining the precursor in an unfolded form, SecB performs a second function that enhances the rate of protein export. SecB might simultaneously block folding and accelerate protein export by stabilizing the particular conformation of the precursor that is most favorable for productive interaction with other components of the export apparatus. Binding to the precursor protein, SecB prevents alternate processes, such as folding and aggregation, and transfers the precursor to the membrane-bound translocon in an appropriate state for rapid and productive translocation. Indeed, using the purified components proOmpA, SecB, and SecA, Hartl et al (1990) have established the sequence of reactions involved in the productive, high-affinity binding of the precursor to inner membrane vesicles. In addition to its role in stabilizing preproteins in solution, SecB prevents membrane association at nonproductive, low-affinity sites. Furthermore, SecB itself has a direct binding affinity for SecA, although the SecA-SecB interaction is weak in solution (Hoffschulte et al 1994) but occurs with high affinity at the membrane surface and is promoted by precursor proteins (Hartl et al 1990). SecA binds the proOmpA-SecB complex (Kd = 60 nM) through direct recognition of both the SecB (Kd = 200 nM) and the signal and mature domains of the precursor protein. This contributes to the accurate targeting of the preprotein-SecB complex to the membrane-bound translocase. The SecB-binding site on SecA is located at the extreme carboxy terminus (22 aminoacyl residues) of the SecA dimer (Fekkes et al 1997), although there is a possibility that SecA has other SecB-binding sites (Fig 3). These findings raise questions regarding the mechanics of the SecA-SecB interaction. For example, does the SecB-binding site on SecA just serve to tether the 2 proteins together at the gateway of translocation sites? Or, does it also serve as a switch to trigger another interaction(s) once SecA is occupied by SecB? What is the outcome of SecA-SecB binding to precursor proteins? Does SecB just deliver the precursor proteins to SecA or does SecB induce SecA to adopt a higher-affinity conformation for the precursor proteins?
Fig 3.
Schematic representation of the sites of interaction in the SecA-SecB-precursor ternary complex. Residues of SecB responsible for SecA-binding interface with the C-terminal 22 aminoacyl residues of SecA. Residues of SecB associated with interactions with the precursor are also shown. Residues of SecB are italicized, and charged residues are in boldface
Site-directed mutagenesis has revealed 2 distinct classes of secB mutants (Kimsey et al 1995) (Fig 3). Substitutions at every second residue within the region encompassing Phe-74, Cys-76, Val-78, or Gln-80 reduce the ability of SecB to form stable complexes with pre-MBP but cause only mild defects in the rate of MBP export. The characteristics of this class of mutants suggest that a primary binding site on SecB for pre-MBP is hydrophobic and contains β-sheet secondary structure. In contrast, substitutions at Asp-20, Glu-24, Leu-75, or Glu-77 cause a severe reduction in the rate of MBP export but do not disrupt SecB–pre-MBP complex formation. These mostly acidic residues may function to modulate the opening of a preprotein binding site on the translocon, allowing both high-affinity preprotein binding and rapid dissociation of SecB-preprotein complexes at the membrane translocation site.
Biochemical approaches have also been used to identify characteristics of SecB-binding sites on precursor proteins. When SecB is in complex with a ligand, it becomes resistant to proteinase K, unlike free SecB. It has been postulated, based on the sensitivity to proteolysis experiments, that the binding of positively charged peptides changes the conformation of SecB (Randall 1992). Saturation of the binding sites induces a further conformational change that fully exposes the hydrophobic site, which is then available to bind hydrophobic regions of polypeptide ligands. Interestingly, sequence analysis of the extreme carboxy terminus of SecA suggests it is strongly electropositive, with a pI value of 9.5–10.8 (Fig 3). Therefore, it is tempting to speculate that the carboxy terminus of SecA is recognized by SecB through electrostatic interactions. Consistent with this idea, mutations at Leu-75 and Glu-77 of SecB result in loss of binding to SecA (Fekkes et al 1998). However, the specific SecA-SecB interaction is difficult to explain merely by electrostatic interactions. Rajapandi and Oliver (1994) have shown that substitution of the conserved Cys-896 and Cys-885 or Cys-887 with serine in the carboxy-terminal tail of SecA inhibits the translocation activity of SecA.
CONCLUDING REMARKS
Some precursor proteins require SecB to accomplish efficient export, whereas others do not. Several investigators have tried to understand why. The most perplexing findings are that there is no obvious rationale for SecB-dependent proteins to be SecB dependent and that SecB-binding sites are distributed evenly throughout both SecB-dependent and SecB-independent proteins. In addition, SecB can bind mature proteins even in the absence of a signal sequence. A prevailing explanation is given by the kinetic partitioning model (Hardy and Randall 1991). It states that selectivity in binding is due to a kinetic partitioning of polypeptides between folding and aggregation and association with SecB. In other words, only when the rate of folding is sufficiently slow does SecB bind to a polypeptide, leading to its translocation. This model, however, raises some questions. For example, how do cytoplasmic proteins and SecB-independent secretory proteins escape SecB binding? According to Hardy and Randall (1991) and Randall et al (1994), the rate constant for association of SecB with precursor proteins is estimated to be about 107 − 108 M−1 s−1, which will give a pseudo–first-order SecB association rate of 4 to 40 s−1, assuming the free (uncomplexed) cellular concentration of SecB is about 0.4 μM (Hardy and Randall 1991). However, the experimentally measured rate constant for association of SecB with bovine pancreas trypsin inhibitor reported by Fekkes et al (1995) is at least 109 M−1 s−1, which will give a pseudo–first-order SecB association rate of 400 s−1 or faster based on the same SecB concentration. Considering that the rate constants for cytoplasmic protein folding are in the range of 5 to 10 s−1 in vitro (Hardy and Randall 1991), it seems possible that SecB could bind efficiently to most cytoplasmic proteins. Therefore, the kinetic partitioning model is not sufficient to support a substrate selectivity role for SecB.
How can nonselective SecB binding lead to selective SecB use? There could be several answers to this question. First, cytoplasmic proteins may be preferentially recognized by other chaperones such as GroEL and DnaK. In a similar manner, SecB-independent proteins may be preferentially recognized by other transport factors such as SRP. These explanations invoke the notion that SecB itself is not selective and imply that SecB-dependent proteins become SecB dependent not because SecB preferentially binds to them but because these proteins are not readily recognized by other chaperones or SRP.
Second, SecB may acquire selectivity by recognizing its substrate in combination with other partner protein(s). The most probable partner is SecA. Even in the absence of precursor proteins, the SecB tetramer has an affinity of about 30 to 250 nM for membrane-bound SecA (Hartl et al 1990; Fekkes et al 1997). Since the concentration of the SecB tetramer in the cell is estimated to be about 1 to 13 μM (Hardy and Randall 1991; Knoblauch et al 1999), it is very likely that membrane-bound SecA is in complex with SecB even in the absence of preproteins. Our cross-linking data involving SecA and SecB (Kim and Kendall, unpublished results) also agree with this idea in that, in that range of SecB (1–13 μM tetramer), SecA-SecB cross-linking is saturated. In vivo, if most of the SecA is in complex with SecB, then precursor proteins should be recognized by both SecA and SecB simultaneously. Since SecB does not have any striking substrate specificity and because it has an ability to bind most unfolded polypeptides, it is more reasonable that SecA has a main role in choosing its substrate by binding to the signal peptide specifically (Miller et al 1998). SecB might keep the precursor protein unfolded by binding to the mature region nonspecifically. This SecA-SecB complex could promote substrate recognition not only because it endows selectivity but also because it could enhance the affinity for precursor proteins. We observed a SecB-induced conformational change of SecA. Consistent with this idea, in the presence of SecB, the SecA ATPase activity is increased (Kim and Kendall, unpublished results). These findings predict that SecB-independent proteins may be well recognized by SecA even in the absence of SecB, suggesting they have a higher affinity for SecA than SecB-dependent proteins. Perhaps, SecB-independent proteins follow a strict cotranslational translocation pathway via SRP because they have a higher affinity for Ffh than SecB. These precursor proteins in complex with SRP could be targeted to SecA with high efficiency even in the absence of SecB. Consistent with this idea, when the hydrophobicity of the signal sequence is increased to optimize Ffh binding, the precursor protein becomes SecB independent, whereas when the hydrophobicity is decreased, the precursor protein becomes SecB dependent (Kim et al 2000). Another possibility is that the SecB-independent proteins are better substrates for other chaperones or SecA itself. Bla seems to fall in this category because it has been reported that GroEL binds to Bla (Bochkareva et al 1988). Interestingly, GroEL binds SecA (Kd = 1 μM) at the cytoplasmic membrane (Bochkareva et al 1998). The specificity of GroEL for precursor proteins may also be decided in the context of SecA rather than by GroEL itself.
Acknowledgments
We thank Alexander Miller for critically reading the manuscript. Work on protein transport in the Kendall laboratory is funded by grants from the National Institute of Health (GM37639), the US Department of Agriculture (99–35304–8095), and the North Atlantic Treaty Organization (CRG 960684).
REFERENCES
- Akiyama Y, Ito K. Export of Escherichia coli alkaline phosphatase attached to an integral membrane protein, SecY. J Biol Chem. 1989;264:437–442. [PubMed] [Google Scholar]
- Altman E, Emr SD, Kumamoto CA. The presence of both the signal sequence and a region of mature LamB protein is required for the interaction of LamB with the export factor SecB. J Biol Chem. 1990;265:18 154–18 168. [PubMed] [Google Scholar]
- Bassilana M, Wickner W. Purified Escherichia coli preprotein translocase catalyzes multiple cycles of precursor protein translocation. Biochemistry. 1993;32:2626–2630. doi: 10.1021/bi00061a021. [DOI] [PubMed] [Google Scholar]
- Bernstein HD, Poritz MA, Strub K, Hoben PJ, Brenner S, Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- Blobel G, Dobberstein B. Transfer of proteins across membranes. J Cell Biol. 1975;67:835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva ES, Lissin NM, Girshovich AS. Transient association of newly synthesized unfolded proteins with the heat-shock GroEL protein. Nature. 1988;336:254–257. doi: 10.1038/336254a0. [DOI] [PubMed] [Google Scholar]
- Bochkareva ES, Solovieva ME, Girshovich AS. Targeting of GroEL to SecA on the cytoplasmic membrane of Escherichia coli. Proc Natl Acad Sci U S A. 1998;95:478–483. doi: 10.1073/pnas.95.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Brundage L, Fimmel CJ, Mizushima S, Wickner W. SecY, SecE and band 1 form the membrane-embedded domain of Escherichia coli preprotein translocase. J Biol Chem. 1992;267:4166–4170. [PubMed] [Google Scholar]
- Brundage L, Hendrick JP, Schiebel E, Driessen AJM, Wickner W. The purified E coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- Cabelli R, Chen LL, Tai PC, Oliver DB. SecA protein is required for secretory protein translocation into E coli membrane vesicles. Cell. 1988;55:683–692. doi: 10.1016/0092-8674(88)90227-9. [DOI] [PubMed] [Google Scholar]
- Chen L, Tai PC. Evidence for the involvement of ATP co-translational protein translocation. Nature. 1987;328:164–166. doi: 10.1038/328164a0. [DOI] [PubMed] [Google Scholar]
- Clemons WM Jr, Gowda K, Black SD, Zwieb C, Ramakrishnan V. Crystal structure of the conserved subdomain of human protein SRP54M at 2.1 Å resolution: evidence for the mechanism of signal peptide binding. J Mol Biol. 1999;292:697–705. doi: 10.1006/jmbi.1999.3090. [DOI] [PubMed] [Google Scholar]
- Collier DN, Bankaitis VA, Weiss JB, Bassford PJ Jr. The antifolding activity of SecB promotes the export of the E coli maltose-binding protein. Cell. 1988;53:273–283. doi: 10.1016/0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- Connolly T, Gilmore R. The signal recognition particle receptor mediates the GTP-dependent displacement of SRP from the signal sequence of the nascent polypeptide. Cell. 1989;57:599–610. doi: 10.1016/0092-8674(89)90129-3. [DOI] [PubMed] [Google Scholar]
- Crooke E, Brundage L, Rice M, Wickner W. ProOmpA spontaneously folds in a membrane assembly competent state which trigger factor stabilizes. EMBO J. 1988;7:1831–1836. doi: 10.1002/j.1460-2075.1988.tb03015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K, Lill R, Crooke E, Rice M, Moore K, Wickner W, Oliver DB. SecA protein, a peripheral protein of the Escherichia coli plasma membrane, is essential for the functional binding and translocation of proOmpA. EMBO J. 1989;8:955–959. doi: 10.1002/j.1460-2075.1989.tb03457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVrije T, Batenburg AM, Jordi W, DeKruijff B. Inhibition of PhoE translocation across Escherichia coli inner-membrane vesicles by synthetic signal peptides suggests an important role of acidic phospholipids in protein translocation. Eur J Biochem. 1989;180:385–392. doi: 10.1111/j.1432-1033.1989.tb14660.x. [DOI] [PubMed] [Google Scholar]
- Diamond DL, Randall LL. Kinetic partitioning. J Biol Chem. 1997;272:28994–28998. doi: 10.1074/jbc.272.46.28994. [DOI] [PubMed] [Google Scholar]
- Douville K, Leonard M, Brundage L, Nishiyama KI, Tokuda H, Mizushima S, Wickner W. Band 1 subunit of preprotein translocase and integral membrane export factor P12 are the same protein. J Biol Chem. 1994;269:18705–18707. [PubMed] [Google Scholar]
- Douville K, Price A, Eichler J, Economou A, Wickner W. SecYEG are the stoichiometric components of preprotein translocase. J Biol Chem. 1995;270:20106–20111. doi: 10.1074/jbc.270.34.20106. [DOI] [PubMed] [Google Scholar]
- Driessen AJM. SecA, the peripheral subunit of the Escherichia coli precursor protein translocase, is functional as a dimer. Biochemistry. 1993;32:13190–13197. doi: 10.1021/bi00211a030. [DOI] [PubMed] [Google Scholar]
- Duong F, Wickner W. Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 1997;16:2756–2768. doi: 10.1093/emboj/16.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman DM, Steitz TA. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell. 1981;23:411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- Fekkes P, de Wit JG, van der Wolk JPW, Kimsey HH, Kumamoto CA, Driessen AJM. Preprotein transfer to the Escherichia coli translocase requires the cooperative binding of SecB and the signal sequence to SecA. Mol Microbiol. 1998;29:1179–1190. doi: 10.1046/j.1365-2958.1998.00997.x. [DOI] [PubMed] [Google Scholar]
- Fekkes P, den Blaauwen T, Driessen AJM. Diffusion-limited interaction between unfolded polypeptides and the Escherichia coli chaperone SecB. Biochemistry. 1995;34:10 078–10 085. doi: 10.1021/bi00031a032. [DOI] [PubMed] [Google Scholar]
- Fekkes P, van der Does C, Driessen AJM. The molecular chaperone SecB is released from the carboxy-terminus of SecA during initiation of precursor protein translocation. EMBO J. 1997;16:6105–6113. doi: 10.1093/emboj/16.20.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freymann DM, Keenan RJ, Stroud RM, Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- Gannon PM, Li P, Kumamoto CA. The mature portion of Escherichia coli maltose-binding protein (MBP) determines the dependence of MBP on SecB for export. J Bacteriol. 1989;171:813–818. doi: 10.1128/jb.171.2.813-818.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada M, Nishiyama KI, Mizushima S, Tokuda H. Reconstitution of an efficient protein translocation machinery comprising SecA and the three membrane proteins, SecY, SecE, and SecG (p12) J Biol Chem. 1994;269:23 625–23 631. [PubMed] [Google Scholar]
- Hanein D, Matlack KES, Jungnickel BJ, Plath K, Kalies KW, Miller KR, Rapoport TA, Akey CW. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- Hardy SJS, Randall LL. A kinetic partitioning model of selective binding of nonnative proteins by the bacterial chaperone SecB. Science. 1991;251:439–443. doi: 10.1126/science.1989077. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Lecker S, Schiebel E, Hendrick JP, Wickner W. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E coli plasma membrane. Cell. 1990;63:269–279. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- Hoffschulte HK, Drees B, Müller M. Identification of a soluble SecA/SecB complex by means of a subfractionated cell-free export system. J Biol Chem. 1994;269:12 833–12 839. [PubMed] [Google Scholar]
- Inouye M, Halegoua S. Secretion and membrane localization of proteins in Escherichia coli. CRC Crit Rev Biochem. 1980;7:339–371. doi: 10.3109/10409238009105465. [DOI] [PubMed] [Google Scholar]
- Izard JW, Doughty MB, Kendall DA. Physical and conformational properties of synthetic idealized signal sequences parallel their biological function. Biochemistry. 1995;34:9904–9912. doi: 10.1021/bi00031a012. [DOI] [PubMed] [Google Scholar]
- Joly JC, Leonard MR, Wickner W. Subunit dynamics in Escherichia coli preprotein translocase. Proc Natl Acad Sci U S A. 1994;91:4703–4704. doi: 10.1073/pnas.91.11.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, McKnight CJ, Gierasch LM. Biophysical studies of signal peptides: implications for signal sequence functions and the involvement of lipid in protein export. J Bioenerg Biomembr. 1990;22:213–232. doi: 10.1007/BF00763166. [DOI] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Walter P, Stroud RM. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- Kim J, Luirink J, Kendall DA. SecB dependence of an exported protein is a continuum influenced by the characteristics of the signal peptide or early mature region. J Bacteriol. 2000;182:4108–4112. doi: 10.1128/jb.182.14.4108-4112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimsey HH, Dagarag MD, Kumamoto CA. Diverse effects of mutation on the activity of the Escherichia coli export chaperone SecB. J Biol Chem. 1995;270:22831–22835. doi: 10.1074/jbc.270.39.22831. [DOI] [PubMed] [Google Scholar]
- Knoblauch NT, Rudiger S, Schonfeld HJ, Driessen AJM, Schneider-Mergener J, Bukau B. Substrate specificity of the SecB chaperone. J Biol Chem. 1999;274:34 219–34 225. doi: 10.1074/jbc.274.48.34219. [DOI] [PubMed] [Google Scholar]
- Kumamoto CA. Escherichia coli SecB protein associates with exported protein precursors in vivo. Proc Natl Acad Sci U S A. 1989;86:5320–5324. doi: 10.1073/pnas.86.14.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto CA. SecB protein: a cytosolic export factor that associates with nascent exported proteins. J Bioenerg Biomembr. 1990;22:337–351. doi: 10.1007/BF00763171. [DOI] [PubMed] [Google Scholar]
- Kumamoto CA. Molecular chaperones and protein translocation across the Escherichia coli inner membrane. Mol Microbiol. 1991;5:19–22. doi: 10.1111/j.1365-2958.1991.tb01821.x. [DOI] [PubMed] [Google Scholar]
- Kumamoto CA, Beckwith J. Mutations in a new gene, secB, cause defective protein localization in Escherichia coli. J Bacteriol. 1983;154:253–260. doi: 10.1128/jb.154.1.253-260.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto CA, Gannon PM. Effects of Escherichia coli secB mutations on pre-maltose binding protein conformation and export kinetics. J Biol Chem. 1988;263:11 554–11 558. [PubMed] [Google Scholar]
- Kumamoto CA, Nault AK. Characterization of the Escherichia coli protein-export gene secB. Gene. 1989;75:167–175. doi: 10.1016/0378-1119(89)90393-4. [DOI] [PubMed] [Google Scholar]
- Kusters R, de Vrije T, Breukink E, de Kruijff B. SecB protein stabilizes a translocation-competent state of purified prePhoE protein. J Biol Chem. 1989;264:20 827–20 830. [PubMed] [Google Scholar]
- Larsen N, Zwieb C. SRP-RNA sequence alignment and secondary structure. Nucleic Acids Res. 1991;19:209–215. doi: 10.1093/nar/19.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecker S, Lill R, Ziegelhoffer T, Georgopoulos C, Bassford PJ Jr, Kumamoto CA, Wickner W. Three pure chaperone proteins of Escherichia coli—SecB, trigger factor and GroEL—form soluble complexes with precursor proteins in vitro. EMBO J. 1989;8:2703–2709. doi: 10.1002/j.1460-2075.1989.tb08411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:259–269. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- Luirink J, ten Hagen-Jongman CC, Van der Weijden CC, Oudega B, High S, Dobberstein B, Kusters R. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO J. 1994;13:2289–2296. doi: 10.1002/j.1460-2075.1994.tb06511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer TH, Menetret JF, Breitling R, Miller KR, Akey CW, Rapoport TA. The bacterial SecY/E translocation complex forms channel-like structures similar to those of the eukaryotic Sec61p complex. J Mol Biol. 1999;285:1789–1800. doi: 10.1006/jmbi.1998.2413. [DOI] [PubMed] [Google Scholar]
- Miller A, Wang L, Kendall DA. Synthetic signal peptides specifically recognize SecA and stimulate ATPase activity in the absence of preprotein. J Biol Chem. 1998;273:11 409–11 412. doi: 10.1074/jbc.273.19.11409. [DOI] [PubMed] [Google Scholar]
- Miller JD, Bernstein HD, Walter P. Interaction of E coli Ffh/4.5S ribonucleoprotein and FtsY mimics that of mammalian signal recognition particle and its receptor. Nature. 1994;367:657–368. doi: 10.1038/367657a0. [DOI] [PubMed] [Google Scholar]
- Montoya G, Svensonn C, Luirink J, Sinning I. Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature. 1997;385:365–368. doi: 10.1038/385365a0. [DOI] [PubMed] [Google Scholar]
- Nishiyama KI, Hanada M, Tokuda H. Disruption of the gene encoding p12 (SecG) reveals the direct involvement and important function of SecG in the protein translocation of Escherichia coli at low temperature. EMBO J. 1994;13:3272–3477. doi: 10.1002/j.1460-2075.1994.tb06628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama KI, Mizushima S, Tokuda H. A novel membrane protein involved in protein translocation across the cytoplasmic membrane of Escherichia coli. EMBO J. 1993;12:3409–3415. doi: 10.1002/j.1460-2075.1993.tb06015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips GJ, Silhavy TJ. The E coli ffh gene is necessary for viability and efficient protein export. Nature. 1992;359:744–746. doi: 10.1038/359744a0. [DOI] [PubMed] [Google Scholar]
- Poritz MA, Bernstein HD, Strub K, Zopf D, Wilhelm H, Walter P. An E coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990;250:1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- Poritz MA, Strub K, Walter P. Human SRP RNA and E coli 4.5S RNA contain a highly homologous domain. Cell. 1988;55:4–6. doi: 10.1016/0092-8674(88)90003-7. [DOI] [PubMed] [Google Scholar]
- Rajapandi T, Oliver D. Carboxy-terminal region of Escherichia coli SecA ATPase is important to promote its protein translocation activity in vivo. Biochem Biophys Res Commun. 1994;200:1477–1483. doi: 10.1006/bbrc.1994.1617. [DOI] [PubMed] [Google Scholar]
- Randall LL. Peptide binding by chaperone SecB: implications for recognition of nonnative structure. Science. 1992;257:241–245. doi: 10.1126/science.1631545. [DOI] [PubMed] [Google Scholar]
- Randall LL, Hardy SJS. Correlation of competence for export with lack of tertiary structure of the mature species: a study in vivo of maltose-binding protein in E coli. Cell. 1986;46:921–928. doi: 10.1016/0092-8674(86)90074-7. [DOI] [PubMed] [Google Scholar]
- Randall LL, Topping TB, Hardy SJS. No specific recognition of leader peptide by SecB, a chaperone involved in protein export. Science. 1990;248:860–863. doi: 10.1126/science.2188362. [DOI] [PubMed] [Google Scholar]
- Randall LL, Topping TB, and Hardy SJS 1994 The basis of recognition of nonnative structure by the chaperone SecB. In: The Biology of Heat Shock Proteins and Molecular Chaperones, ed Morimoto RI, Tissieres A, Georgopoulos C. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 285–298. [Google Scholar]
- Ribes V, Römisch K, Giner A, Dobberstein B, Tollervey D. E coli 4.5S RNA is part of a ribonucleoprotein particle that has properties related to signal recognition particle. Cell. 1990;63:591–600. doi: 10.1016/0092-8674(90)90454-m. [DOI] [PubMed] [Google Scholar]
- Römisch K, Webb J, Lingelbach K, Gausepohl H, Dobberstein B. GTPase domain of the 54-kD subunit of the mammalian signal recognition particle is required for protein translocation but not for signal sequence binding. J Cell Biol. 1990;111:1793–1802. doi: 10.1083/jcb.111.5.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz PJ, Riggs PD, Jacq A, Fath MJ, Beckwith J. The secE gene encodes an integral membrane protein required for protein export in Escherichia coli. Genes Dev. 1989;3:1035–1044. doi: 10.1101/gad.3.7.1035. [DOI] [PubMed] [Google Scholar]
- Simon SM, Blobel G. A protein-conducting channel in the endoplasmic reticulum. Cell. 1991;65:371–380. doi: 10.1016/0092-8674(91)90455-8. [DOI] [PubMed] [Google Scholar]
- Simon SM, Blobel G. Signal peptides open protein-conducting channels in. E coli. Cell. 1992;69:677–684. doi: 10.1016/0092-8674(92)90231-z. [DOI] [PubMed] [Google Scholar]
- Topping TB, Randall LL. Determination of the binding frame within a physiological ligand for the chaperone SecB. Protein Sci. 1994;3:730–736. doi: 10.1002/pro.5560030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent QA, Kendall DA, High S, Kusters R, Oudega B, Luirink J. Early events in preprotein recognition in E coli: interaction of SRP and trigger factor with nascent polypeptides. EMBO J. 1995;14:5494–5505. doi: 10.1002/j.1460-2075.1995.tb00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent QA, Scotti PA, High S, et al. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 1998;17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G, Blomberg C. Trans-membrane translocation of proteins. Eur J Biochem. 1979;97:175–181. doi: 10.1111/j.1432-1033.1979.tb13100.x. [DOI] [PubMed] [Google Scholar]
- Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Blobel G. Cytosolic factor purified from Escherichia coli is necessary and sufficient for the export of a preprotein and is a homotetramer of SecB. Proc Natl Acad Sci U S A. 1989;86:2728–2732. doi: 10.1073/pnas.86.8.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Hayashi S, Wu HC. Synthesis and export of the outer membrane lipoprotein in Escherichia coli mutants defective in generalized protein export. J Bacteriol. 1988;170:4001–4007. doi: 10.1128/jb.170.9.4001-4007.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss JB, Ray PH, Bassford PJ Jr. Purified SecB protein of Escherichia coli retards folding and promotes membrane translocation of the maltose-binding protein in vitro. Proc Natl Acad Sci U S A. 1988;85:8978–8982. doi: 10.1073/pnas.85.23.8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Gierasch LM. Domain interactions in E coli SRP: stabilization of M domain by RNA is required for effective signal sequence modulation of NG domain. Mol Cell. 1997;1:79–87. doi: 10.1016/s1097-2765(00)80009-x. [DOI] [PubMed] [Google Scholar]
- Zorf D, Bernstein HD, Walter P. The 54-kD protein of signal recognition particle contains a methionine-rich RNA binding domain. J Cell Biol. 1993;120:1113–1121. doi: 10.1083/jcb.111.5.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]