INTRODUCTION
A number of conditions that are damaging to the proteins in cells can trigger the increased expression of several highly conserved proteins that are referred to as heat shock or heat stress proteins (Hsps). Correspondingly, these proteins protect essential cellular processes that are dependent on the state of cellular proteins. Most, if not all, of these proteins are also expressed in the absence of stress and have been shown to perform critical functions in the nonstressed cell. The functions of these proteins include acting as molecular chaperones, which can prevent untoward interactions between unfolded proteins that may take place in several major cellular processes, such as protein transport, translation, and protein folding. Additionally, they can act to inhibit the irreversible aggregation of denatured proteins and, in some instances, function in the refolding of denatured proteins (Gething and Sambrook 1992; Becker and Craig 1994). It is well understood that the major Hsp families are evolutionarily conserved. The Hsps of mammalian cells can be classified into several families of sequence-related proteins, which are designated by their molecular sizes (in kilodaltons). In order of molecular size, the major families are as follows: the Hsp25/Hsp27/Hsp28 family (the small stress proteins), the Hsp40 family, the Hsp60 family, the Hsp70 family, the Hsp90 family, and the Hsp110/SSE family. The Hsp110 family does not include Hsp104, which is clearly distinguishable from Hsp110 family members and appears to be restricted to lower eukaryotes.
The stress proteins of eukaryotes are present in all the major cellular compartments. The Hsps are principally found in the cytoplasm, nucleus, and mitochondria. The chaperones in the endoplasmic reticulum (ER) are functionally and structurally related to the Hsps but are induced by stresses that disrupt the function of the ER and cause unfolded protein to accumulate in the lumen (eg, glucose starvation, oxygen deprivation, or calcium ionophores) rather than by heat shock. Because they were first identified as being induced by glucose starvation, these stress proteins are known as glucose-regulated proteins (Grps). The principal Grps of mammals are Grp78, Grp94, and Grp170. These stress proteins have been shown to bind to newly synthesized, unfolded, and/or incompletely glycosylated proteins in the lumen of the ER and to be involved in peptide import. Grp78 is homologous to cytoplasmic Hsp70, whereas Grp94 is homologous to Hsp90. This suggested to us that Grp170 might be homologous to Hsp110. Later we will show that this is partially correct.
Curiously, despite their relative abundance and frequent appearance in numerous studies over many years, Hsp110 and Grp170 remained almost entirely ignored into the 1990s. These large stress proteins had not been cloned as of a few years ago, whereas 200 to 300 complete and partial Hsp70 sequences existed in the databases. A large number of sequences for members of other major Hsp families were also available. In the early 1990s, a number of sequences coding for large stress proteins were cloned. These proteins were distant relatives of the Hsp70 family, ie, being much more diverged from the Hsp70s than any previously identified sequence in the database. Subsequently, by cloning and sequencing complementary DNA (cDNA) coding for the Hsp110 and Grp170 of mammals, we identified 2 new groups of stress proteins related to the Hsp70 family (Lee-Yoon et al 1995; Chen et al 1996). Although Hsp110 and Grp170 sequences are related to Hsp70 family members, it is important to recognize that proteins in the Hsp110 or Grp170 families are not Hsp70 family members any more than Hsp70s are Grp170 or Hsp110 family members. Rather they comprise a group that might best be described as the Hsp70-Hsp110-Grp170 superfamily. We review herein aspects of the structural, functional, and evolutionary relations between these large and distant relatives of the Hsp70 family. Another review of this area has been recently published (Gething 1997).
Hsp110/SSE SUBFAMILY OF THE Hsp70/DnaK SUPERFAMILY
Considering the similarities between Hsp110, Grp170, and members of the Hsp70 family (see below), it is surprising that the relations of these large stress proteins with the Hsp70 family had not been identified much earlier. Hsp70 cDNAs or antibodies have been frequently used for identification of new members of this family, and this approach failed to detect Hsp110 and Grp170 sequences. Although numerous Hsp70 family antibodies often cross-react with other Hsp70 family members, they do not (to date) cross react with Hsp110 or Grp170. Conversely, antibodies now available for Hsp110 or Grp170 do not cross-react with Hsp70s or with one another.
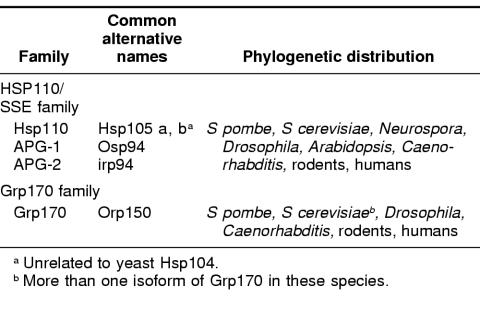
The Hsp70 family has been considered to be one of the most highly conserved gene families in biology (Boorstein et al 1994), with all members being of approximately the same size and having limited sequence divergence. However, several reports (Foltz et al 1993; Fathallah et al 1993; Mukai et al 1993; Lee-Yoon et al 1995) described large members of the Hsp70 family that were far more diverged in sequence than any of the previously described Hsp70 proteins. The fact that these large and diverged relatives of the Hsp70 family were themselves interrelated was not apparent until the cloning mammalian Hsp110, when it became evident that Hsp110 and the other above sequences comprised a statistically significant, distinct Hsp70 subfamily (Lee-Yoon et al 1995). At the time of this writing, several more Hsp110-related sequences have been reported (Lee-Yoon et al 1995; Morozov et al 1995; Yasuda et al 1995; Storozhenko et al 1996; Mukai et al 1993; Kaneko et al 1997a, 1997c; Kojima et al 1996). The Genbank database now includes Hsp110 proteins from hamster, mouse, humans, Arabidopsis, Drospophila, 2 species of sea urchin, Caenorhabitis elegans, Saccharomyces cerevisiae, and Neurospora (Table 1). No prokaryotic member has yet been identified. As would be predicted, based on these facts, a zoo blot analysis, using a probe containing the conserved regions (not found in other stress proteins) of the subfamily, shows that similar sequences are present in the genomes of every organism examined from yeast to humans (Lee-Yoon et al 1995). We have previously suggested that this stress protein family be identified as the Hsp110/SSE family, both as a result of the long-used (Hsp) term in mammalian cells and the identification of the yeast members and their designation as stress seventy (SS) (ie, the yeast stress 70 protein, SSE1, which is a member of the subfamily that we have defined).
Table 1.
Mammalian members of the Hsp110/SSE and Grp170 stress protein families
The Hsp110 protein has been noted in a vast number of studies using many different cell types (eg, Levinson et al 1980; Hightower et al 1980; Subjeck and Sciandra 1982, Subjeck et al 1982a; Landry et al 1982; Welch et al 1983; Tomasovic et al 1983). After Hsp70 and Hsp90, Hsp110 is the third or fourth most abundant Hsp in most mammalian cell lines and tissues. In CHO cells, Hsp110 accounts for 0.7% of total cell protein after heat shock compared with 3.2% for Hsc70 plus Hsp70 and 1.2% for Hsp90 (Subjeck et al 1982b). Hsp110 expression in mouse EMT6 cells is comparable to that in CHO cells, whereas it is less strongly expressed in mouse embryo 10T 1/2, MCA, Rat-1, and HeLa cells (in approximate decreasing order of expression). Although its expression can vary significantly, Hsp110 remains an easily detected Hsp in mammalian cell lines. In parallel with its varied expression in cell lines, Hsp110 expression in different murine tissues also varies significantly (Lee-Yoon et al 1995). The constitutive expression of Hsp110 is lowest in heart and skeletal muscle, whereas it is strongly expressed in liver and brain (Lee-Yoon et al 1995; Yasuda et al 1995). Indeed, the expression of Hsp110 in normothermic (ie, control) brain is comparable with that of heat-shocked CHO cells (ie, making it approximately 0.7% of total brain protein mass) (Oh et al 1997). This would indicate that Hsp110 is one of the most abundant proteins in the brain. It is also readily inducible in all murine tissues examined and often very strongly inducible in human tumors and lymphoid tissues. The Hsp110 (Hsp105) genomic sequence from mouse has been isolated and characterized (Yasuda et al 1999), including about 1.2 kb of the 5′-flanking region. The transcription initiation site was located 165 bp upstream of the ATG translation initiation codon. The promoter was found to contain a TATA box, a CAAT box, an inverted CAAT box, and 2 GC boxes. Two heat shock element sequences were found as 4 nGAAn repeats at nucleotides −64 and −128. Promoter analysis defined a minimal region, which contained the 2 consensus heat shock element sequences, as active in response to heat shock and also sufficient for constitutive expression of the gene. The genetic mechanisms that determine tissue-specific levels of expression for Hsp110 have not been discovered.
MODELING Hsp110 BY HOMOLOGY TO Hsp70/DnaK
The Hsp70/DnaK family of proteins has been well studied and contains members in all eukaryotes and all eubacteria (DnaK) (Becker and Craig 1994). Much can be inferred about the structure and function of Hsp110 from a comparison of its sequence with that of Hsp70/DnaK. To construct targeted mutations of the Hsp110 cDNA, we built a model of Hsp110 based on its sequence similarity with the DnaK family of stress proteins (Oh et al 1999). The model (Fig 1B) is shown diagrammatically and is compared with a similar diagram of DnaK (Fig 1A) based on an x-ray structure determination (Zhu et al 1996). Residues 1–394 of Hsp110 (designated as adenosine triphosphatase [ATPase] in Fig 1) show 34% identity in amino acid sequence to the same region of DnaK. This region in DnaK is responsible for adenosine triphosphate (ATP) binding. From amino acid 394 to amino acid 509, Hsp110 is predicted to exhibit 7 β-strands (the β-sheet domain or B in Fig 1). This region demonstrates some sequence similarity to and structurally aligns with the corresponding region of the DnaK. It is this region of DnaK that represents its peptide-binding domain and consists of 8 major β-strands arranged as a β-sandwich. The following 98 amino acids of Hsp110 are composed of a number of negatively charged residues that computer analysis fails to predict as having any obvious secondary structure. This region is, therefore, referred to as the loop domain (residues 510–608 or domain L). Finally, distal to the loop domain the C-terminal residues of Hsp110 are predicted to form a series of α-helices (residues 608–858 or domain H). Although there is little sequence identity in this region between Hsp110 and DnaK (or Hsc70), DnaK shows 3 large helices that make up a helix-turn-helix structure. Figure 1B correspondingly arranges the helices of Hsp110. It is in this region that the Hsp110 family members exhibit a high degree of sequence homology among themselves, thereby providing features diagnostic for this family (Lee-Yoon et al 1995). A small degree of sequence similarity can be found between Hsp110 and DnaK families in certain of these helices, specifically helix B and helix C. The first motif we designate as “Magic” (LEKERNDAKNAVEECVY), which begins at residue 626 in Hsp110 CHO and ends at residue 644. The second motif, “TedWlyee” (TEDWLYEEGEDQAKQAY), begins with residue 673 and ends at 689. Versions of these motifs also appear in many non–stress proteins, including parasite surface antigens, viral polyproteins, muscular and cytoskeletal proteins, and a large number of proteins with unknown function. Since many of the proteins containing these motifs specifically bind to other proteins, it is tempting to suggest that they are involved in substrate or cochaperone recognition and binding to Hsp110/SSE proteins. In DnaK, the β-sandwich serves as the peptide-binding site, whereas the helix-turn-helix structure (lid), poised above the β-sandwich, is thought to regulate entry and/or exit of the peptide substrate (Zhu et al 1996). The conserved expanded version of this lid in the Hsp110 family may represent elaboration on the functional properties of the homologous DnaK structure.
Fig 1.
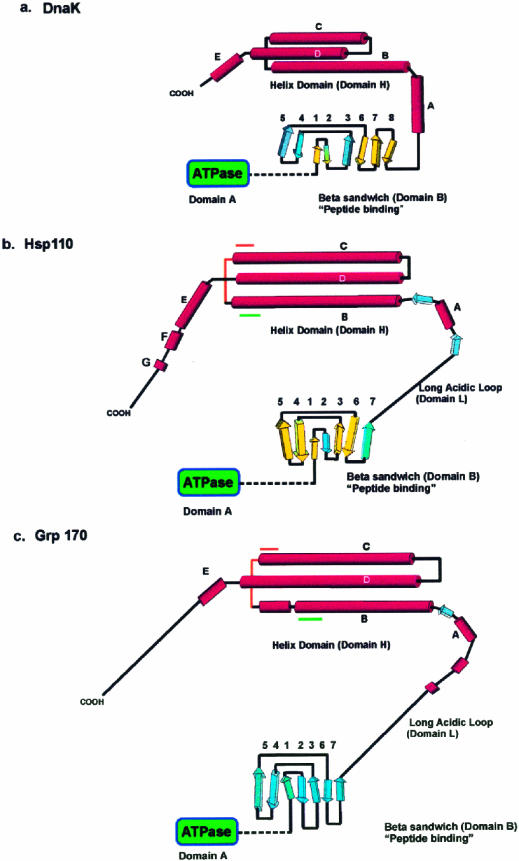
Models for the fold of Hsp110 and Grp170 based on the structure of DnaK (modified from Oh et al 1999). (A) The secondary structure and fold of the DnaK peptide-binding domains based on the x-ray crystallographic structure reported by Zhu et al (1996). The predicted secondary structure and proposed fold for (B) Hsp110 and (C) Grp170, beginning at residue 390, are diagrammatically represented. The domains (A, B, L, and H), as defined in the text, are indicated on the diagram. The β-strands in DnaK and Hsp110 β-domains, sharing both structural (PHD) and significant sequence similarity by Matchbox alignment (Depiereux et al 1997), appear in yellow. Although there is sequence similarity between the β-domains of Hsp110 and Grp170, there is not enough structural and sequence similarity to allow us to identify homologous strands. The predicted helical segments in Hsp110 and Grp170 are shown arranged in a helix-turn-helix structure, as suggested by the helical domain in DnaK. The positions of the conserved sequence motifs (Magic and TedWylee) are indicated by green and orange lines
Lastly, all Hsp110s possess a C-terminal domain (DLD, DVD, etc) that is highly similar to the EEVD exhibited by Hsp70s. Curiously, this C-terminal motif is also observed at the C-terminus of many Hsp90 family proteins.
FUNCTIONAL PROPERTIES OF Hsp110
Between the time Hsp110 was first recognized as a Hsp in the early 1980s and its cloning (Lee-Yoon et al 1995) in 1995, very little had been learned about its functional properties. However, one very early study using an antibody prepared against Hsp110 suggested that it preferentially localized in nuclei and nucleoli of tissue culture cells, especially in proliferating cells or after heat shock (Subjeck et al 1983). However, other antibodies prepared against Hsp110 suggest that it is primarily cytoplasmic and becomes nuclear after heating (Hatayama et al 1994; Lee-Yoon et al 1995) and does not localize in nucleoli or does so only in the periphery. Although a general nuclear localization after heat shock or on stimulation of proliferation seems to be on solid ground, more detailed localization studies, including preheating and postheating studies using different antibodies, are needed to verify a more specific localization.
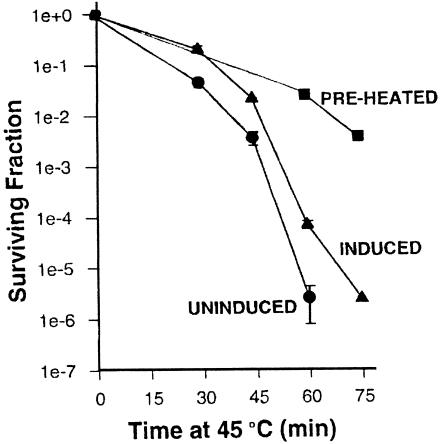
Although other Hsps were intensely studied with respect to function and chaperoning properties, Hsp110 has lagged far behind. Within the last few years, in vivo and in vitro chaperoning functions for Hsp110 have finally been demonstrated. Recently, Oh et al (1997) have shown that Hsp110 confers thermal tolerance to cells when overexpressed in them and that it can prevent aggregation of denatured proteins in vitro. Overexpression of Hsp110 alone in Rat-1 cells provided about a 2-log better survival at 45°C for 60′ than that of control cells, whereas the fully induced heat shock response yields approximately a 3-log better survival (Fig 2). Similar results were obtained with HeLa cells. The ability of Hsp110 to provide thermal tolerance in vivo is presumably mediated through its chaperoning activity detected in vitro. Oh et al (1997) conducted a series of peptide-binding experiments on Hsp110 and Hsc70 in which the ability of these chaperones to bind (hold) and fold luciferase, under various conditions, was compared. The following conclusions emerge from these in vitro binding experiments: (1) Hsp110 is significantly more efficient at stabilizing heat-denatured luciferase than is Hsc70; (2) Hsp110 cannot by itself refold luciferase; and (3) luciferase, bound to Hsp110, can refold in a reticulocyte lysate. Similar results have been obtained by Brodsky et al (1999), who have shown that yeast Hsp110 (Sse1p) prevents thermal aggregation of luciferase and holds it in a folding competent state. In this case, yeast cytosol containing an ATP-generating system and an active Hsp70 are required for refolding. These studies suggest that, although Hsp110 holds luciferase in a folding competent state, other chaperones (specifically Hsp70) are required for refolding.
Fig 2.
Overexpression of Hsp110 in cultured mammalian cells increases thermal tolerance. Hamster Hsp110 was placed under the control of a tet-off inducible promoter. When Hsp110 was expressed at heat shock levels (induced), partial resistance to 45°C heat shocks of varying duration was observed relative to uninduced control cells. Full thermotolerance is induced by preheating. Modified from Oh et al (1997)
Recent studies have shown that in addition to their peptide-binding properties, Hsp110 and Hsp70 preferentially bind AU-rich regions of RNA in vitro. It was proposed that Hsp110 and Hsp70 have in vivo RNA chaperoning properties, specifically binding 3′ UTR ARE (untranslated regions containing AU-rich instability determinant elements) of messenger RNA (mRNA), perhaps regulating mRNA degradation and/or translation of lymphokine and other short-lived messages (Henics et al 1999).
STRUCTURE-FUNCTION RELATIONS FOR Hsp110
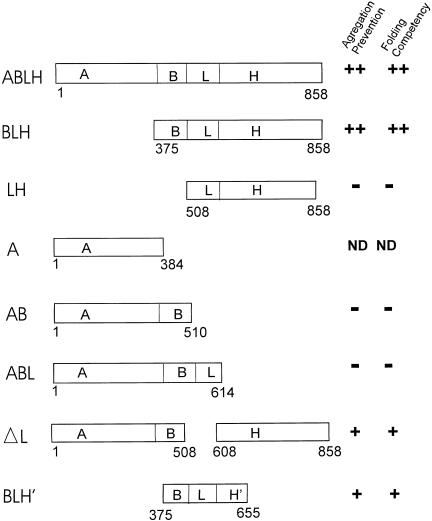
Progress has been made in identifying the structures in our model of Hsp110 (presented above), which can account for the functional properties just discussed. Based on the similarities between the predicted structural elements of Hsp110 and the actual structure of DnaK, we constructed various deletion mutants (Oh et al 1999). These mutants were constructed to include the specific domains defined above. Figure 3 shows a schematic diagram of these deletion mutants. Two assays were used to identify the functional domains of Hsp110 required for chaperoning activity: (1) the ability of the mutants to prevent luciferase aggregation induced by heat treatment (ie, holding) and (2) the ability of mutants to maintain the denatured luciferase in a folding competent state (ie, to recover activity). As shown in Figure 3, mutant BLH (ie, containing only the β-sheet peptide-binding domain [B], loop [L], and α-helical domain [H]) is able to prevent the aggregation of heat-denatured luciferase as efficiently as does wild type Hsp110. However, mutant LH (domain B removed) does not prevent luciferase aggregation. This strongly argues that domain B–containing amino acids from 375 to 507 (β-sheet domain) contains the substrate-binding domain, in agreement with sequence alignment and secondary structure predictions. This also indicates that the ATP-binding domain is dispensable for this function. However, mutant AB (containing only ATP-binding and β-sheet peptide-binding domains) was entirely nonfunctional, indicating that in addition to the B domain, more C-terminal domains of Hsp110 are essential for its ability to stabilize heat-denatured luciferase. Additionally, studies of this type demonstrated that the loop domain (L), although not essential for the holding activity of Hsp110, was a significant factor in the efficiency with which Hsp110 stabilizes luciferase in a folding competent form. The deletion of L from Hsp110 renders it more comparable in its holding efficiency to Hsc70 and makes it structurally more similar to Hsc70, which also lacks this type of loop domain. Finally, a truncated form of Hsp110 containing the first part of the lid (between amino acids 614 and 655) shows 50% activity in holding heated luciferase compared with the wild type protein, strongly suggesting that the complete lid domain is needed for complete functionality. The ability of each of these mutants to then refold the bound and heat-denatured luciferase on the addition of rabbit reticulocyte lysate directly correlated with the holding ability of each of the mutants. The studies detailed above provide the first view of structure-function relations for Hsp110.
Fig 3.
Analysis of Hsp110 deletion mutants. Schematic diagram illustrating the deletion mutants of Hsp110. The domains examined are as follows: A, ATP-binding domain; B, β-sheet (peptide-binding) domain; L, acidic loop; H, α-helical domain; H′, first 2 α-helices of H domain. ND, not determined. Modified from Oh et al (1997)
The observation that the ATP-binding domain of Hsp110 is not required for its holding functions in vitro is counterintuitive. In fact, Hsp110 does not bind ATP in vitro, despite the conservation of its ATP-binding domain, whereas the isolated ATP-binding domain (A) does (Oh et al 1999). The existence of a conserved ATP-binding domain and the ability of the isolated domain to bind ATP in vitro Hsp110 argues strongly that Hsp110 should, in fact, exhibit ATP binding in vivo. Moreover, the apparent constraint on ATP binding to Hsp110 in vitro might be an indication that Hsp110 has regulatory features that differ from the present models for Hsp70, in which the binding and hydrolysis of ATP are thought to regulate access to the peptide-binding site (Zhu et al 1996). If so, then the binding and putative hydrolysis of ATP could be expected to have implications for the unique specificity of Hsp110 holding and (possibly) folding functions and/or interactions with cochaperones. Thus, it appears that Hsp110 binds to substrate in a way similar to that of DnaK, but the details of this chaperone's substrate specificity and regulation are probably different from those of Hsp70. Perhaps Hsp110 requires a second cochaperone in addition to a DnaJ homologue, the purpose of which would be to physically open the ATP-binding domain to allow ATP binding. In fact, Hsp110's ability to function cooperatively in the folding of luciferase with Hdj-1 in the presence of Hsc70, as discussed above, indicates that an active interaction between Hsp110 and Hsc70/Hdj-1 does occur in vitro (Oh et al 1997).
NATIVE STRUCTURE OF Hsp110 AND INTERACTIONS WITH OTHER CHAPERONES
Recent efforts to examine the native interactions of Hsp110 in situ have indicated that it may not act alone in the mammalian cytosol. A study by Ishihara et al (1999) showed that Hsp110 (referred to as Hsp105 in their study) exists in large multiprotein complexes that contain multiple copies of Hsp110 and Hsc70 in FM3A cells. Similar studies (Wang et al 2000) also show that, on immunoprecipitation with anti-Hsp110 antisera, the principal coimmunoprecipitated species is Hsc70. This has been observed in several cell types and murine tissues. Although less obvious, Hsp25 (the mammalian small Hsp) can also be detected to interact with the Hsp110-Hsc70 complex. In agreement with Ishihara et al (1999), purification of native Hsp110 using ion exchange and size exclusion chromatography indicates that this Hsp110 complex exists as a 400 kDa to 700 kDa native composite. It was also found (Wang et al 2000) that these 3 chaperones spontaneously form a large molecular complex in vitro. Moreover, this complex forms in the absence of an added substrate, but substrate (luciferase) can be induced to migrate from outside the complex into the complex by application of a heat stress forming (minimally) a Hsp110-Hsc70-Hsp25-luciferase composite in vivo. The migration of heat-denatured luciferase into this fraction following heat shock in vitro argues for an active chaperoning activity of the complex in vivo. One suggestion is that Hsc70 may specifically bind to Hsp110, which is the more efficient holding chaperone, in a manner that allows transfer of substrate from Hsp110 to Hsc70, with subsequent folding in conjunction with DnaJ homologues and other chaperones. It is also possible that Hsp110 may itself interact with DnaJ proteins in folding with the addition of a yet to be identified chaperone that acts to open the Hsp110 ATP-binding domain as suggested above. Furthermore, overexpression of SSE1 in yeast has been found to suppress the thermosensitivity associated with the ydj1-151 mutation (J. Goeckeler and J. L. Brodsky, unpublished results), suggesting that Sse1p and Ydj1p interact. The role of Hsp25 in this complex is unknown, but it does not appear to assist Hsp110-Hsc70-Hdj-1 in folding.
The fact that these 3 chaperones interact may represent a general phenomenon. Plesofsky-Vig and Brambl (1998) have recently shown that the small Hsp of Neurospora crassa, Hsp30, binds to 2 cellular proteins, Hsp70 and Hsp88. Cloning and analysis of Hsp88 have shown that it represents the Hsp110 of N crassa, suggesting that the interactions described above are phylogenetically conserved.
It has recently been shown that SSE1 encoding a yeast member of the Hsp110 family is required for the function of exogenous glucocorticoid receptor and physically associates with the Hsp90 in yeast (Liu et al 1999). Although interaction of Hsp110 with Hsp90 in mammalian cells was not detectable by a coimmunoprecipitation assay, it is possible that the amount of Hsp90 and Hsp110 involved in a functional complex is so small that it goes undetected using this type of approach.
THE MAMMALIAN MEMBERS OF THE Hsp110/SSE FAMILY: AN OVERVIEW
Hsp110 has been found in all vertebrates examined but has been characterized best in mammals. Mammals (mouse, hamster, and humans) appear to have multiple isoforms. There are, however, interesting variants that have been reported (see Table 1).
Fujita and coworkers (Kaneko et al 1997a, 1997c) have cloned and identified other Hsp110-related cDNAs and have designated them APG-1 and APG-2 (A: ATP-binding domain, P: peptide-binding domain, G: germ cell derived) from both mouse and humans. Presumably, similar proteins will be found in other mammals. Although these proteins are primarily expressed in the gonads, they are also expressed at lower levels in all tissues of the mouse. APG-1 is found in highest concentration in testis, particularly in germ cells. The expression level of APG-1 increases with the maturation of germ cells, although it is not heat inducible in germ cells, suggesting the involvement of APG-1 in normal germ cell development (Kaneko et al 1997b). Conversely, APG-1 is heat inducible in somatic cells; however, the optimal heat condition for the induction is different from that of Hsp70 (stronger induction is observed by a shift from 32°C to 39°C rather than by a shift from 32°C to 42°C or the traditional shift from 37°C to 42°C). This preference of heat induction is also observed in the induction of Hsp110 (Kaneko et al 1997c). Although the biological meaning of a shift from 32°C to 39°C remains unknown, the difference in the optimal heat conditions may reflect certain differential roles between Hsp110/APG-1 and Hsp70 during heat response. APG-1 is also known as Osp94 (Kojima et al 1996), because it was cloned as a cDNA to one of the mRNAs that is up-regulated by osmotic stress in the inner medulla of the mouse kidney. In addition, Santos et al (1998) have shown that both Hsp110 and APG-1 (Osp94) are induced by hyperosmotic sodium chloride stress. Interestingly, these authors have demonstrated that heat shock and hyperosmotic stress are cross-tolerant stresses.
The Hsp110 isoform, APG-2, is highly expressed in both ovarian and testicular tissue of the mouse. It is also expressed to a lesser extent in somatic tissues. APG-2 is not heat inducible by either traditional temperature shifts or shifts that induce APG-1. Yagita et al (1999) have cloned a protein from rat that is >90% identical to APG-2, which is constitutively expressed in the brain (localized in the hippocampus) and is clearly enhanced 4–24 hours after a transient ischemia event. They have designated this protein as irp94. A putative 77-kDa protein, HSP70RY, whose cDNA was first isolated from transformed lymphocytes, was one of the first Hsp110/SSE sequences to appear in the Genbank (Fathallah et al 1993). This cDNA represented the APG-2 coding sequence truncated at the 3′ end. However, polymerase chain reaction–based cloning failed to isolate the genomic sequence corresponding to HSP70RY cDNA (Nonoguchi et al 1999). Whether HSP70RY is derived from the particular transformed lymphocytes or is an artificial product that results from cloning and/or sequencing anomalies is currently unknown.
Hsp110 is also denoted as Hsp105 (Hatayama et al 1994; Honda et al 1989). In mouse FM3A cells, 2 alternative forms, 105 α and 105 β, are observed, with 105 β having 43 fewer amino acids than 105 α. It has been proposed that the smaller version is a result of alternative splicing (Yasuda et al 1995). The smaller version is only observed by a continuous heat shock at 42°C, whereas the larger form (murine Hsp110) is preferentially induced after recovery from heat shock at 42°C or higher. Interestingly, the amino acids deleted in the smaller version are in the long acidic domain of full-length Hsp110. The differential roles played by these 2 versions of Hsp110 remain unknown.
Of significant interest is the fact that Hsp110 can also be induced by the human papilloma virus oncoprotein, E7, which is a viral transcription factor (Morozov et al 1995). The significance of this observation is not yet known. That this induction requires the presence of the E7 conserved region 2, which is essential for the binding of E7 to retinoblastoma family proteins, suggests that Hsp110 induction may be coordinated with the cell cycle.
The fact that a multiplicity of genes exists for each family (Hsp70 and Hsp110) further argues for important differences in the general functions of each. Indeed, a different requirement for Hsp110 and Hsc70 in cells is suggested by the fact that they are differentially expressed in different tissues and different regions of the same tissue (eg, brain; Hylander et al 2000; Xue et al 1998). Specifically, mammalian cerebellum expresses little Hsp110, whereas expression in other brain regions is highly abundant (Hylander et al 2000). Curiously, the cerebellum is specifically sensitive to heat stoke and alcohol-associated toxic effects (Manto 1996 ; Albukrek et al 1997).
THE NONMAMMALIAN MEMBERS OF THE Hsp110 SUBFAMILY
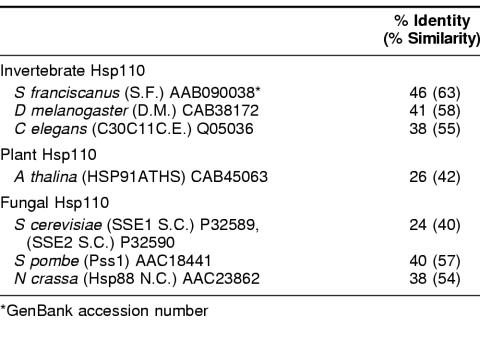
Table 2 shows the percentage of identity and similarity between Hsp110 family members from a diverse set of nonmammalian eukaryotes: 3 invertebrates, a vascular plant, baker's yeast, and Neurospora. Considerably less sequence diversity exists in a similar phylogenetic sampling of Hsp70/DnaK family sequences, which generally show 45% identity or better (Boorstein et al 1994). The yeast members of the Hsp110 group are well studied. SSE1 and SSE2 sequences were among the first Hsp110/SSE sequences to appear in the databases (Mukai et al 1993). These proteins show only about 25% identity to CHO Hsp110 and have a calculated size of 77.4 kDa, but their sequence organization is similar to that of other family members (Lee-Yoon et al 1995). Sse1p is primarily an inducible protein, whereas Sse2p is primarily constitutive. Constitutive mRNA levels for SSE2 are higher than for SSE1. Induction of SSE2 mRNA results from a shift from 23°C to 37°C.
Table 2.
Percentage of identity and similarity between CHO Hsp110 and selected nonmammalian Hsp110s
A loss of function mutation in the SSE1 gene that causes slower growth at all temperatures (20°C to 27°C) has been characterized (Mukai et al 1993), whereas mutations disrupting SSE2 have no effect on growth (However, overexpression of Sse2p from a multicopy plasmid can suppress the slow growth effect of a defective SSE1 gene) (Shirayama et al 1993). Overexpression of Sse1p (also known as MSI3) can suppress temperature-sensitive defects in the Ras signaling pathway of S cerevisiae. Specifically, overexpression of Sse1p suppresses temperature-sensitive mutations of the Ira1 gene, which is involved in activating guanosine 5′-triphosphatase or mutants in the gene Bcy1, which encodes the regulatory subunit of the cyclic adenosine 3′:5′-monophosphate–dependent protein kinase in yeast (Tanaka et al 1990; Tortora et al 1984). Whether this suppression of phenotype is mediated by specific interactions with the proteins encoded by the mutant genes (Ira1, Bcy) or is a result of general chaperoning by Sse1p is not known. However, a recent report from Liu et al (1999) shows that Sse1p is found in the Hsp90 complex and acts as a specific cochaperone. Since many kinases involved in signaling pathways require the function of the Hsp90 chaperoning complex for their activity, the suppression of Ira1 and Bcy mutant phenotypes by overexpression of SSE1 may be related to improved chaperoning of these proteins by the complex. The Hsp110/SSE member in S pombe has recently been cloned and sequenced (Chung et al 1998) and is designated as Pss1. The pss1+ gene product of the fission yeast S pombe has sequence homology to the (S cerevisiae) Hsp Sse1p/Msi3p (43% identity). It functionally complements the growth defect of a mutant ras gene, a synthetic lethal mutant of ras1+, which shows severe retardation of growth and a cell aggregation phenotype. Disruption of the pss1+ gene confers a temperature-sensitive growth phenotype but also causes an increase in cell survival thermotolerance in S pombe. Liu et al (1999) have shown that Sse1p is necessary for the proper folding of heat shock factor in S cerevisiae and that the entire compliment of Hsps is induced by disruption of SSE1 function. Similarly, the disruption of pss1+ may induce thermal survival in S pombe through induction of a heat shock response. The transcript level of pss1+ is moderate during steady-state growth at 25°C and increases a fewfold on shifting to 42°C. Transcription of pss1+ also increases in nitrogen-starvation conditions. These observations suggest that Pss1 provides the Hsp110 function for S pombe.
Hsp88 of Neurospora was isolated as 1 of 2 proteins that specifically bound to Neurospora Hsp30 (described above, Plesofsky-Vig and Brambl 1990). Its cDNA was sequenced, and its predicted amino acid sequence has been determined. Although the protein is only 88 kDa, its sequence organization is typical of the Hsp110/SSE family. The authors have identified motifs characteristic of an ATPase domain at the N-terminus of the protein, a putative β-strand region, and C-terminal motifs characteristic of Hsp110.
Hsp110/SSE FAMILY MEMBERS FROM INVERTEBRATES
There are currently 3 invertebrate phyla represented in the Hsp110/SSE family sequences from invertebrates in the Genbank database. These are from Caenorabditis (Aschelminthes), Drosophila (Arthropoda), and 2 sea urchin species (Echinodermata). The Drosophila and Caenorabditis members are represented by cloned sequences that have not been well studied, whereas the sea urchin members (especially the proposed sea urchin egg receptor for sperm) have been studied in some detail.
A 70-kDa peptide isolated from protease-digested, dejellied S purpuratus eggs was found to bind to sperm and inhibit fertilization (Foltz and Lennarz 1990). A cDNA coding for a protein that contains this fragment was isolated by screening an oocyte-ovary cDNA library. Other overlapping clones were identified and assembled into a putative full-length cDNA. The N-terminus consisted of a putative signal peptide, whereas the two-thirds of this full cDNA distal to the signal sequence predicted a polypeptide that has a high degree of identity to Hsp110. The C-terminal third contained a putative trans-membrane domain and a proposed C-terminal cytoplasmic domain. Mauk et al (1997) subsequently cloned a cDNA coding for a 97-kDa protein from S franciscanus ovaries that is 96% identical to the N-terminal half of the putative receptor for sperm (in the region between the signal peptide and the trans-membrane domain). This protein shows 45% identity to CHO Hsp110. Mauk et al have designated this predicted protein to be the Hsp110 of S franciscanus. These authors have not been able to detect any other Hsp110 family-related sequences in S franciscanus cDNAs. In the light of the findings of Mauk et al, Just and Lennarz (1997) re-evaluated the egg receptor sequence. They discovered cloning artifacts that resulted in their eliminating both the N-terminal signal peptide and the trans-membrane domain originally described. The corrected sequence is 889 amino acids in length and is 48% identical to CHO Hsp110. Giusti et al (1997) re-examined the surface localization of the receptor. They used several well-characterized antibodies against the putative receptor protein, against the 70-kDa fragment of that protein, and also against synthetic peptides based on the corrected sequence described above. By using these antibodies in confocal immunofluorescence microscopy and differential extraction protocols, they were able to show that at least a portion of the putative receptor protein lies on the surface of the sea urchin egg. More recent work by Hirohashi and Lennarz (1998a, 1998b) has provided evidence that the sperm receptor lies within the glycoprotein-rich viteline layer on the surface of the egg and confirmed that it contains, within its N-terminal half, specific binding sites for sperm. Stears and Lennarz (1997) identified the residues in the sperm receptor that are responsible for binding spermatozoa to the egg. This region corresponds to the first 2 or 3 β-strands in the peptide-binding domain of Hsp110.
The Hsp110/SSE subfamily members from Drosophila and Caenorhabditis should be of great interest for further study, since these invertebrates have long served as model multicellular organisms. Their genetics, development, and molecular biology are relatively well understood.
Grp170: A PUTATIVE COUNTERPART TO Hsp110 IN THE ER
Three major members of the Grp family are known: Grp78, Grp94, and Grp170. All of these are retained ER proteins (Lin et al 1993). Grp170 has been the least studied of this group (Olden et al 1979; Sciandra and Subjeck 1983; Shen et al 1987). Grps have been long characterized as inducible by a variety of conditions, such as glucose starvation, calcium ionophores, low pH, a variety of reducing conditions, and other stresses that disrupt the function of the ER (see references in Lin et al 1993). Possibly, the most physiologically significant condition that induces the Grp set is exposure to anoxia, possibly creating a reducing condition within the cell as a direct result of the absence of oxygen (Sciandra et al 1984).
As with Hsp110, several large Grp170 cDNAs have been recently cloned and analyzed. Some of the mammalian Grp170 sequences have also been referred to as orp150. We will unify terms here and use the original and broadly recognized Grp nomenclature.
Sequence analysis of Grp170 has suggested that it, like Hsp110, is a highly diverged relative of the Hsp70 family. However, although they have a somewhat greater degree of sequence similarity to the Hsp110/SSE family sequences than to members of the Hsp70 family, the Grp170s are essentially as diverged from the Hsp110 family of proteins as they are from the Hsp70s (Chen et al 1996; Craven et al 1997).
When members of the 110/SSE group, the Hsp70/DnaK group, and the Grp170 group are aligned by CLUSTAL W (Higgins and Sharp 1988), bootstrapped with SEQEBOOT and a phylogenetic tree is constructed using PHYLIP (Felsenstein 1989), the 3 families of proteins form an unrooted tree in which most bootstrapped tree branches are in agreement (Fig 4).
Fig 4.
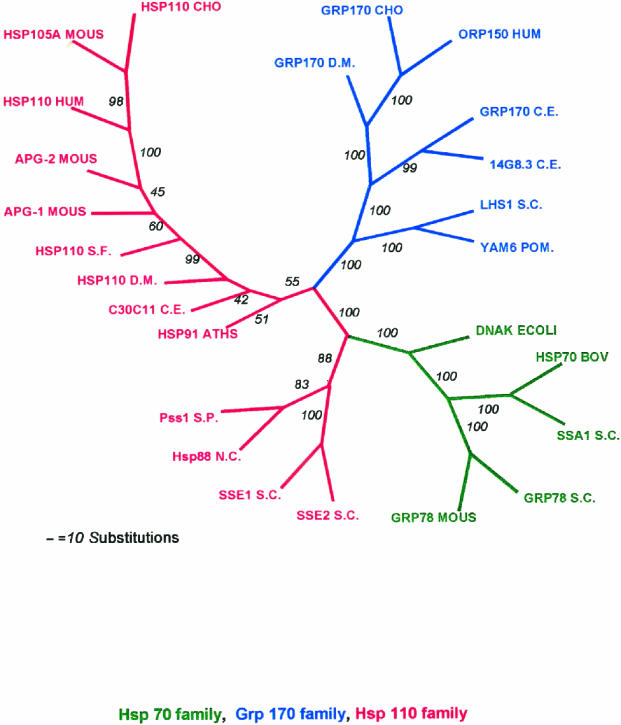
Phylogenetic tree for Hsp70 superfamily. Eleven members of the Hsp110/SSE family and 9 members of the Grp170 family were aligned with 5 members of the Hsp70/DnaK family using the CLUSTAL W program, and the alignments were analyzed by programs in PHYLIP (Phylogeny Inference Package) version 3.57c. The alignments were bootstrapped with SEQBOOT to generate 100 sets of alignment data. The data sets were used to generate 100 phylogenetic trees by use of the POTOPARS (maximum parsimony) program. A consensus tree was determined by use of the program CONSENSE. The tree is displayed as unrooted, with the italicized numbers indicating the number of trees in which the consensus bifurcation distal to the number shown was represented in the total set of 100 trees. Abbreviations and sequences used are the same as in Tables 2 and 3, except the following additional sequences were used in the tree: HSP110CHO (Q60446) Chinese hamster, APG-1 MOUS (D49482) mouse, APG-2 MOUS (Q61316) mouse, YAM6 S. POM (Q10061) S pombe, 14G8.3 C.E. (CAA91809) C elegans, SSA1 S.C. (6319314), S cerevisiae Grp78S.C.; KAR2 (6322426) S cerevisiae, HSP70BOV (P34933) bovine
The Hsp110s of the yeasts and Neurospora appear to be more closely allied with the Hsp70/DnaK family than are the Grp170s and the Hsp110s of higher eukaryotes. However, as discussed above, the smaller Hsp110 yeast and Neurospora sequences still possess the identifying structural properties of the Hsp110s and are, therefore, categorized as such. Both Grp170 and Hsp110 members appear to be present in all eukaryotes examined to date. The presence of large Hsp70-like stress proteins in all major eukaryotic taxa suggests their single origin in primitive eukaryotes. Although convergent evolution could explain the origin of these proteins, their structural, functional, and regulatory similarities to the Hsp70/DnaK family proteins suggest a common ancestor. However, it is also difficult to rule out the possibility of lateral gene transfers as a mode of acquisition for these genes (Doolittle 1999).
Grp170 DOMAIN STRUCTURE: RELATIONSHIP TO Hsp110
Further examination of the relationship between Grp170 and members of both the DnaK and Hsp110 families indicate that, although Grp170 is only distantly related to the Hsp110 family, the domain structure of the Grp170 sequence in its carboxyl terminal one-third is clearly more like Hsp110 than members of the DnaK/Hsp70 group. This is most evident in the long loop domain, which is characteristic of these proteins (Chen et al 1996; Craven et al 1997). Although some predictions suggest some short helical structures in the Grp170 sequence from residue 553–688, most of the sequence is predicted to be loop. The sequences of these loop domains are divergent in members of the Hsp110 family and the Grp170 family. Distal to this loop domain, Grp170 (residues 700–900) shares 19% identity with the carboxyl terminal 200 amino acids of bovine Hsc70 and 18% identity with the corresponding segment of Grp78 but exhibits 40% identity with Hsp110 (residues 600–800). This region of the Hsp110/SSE family contains a pattern of conserved sequences, including Magic and TedWylee, which define this group (Lee-Yoon et al 1995 and described above). Secondary structure predictions of Grp170 suggest that the overall organization of Grp170 is also similar to that of Hsp110 (Fig 1C): it contains an apparent ATPase domain, β-strand domain, and long loop followed by a helical domain. The expansion of Grp170 relative to Hsp110 occurs mainly in the central loop domain; however, insertions have also occurred in the loops of the β-sandwich domain and in the C-terminal random coil (Fig 1 B,C).
Based on the above structural relations, we would predict that the various common domains between the Hsp and Grp members of the large Hsp70-like chaperones would carry out similar functions (peptide binding, ATP binding, regulation, etc). However, the degree of sequence divergence between these 2 groups is too large to make these predictions without considerable caution. For example, although wild type Hsp110 binds ATP poorly, Grp170 is an avid ATP binder (Chen et al 1996; Oh et al 1999). An analysis by targeted deletions based on this model for Grp170, as done with Hsp110, would answer many questions about Grp170 function.
FUNCTIONS OF Grp170
In the case of Hsp110, we have a good idea of its structure-function relations with respect to its chaperoning abilities but have only glimpses of its cellular and physiological importance. The situation with Grp170 is reversed. Grp170 inhibits aggregation of denatured protein (Chen et al, unpublished data). However, nothing else concerning its chaperoning activity is known.
Several lines of investigation have given insight into some of its biological functions. Most specifically, the induction of Grp170 (as well as other Grps) by anoxia, low pH, and glucose starvation, all conditions associated with the ischemic state, has important ramifications in several areas of pathophysiology.
One line of investigation concerns the induction of Grp170 under physiological stress and its potential protective functions. Cai et al (1993) demonstrated that Grps (including Grp170) were induced in radiation induced fibrosarcoma (RIF) murine tumors during tumor growth as a function of the development of ischemia and necrosis as tumors increased in size. A role in protection of ER function under anoxic and ischemic stress was suggested based on survival of cancer cells detected in the visibly necrotic material of the large tumors expressing Grp170. A different but somewhat parallel observation describes an elevated expression of Grp170 in human breast tumors (Yoshitane et al 1998) compared with normal breast tissue. Another area where anoxia and ischemia are of great importance is in the response of brain tissue to stroke and myocardial infarction. Kuwabara et al (1996) demonstrated the induction of Grp170 in cultured rat astrocytes exposed to hypoxia and in ischemic astrocytes in mouse brain. In addition to astrocytes, Kohji et al (1998) further demonstrated the induction of Grp170 in mouse neurons 1 hour after middle cerebral artery occlusion. Other studies suggest roles for Grp170 in human atherosclerotic plaques (Yoshitane 1996), hepatic response to caloric intake, ie, diet (Dhahbi et al 1997), and resistance to some cancer chemotherapeutic agents (Shen et al 1987).
Many of these reports focus specifically on Grp170; however, it is likely that most observations concerning physiological responses and protective phenomenon reflect the coordinated functions of the entire set of Grp stress proteins. To understand the specific role(s) of individual proteins, such as Grp170, requires dissection of protein folding and quality control pathways in vivo and in vitro rather than tissue level observations.
At this time, limited information concerning Grp170 and its intracellular functions is available. Lin et al (1993) first clearly described Grp170 as being an ER resident glycoprotein. Subsequently, it was demonstrated that it has a C-terminal KNDEL sequence that is responsible for its ER retention (Naved et al 1995; Chen et al 1996). Coimmunoprecipitation studies indicated that Grp170 interacts with the other major Grps, Grp78 and Grp94. Additionally, Grp170 was found to interact in vivo with immunoglobulin chains in 4 different B-cell hybridomas. On the basis of these data, it was suggested that Grp170 played a role in immunoglobulin folding and assembly in conjunction with Grp78 and Grp94 (Lin et al 1993). Melnick et al (1994) have independently demonstrated an interaction among Grp170, Grp78, Grp94, and immunoglobulin light chain. Kuznetsov et al (1997) have shown that Grp170 and some other Grps complex with thyroglobulin, a major protein secreted by thyroid epithelial cells. Therefore, it is likely that Grp170's interactions in the ER are not limited to immunoglobulin chain and the other Grps. The S cerevisiae Grp170 is referred to as Lhs1p. Saris et al (1997) have shown that a marker enzyme, inactivated in the yeast ER by high temperature, requires Grp170 (Lhs1p) for solubilization and refolding. This observation suggests that Grp170/Lhs1p is required for refolding of denatured protein in the ER and for protection of these proteins from proteolysis in S cerevisiae subjected to heat stress.
Although the above studies suggest a role in protein folding and/or assembly, a possible function for Grp170 in import of proteins and peptides has also been described. Dierks et al (1996) removed ATP-binding proteins from a microsomal extract by passage over an ATP-agarose column. The bound fraction consisted of the Grps and other proteins, including Grp170. Grp170 was found to be the most efficient ATP-binding protein in the extract. Microsomes (proteoliposomes) reconstituted with the full extract imported protein, whereas microsomes containing the extract that had been depleted in ATP-binding proteins did not. Addition of purified Grp78 (previously believed to be the required ATPase for import) as a single ATP-binding component for reconstitution of microsomes from an extract did not restore import function to proteoliposomes depleted of ATP-binding proteins. It was, therefore, suggested that Grp170 might be the ATPase responsible for efficient import of proteins from the cytosol into the ER. More recently, Spee et al (1999) identified Grp170 as an ER chaperone involved with peptide transport into the ER via the transporter associated with antigen processing. The relation of Grp170 to the transporter associated with antigen processing function suggests that Grp170 may be involved in the antigen presentation pathway. Grp170 has also been shown to be a major calcium-binding protein in the ER (Naved et al 1995).
SUMMARY
Both the Grp170 and Hsp110 families represent relatively conserved and distinct sets of stress proteins, within a more diverse category that also includes the Hsp70s. All of these families are found in a wide variety of organisms from yeasts to humans. Although Hsp110s or Grp170s are not Hsp70s any more than Hsp70s are Hsp110s or Grp170s, it is still reasonable to refer to this combination of related families as the Hsp70 superfamily based on arguments discussed above and since no obvious prokaryotic Hsp110 or Grp170 has yet been identified.
These proteins are related to their counterparts in the Hsp70/Grp78 family of eukaryotic stress proteins but are characterized by significantly larger molecular weights. The members of the Grp170 family are characterized by C-terminal ER retention sequences and are ER localized in yeasts and mammals. As a Grp, Grp170 is recognized to be coregulated with other major Grps by a well-known set of stress conditions, sometimes referred to as the unfolded protein response (Kozutsumi et al 1988; Nakaki et al 1989). The Hsp110 family members are localized in the nucleus and cytoplasm and, with other major Hsps, are also coregulated by a specific set of stress conditions, most notably including hyperthermic exposures. Hsp110 is sometimes called Hsp105, although it would be preferable to have a uniform term.
The large Hsp70-like proteins are structurally similar to the Hsp70s but differ from them in important ways. In both the Grp170 and Hsp110 families, there is a long loop structure that is interposed between the peptide-binding β-domain and the α-helical lid. In the Hsp110 family and Grp170, there are differing degrees of expansion in the α-helical domain and the addition of a C-terminal loop. This gives the appearance of much larger lid domains for Hsp110 and Grp170 compared with Hsp70. Both Hsp110 and Grp170 families have relatively conserved short sequences in the α-helical domain in the lid, which are conserved motifs in numerous proteins (we termed these motifs Magic and TedWylee as discussed earlier). The structural differences detailed in this review result in functional differences between the large (Grp170 and Hsp110) members of the Hsp70 superfamily, the most distinctive being an increased ability of these proteins to bind (hold) denatured polypeptides compared with Hsc70, perhaps related to the enlarged C-terminal helical domain. However, there is also a major difference between these large stress proteins; Hsp110 does not bind ATP in vitro, whereas Grp170 binds ATP avidly.
The role of the Grp170 and Hsp110 stress proteins in cellular physiology is not well understood. Overexpression of Hsp110 in cultured mammalian cells increases thermal tolerance. Grp170 binds to secreted proteins in the ER and may be cooperatively involved in folding these proteins appropriately. These roles are similar to those of the Hsp70 family members, and, therefore, the question arises as to the differential roles played by the larger members of the superfamily. We have discussed evidence that the large members of the superfamily cooperate with members of the Hsp70 family, and these chaperones probably interact with a large number of chaperones and cochaperones in their functional activities. The fundamental point is that Hsp110 is found in conjunction with Hsp70 in the cytoplasm (and nucleus) and Grp170 is found in conjunction with Grp78 in the ER in every eucaryotic cell examined from yeast to humans. This would strongly argue that Hsp110 and Grp170 exhibit functions in eucaryotes not effectively performed by Hsp70s or Grp78, respectively. Of interest in this respect is the observation that all Hsp110 loss of function or deletion mutants listed in the Drosophila deletion project database are lethal. The important task for the future is to determine the roles these conserved molecular chaperones play in normal and physiologically stressed cells.
Table 3.
Percentage of identity and similarity between selected members of the Grp170 subfamily with respect to the sequence of Grp170 from CHO cells
REFERENCES
- Albukrek D, Bakon M, Moran DS, Faibel M, Epstein Y. Heat-stroke-induced cerebellar atrophy: clinical course, CT and MRI findings. Neuroradiology. 1997;39:195–197. doi: 10.1007/s002340050392. [DOI] [PubMed] [Google Scholar]
- Becker J, Craig EA. Heat-shock proteins as molecular chaperones. Eur J Biochem. 1994;219:11–23. doi: 10.1007/978-3-642-79502-2_2. [DOI] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Werner ED, Dubas ME, Goecker JL, Kruse KB, Mc Cracken AA. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J Biol Chem. 1999;274:3453–3460. doi: 10.1074/jbc.274.6.3453. [DOI] [PubMed] [Google Scholar]
- Cai JW, Henderson BW, Shen JW, Subjeck JR. Induction of glucose regulated proteins during growth of a murine tumor. J Cell Physiol. 1993;154:229–237. doi: 10.1002/jcp.1041540204. [DOI] [PubMed] [Google Scholar]
- Chen X, Easton D, Oh HJ, Lee-Yoon D, Liu X, Subjeck J. The 170 kDa glucose regulated stress protein is a large HSP70-, HSP110-like protein of the endoplasmic reticulum. FEBS Lett. 1996;380:68–72. doi: 10.1016/0014-5793(96)00011-7. [DOI] [PubMed] [Google Scholar]
- Chung KS, Hoe KL, Kim KW, Yoo HS. Isolation of a novel heat shock protein 70-like gene, pss1+ of Schizosaccharomyces pombe homologous to 110/SSE subfamily. Gene. 1998;210:143–150. doi: 10.1016/s0378-1119(98)00061-4. [DOI] [PubMed] [Google Scholar]
- Craven RA, Tyson JR, Stirling CJ. A novel subfamily of Hsp70s in the endoplasmic reticulum. Trends Cell Biol. 1997;7:277–282. doi: 10.1016/S0962-8924(97)01079-9. [DOI] [PubMed] [Google Scholar]
- Depiereux E, Baudoux G, Briffeuil P, Reginster I, DeBolle X, Vinals C, Ferytmans E. Match-Box server: a multiple sequence alignment tool placing emphasis on reliability. Comput Appl Biosci. 1997;13:249–256. doi: 10.1093/bioinformatics/13.3.249. [DOI] [PubMed] [Google Scholar]
- Dhahbi JM, Mote PL, Tillman JB, Walford RL, Spindler SR. Dietary energy tissue-specifically regulates endoplasmic reticulum chaperone gene expression in the liver of mice. J Nutr. 1997;127:1758–1764. doi: 10.1093/jn/127.9.1758. [DOI] [PubMed] [Google Scholar]
- Dierks T, Volkmer J, Schlenstedt G, et al. A microsomal ATP-binding protein involved in efficient protein transport into the mammalian endoplasmic reticulum. EMBO J. 1996;15:6931–6942. [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- Fathallah DM, Cherif D, Dellagi K, Arnaout MA. Molecular cloning of a novel human Hsp70 from a B cell line and its assignment to chromosome 5. J Immunol. 1993;151:810–813. [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP—Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Foltz KR, Lennarz WJ. Purification and characterization of an extracellular fragment of the sea urchin egg receptor for sperm. J Cell Biol. 1990;111:2951–2159. doi: 10.1083/jcb.111.6.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz KR, Partin JS, Lennarz WJ. Sea urchin egg receptor for sperm: sequence similarity of binding domain and Hsp 70. Science. 1993;259:1421–1425. doi: 10.1126/science.8383878. [DOI] [PubMed] [Google Scholar]
- Gething MJ. ed. 1997 Guidebook to Molecular Chaperones and Protein-Folding Catalysts. Oxford University Press, Oxford. [Google Scholar]
- Gething MJ, Sambrook JF. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Giusti AF, Hoang KM, Foltz KR. Surface localization of the sea urchin egg receptor for sperm. Dev Biol. 1997;184:10–24. doi: 10.1006/dbio.1997.8507. [DOI] [PubMed] [Google Scholar]
- Hatayama T, Yasuda K, Nishiyama E. Characterization of high molecular mass heat shock proteins and 42°C specific heat shock proteins of murine cells. Biochem Biophys Res Comm. 1994;204:357–365. doi: 10.1006/bbrc.1994.2467. [DOI] [PubMed] [Google Scholar]
- Hatayama T, Yasuda K, Yasuda K. Association of HSP105 with HSC70 in high molecular mass complexes in mouse FM3A cells. Biochem Biophys Res Commun. 1998;248:95–401. doi: 10.1006/bbrc.1998.8979. [DOI] [PubMed] [Google Scholar]
- Henics T, Nagy E, Oh HJ, Csermely P, von Gabain A, Subjeck JR. Mammalian Hsp70 and Hsp110 proteins bind to RNA motifs involved in mRNA stability. J Biol Chem. 1999;274:17 318–17 324. doi: 10.1074/jbc.274.24.17318. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:273–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Hightower LE. Cultured animal cells exposed to amino acid analogues or puromycin rapidly synthesize several polypeptides. J Cell Physiol. 1980;102:407–427. doi: 10.1002/jcp.1041020315. [DOI] [PubMed] [Google Scholar]
- Hirohashi N, Lennarz WJ. The 350-kDa sea urchin egg receptor for sperm is localized in the vitelline layer. Dev Biol. 1998a;204:305–315. doi: 10.1006/dbio.1998.9015. [DOI] [PubMed] [Google Scholar]
- Hirohashi N, Lennarz WJ. Sperm-egg binding in the sea urchin: a high level of intracellular ATP stabilizes sperm attachment to the egg receptor. Dev Biol. 1998b;201:270–279. doi: 10.1006/dbio.1998.8984. [DOI] [PubMed] [Google Scholar]
- Honda K, Hatayama T, Yukioka M. Common antigenicity of mouse 42 degrees C-specific heat-shock protein with mouse Hsp 105. Biochem Biophys Res Comm. 1989;160:60–66. doi: 10.1016/0006-291x(89)91620-3. [DOI] [PubMed] [Google Scholar]
- Hylander B. The distribution and localization of Hsp110 in brain. Brain Res. 2000;869:49–55. doi: 10.1016/s0006-8993(00)02346-5. [DOI] [PubMed] [Google Scholar]
- Ishihara K, Yasuda K, Hatayama T. Molecular cloning, expression and localization of human 105kDa heat shock protein, Hsp105. Biochim Biophys Acta. 1999;1444:138–142. doi: 10.1016/s0167-4781(98)00254-1. [DOI] [PubMed] [Google Scholar]
- Just ML, Lennarz WJ. Reexamination of the sequence of the sea urchin egg receptor for sperm: implications with respect to its properties. Dev Biol. 1997;184:25–30. doi: 10.1006/dbio.1997.8504. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Kimura T, Kishishita M, Noda Y, Fujita J. Cloning of Apg-2 encoding a novel member of heat shock protein 110 family. Gene. 1997a;189:19–24. doi: 10.1016/s0378-1119(96)00807-4. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Kimura T, Nishiyama H, Noda Y, Fujita J. Developmentally regulated expression of APG-1, a member of heat shock protein 110 family in murine male germ cells. Biochem Biophys Res Comm. 1997b;223:113–116. doi: 10.1006/bbrc.1997.6410. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Nishiyama H, Nonoguchi K, Higashitsuji H, Kishishita M, Fujita J. A novel Hsp110-related gene, Apg-1, that is abundantly expressed in the testis responds to a low temperature heat shock rather than the traditional elevated temperatures. J Biol Chem. 1997c;272:2640–2645. doi: 10.1074/jbc.272.5.2640. [DOI] [PubMed] [Google Scholar]
- Kohji M, Tomohiro M, Hiroyuki N, Toshio T, Keisuke K, Yoshitane T, Minoru S, Satoshi O. Marked, sustained expression of a novel 150-kDa oxygen-regulated stress protein, in severely ischemic mouse neurons. Mol Br Res. 1998;60:98–106. doi: 10.1016/s0169-328x(98)00174-0. [DOI] [PubMed] [Google Scholar]
- Kojima R, Randall J, Brenner BM, Gullans SR. Osmotic stress protein 94 (Osp94): a new member of the HSP110/SSE gene subfamily. J Biol Chem. 1996;271:12327–12332. doi: 10.1074/jbc.271.21.12327. [DOI] [PubMed] [Google Scholar]
- Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. The presence of mal folded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- Kuwabara K, Matsumoto M, Ikeda J, et al. Purification and characterization of a novel stress protein, the 150-kDa oxygen-regulated protein (ORP150), from cultured rat astrocytes and its expression in ischemic mouse brain. J Biol Chem. 1996;271:5025–5032. doi: 10.1074/jbc.271.9.5025. [DOI] [PubMed] [Google Scholar]
- Kuznetsov G, Chen LB, Nigam SK. Multiple molecular chaperones complex with misfolded large oligomeric glycoproteins in the endoplasmic reticulum. J Biol Chem. 1997;272:3057–3063. doi: 10.1074/jbc.272.5.3057. [DOI] [PubMed] [Google Scholar]
- Landry J, Bernier D, Chretien P, Nicole LM, Tanguay RM, Marceau N. Synthesis and degradation of heat shock proteins during development and decay of thermotolerance. Cancer Res. 1982;42:2457–2461. [PubMed] [Google Scholar]
- Lee-Yoon D, Easton D, Murawski M, Burd R, Subjeck JR. Identification of a major subfamily of large Hsp70-like proteins through the cloning of the mammalian 110-kDa heat shock protein. J Biol Chem. 1995;270:15 725–15 733. doi: 10.1074/jbc.270.26.15725. [DOI] [PubMed] [Google Scholar]
- Levinson W, Oppermann H, Jackson J. Transition series metals and sulfhydryl reagents induce the synthesis of four proteins in eukaryotic cells. Biochim Biophys Acta. 1980;606:170–180. doi: 10.1016/0005-2787(80)90108-2. [DOI] [PubMed] [Google Scholar]
- Lin HY, Masso-Welch P, Di YP, Cai JW, Shen JW, Subjeck JR. The 170-kDa glucose-regulated stress protein is an endoplasmic reticulum protein that binds immunoglobulin. Mol Biol Cell. 1993;4:1109–1119. doi: 10.1091/mbc.4.11.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XD, Morano KA, Thiele DJ. The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J Biol Chem. 1999;274:26 654–26 660. doi: 10.1074/jbc.274.38.26654. [DOI] [PubMed] [Google Scholar]
- Manto MU. Case report: isolated cerebellar dysarthria associated with a heat stroke. Clin Neurol Neurosurg. 1996;98:55–56. doi: 10.1016/0303-8467(95)00089-5. [DOI] [PubMed] [Google Scholar]
- Mauk R, Jaworski D, Kamei N, Glabe CG. Identification of a 97-kDa heat shock protein from S. franciscanus ovaries with 94% amino acid identity to the S. purpuratus egg surface receptor for sperm. Dev Biol. 1997;184:31–37. doi: 10.1006/dbio.1997.8512. [DOI] [PubMed] [Google Scholar]
- Melnick J, Dul JL, Argon Y. Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature. 1994;370:373–375. doi: 10.1038/370373a0. [DOI] [PubMed] [Google Scholar]
- Morozov A, Subjeck J, Raychaudhuri P. HPV16 E7 oncoprotein induces expression of a 110 kDa heat shock protein. FEBS Lett. 1995;371:214–218. doi: 10.1016/0014-5793(95)00884-c. [DOI] [PubMed] [Google Scholar]
- Mukai H, Kuno T, Tanaka H, Hirata D, Miyakawa T, Tanaka C. Isolation and characterization of SSE1 and SSE2, new members of the yeast Hsp70 multigene family. Gene. 1993;132:57–66. doi: 10.1016/0378-1119(93)90514-4. [DOI] [PubMed] [Google Scholar]
- Nakaki T, Deans RJ, Lee AS. Enhanced transcription of the 78,000-dalton glucose-regulated protein (GRP78) gene and association of GRP78 with immunoglobulin light chains in a nonsecreting B-cell myeloma line (NS-1) Mol Cell Biol. 1989;9:2233–2238. doi: 10.1128/mcb.9.5.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonoguchi K, Itoh K, Xue JH, et al. Cloning of human cDNAs for Apg-1 and Apg-2, members of the Hsp110 family, and chromosomal assignment of their genes. Gene. 1999;237:21–28. doi: 10.1016/s0378-1119(99)00325-x. [DOI] [PubMed] [Google Scholar]
- Oh HJ, Chen X, Subjeck JR. Hsp110 protects heat-denatured proteins and confers cellular thermoresistance. J Biol Chem. 1997;272:31 636–31 640. doi: 10.1074/jbc.272.50.31636. [DOI] [PubMed] [Google Scholar]
- Oh HJ, Easton D, Murawski M, Kaneko Y, Subjeck JR. The chaperoning activity of Hsp110: identification of functional domains by use of targeted deletions. J Biol Chem. 1999;274:15 712–15 718. doi: 10.1074/jbc.274.22.15712. [DOI] [PubMed] [Google Scholar]
- Olden K, Pratt RM, Jaworski C, Yamada KM. Evidence for role of glycoprotein carbohydrates in membrane transport: specific inhibition by tunicamycin. Proc Natl Acad Sci U S A. 1979;76:791–795. doi: 10.1073/pnas.76.2.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesofsky-Vig N, Bramble R. Characterization of an 88-kDa heat shock protein of Neurospora crassa that interacts with Hsp30. J Biol Chem. 1998;273:11 335–11 341. doi: 10.1074/jbc.273.18.11335. [DOI] [PubMed] [Google Scholar]
- Santos BC, Chevaile A, Kojima R, Gullans SR. Characterization of the Hsp110/SSE gene family response to hyperosmolality and other stresses. Am J Physiol. 1998;274:F1054–1061. doi: 10.1152/ajprenal.1998.274.6.F1054. [DOI] [PubMed] [Google Scholar]
- Saris N, Holkeri H, Craven RA, Stirling CJ, Makarow M. The Hsp70 homologue Lhs1p is involved in a novel function of the yeast endoplasmic reticulum, refolding and stabilization of heat-denatured protein aggregates. J Cell Biol. 1997;137:813–824. doi: 10.1083/jcb.137.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciandra JJ, Subjeck JR. The effects of glucose on protein synthesis and thermosensitivity in Chinese hamster ovary cells. J Biol Chem. 1983;258:12091–12093. [PubMed] [Google Scholar]
- Sciandra JJ, Subjeck JR, Hughes CS. Induction of glucose-regulated proteins during anaerobic exposure and of heat-shock proteins after reoxygenation. Proc Natl Acad Sci U S A. 1984;81:4843–4847. doi: 10.1073/pnas.81.15.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Hughes C, Chao C, Cai J, Bartels C, Gessner T, Subjeck J. Coinduction of glucose-regulated proteins and doxorubicin resistance in Chinese hamster ovary cells. Proc Natl Acad Sci U S A. 1987;84:3278–3282. doi: 10.1073/pnas.84.10.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Kawakami K, Matsui Y, Tanaka K, Toh-e A. MSI3, a multicopy suppressor of mutants hyperactivated in the RAS-cAMP pathway, encodes a novel Hsp70 protein of. Saccharomyces cerevisiae. Mol Gen Genet. 1993;240:323–332. doi: 10.1007/BF00280382. [DOI] [PubMed] [Google Scholar]
- Spee P, Subjeck J, Neefjes J. Identification of novel peptide binding proteins in the endoplasmic reticulum: ERp72, calnexin, and grp170. Biochemistry. 1999;38:10559–10566. doi: 10.1021/bi990321r. [DOI] [PubMed] [Google Scholar]
- Stears RL, Lennarz WJ. Mapping sperm binding domains on the sea urchin egg receptor for sperm. Dev Biol. 1997;187:200–208. doi: 10.1006/dbio.1997.8608. [DOI] [PubMed] [Google Scholar]
- Storozhenko S, De Pauw P, Kushnir S, Van Montagu M, Inze D. Identification of an Arabidopsis thaliana cDNA encoding a HSP70-related protein belonging to the HSP110/SSE1 subfamily. FEBS Lett. 1996;390:113–118. doi: 10.1016/0014-5793(96)00640-0. [DOI] [PubMed] [Google Scholar]
- Subjeck JR, Sciandra JJ 1982 . In: Heat Shock: From Bacteria to Man, ed Schlesinger M, Ashburner M, Tissieres A, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, 405–411. [Google Scholar]
- Subjeck JR, Sciandra JJ, Chao CF, Johnson RJ. Heat shock proteins and biological response to hyperthermia. Br J Cancer. 1982a;45:127–131. [PMC free article] [PubMed] [Google Scholar]
- Subjeck JR, Sciandra JJ, Johnson RJ. Heat shock proteins and thermotolerance; a comparison of induction kinetics. Br J Radiol. 1982b;55:579–584. doi: 10.1259/0007-1285-55-656-579. [DOI] [PubMed] [Google Scholar]
- Subjeck JR, Shyy T, Shen J, Johnson RJ. Association between the mammalian 110,000-dalton heat-shock protein and nucleoli. J Cell Biol. 1983;97:1389–1395. doi: 10.1083/jcb.97.5.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Nakafuku M, Satoh T, Marshall MS, Gibbs JB, Matsumoto K, Kaziro Y, Toh-e A. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 1990;60:803–807. doi: 10.1016/0092-8674(90)90094-u. [DOI] [PubMed] [Google Scholar]
- Tomasovic SP, Steck PA, Heitzman D. Heat-stress proteins and thermal resistance in rat mammary tumor cells. Radiat Res. 1983;95:399–413. [PubMed] [Google Scholar]
- Tortora P, Burlini N, Caspani G, Guerritore A. Studies on glucose induced inactivation of gluconeogenetic enzymes in adenylate cyclase and cAMP-dependent protein kinase yeast mutants. Eur J Biochem. 1984;145:543–548. doi: 10.1111/j.1432-1033.1984.tb08590.x. [DOI] [PubMed] [Google Scholar]
- Wang X-Y, Chen X, Oh HJ, Repasky E, Kazim L, Subjeck JR. Characterization of native interaction of Hsp110 with Hsp25 and hsc70. FEBS Lett. 2000;465:98–102. doi: 10.1016/s0014-5793(99)01733-0. [DOI] [PubMed] [Google Scholar]
- Welch WJ, Garrels JI, Thomas GP, Lin JJ, Feramisco JR. Biochemical characterization of the mammalian stress proteins and identification of two stress proteins as glucose- and Ca2+-ionophore-regulated proteins. J Biol Chem. 1983;258:7102–7111. [PubMed] [Google Scholar]
- Xue JH, Fukuyama H, Nonoguchi K, et al. Induction of Apg-1, a member of the heat shock protein 110 family, following transient forebrain ischemia in the rat brain. Biochem Biophys Res Comm. 1998;247:796–801. doi: 10.1006/bbrc.1998.8894. [DOI] [PubMed] [Google Scholar]
- Yagita Y, Kitagawa K, Taguchi A, et al. Molecular cloning of a novel member of the HSP110 family of genes, ischemia-responsive protein 94 kDa (irp94), expressed in rat brain after transient forebrain ischemia. J Neurochem. 1999;72:1544–51. doi: 10.1046/j.1471-4159.1999.721544.x. [DOI] [PubMed] [Google Scholar]
- Yasuda K, Ishihara K, Nakashima K, Hatayama T. Genomic cloning and promoter analysis of the mouse 105-kDa heat shock protein (HSP105) gene. Biochem Biophys Res Commun. 1999;256:75–80. doi: 10.1006/bbrc.1999.0283. [DOI] [PubMed] [Google Scholar]
- Yasuda KA, Nakai T, Hatayama T, Nagata K. Cloning and expression of murine high molecular mass heat shock proteins, HSP 105. J Biol Chem. 1995;270:29178–29723. doi: 10.1074/jbc.270.50.29718. [DOI] [PubMed] [Google Scholar]
- Yoshitane T, Keisuke K, Seiichi H, et al. 150-kD oxygen-regulated protein is expressed in human atherosclerotic plaques and allows mononuclear phagocytes to withstand cellular stress on exposure to hypoxia and modified low density lipoprotein. J Clin Invest. 1996;98:1930–1941. doi: 10.1172/JCI118994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitane T, Keisuke K, Seiichi H, et al. Expression of the 150-kd oxygen-regulated protein in human breast cancer. Lab Invest. 1998;78:699–706. [PubMed] [Google Scholar]
- Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]