Abstract
DnaJ-like proteins are defined by the presence of an approximately 73 amino acid region termed the J domain. This region bears similarity to the initial 73 amino acids of the Escherichia coli protein DnaJ. Although the structures of the J domains of E coli DnaJ and human heat shock protein 40 have been solved using nuclear magnetic resonance, no detailed analysis of the amino acid conservation among the J domains of the various DnaJ-like proteins has yet been attempted. A multiple alignment of 223 J domain sequences was performed, and the levels of amino acid conservation at each position were established. It was found that the levels of sequence conservation were particularly high in ‘true’ DnaJ homologues (ie, those that share full domain conservation with DnaJ) and decreased substantially in those J domains in DnaJ-like proteins that contained no additional similarity to DnaJ outside their J domain. Residues were also identified that could be important for stabilizing the J domain and for mediating the interaction with heat shock protein 70.
INTRODUCTION
The presence of a so-called J domain in a protein defines it as a DnaJ-like protein. The J domain of DnaJ-like proteins is involved in regulating the adenosine triphosphatase (ATPase) activity of heat shock protein 70 (Hsp70) by stimulating adenosine triphosphate (ATP) hydrolysis (Cheetham and Caplan 1998). In general, Hsp70 proteins have a low rate of ATP hydrolysis, and the ATP-binding form of Hsp70 has a low affinity for unfolded proteins, leading to a poor chaperone effect. However, in the presence of a DnaJ-like protein, the hydrolysis of ATP is enhanced. When this activity is enhanced, Hsp70 is in the adenosine 5′-diphosphate–bound state and has a higher affinity for substrate and can, therefore, act more efficiently as a chaperone protein. DnaJ-like proteins, therefore, act as cochaperone proteins by aiding the chaperone function of Hsp70s.
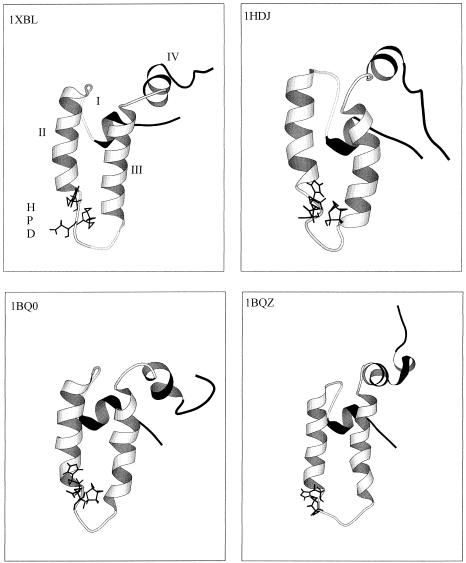
The J domain is a region that bears similarity to the initial 73 amino acids of the Escherichia coli protein DnaJ (Bardwell et al 1986). This region is believed to be the major site of the interaction between DnaJ-like proteins and Hsp70s. The structures of 2 J domains have been determined using nuclear magnetic resonance. These are from DnaJ in E coli and Hsp40 (HDJ1) in humans (Pellechia et al 1996; Qian et al 1996; Huang et al 1999). The structures contain 4 α-helices, with a loop region containing a highly conserved tripeptide of histidine, proline, and aspartic acid residues (HPD motif) located between helices II and III. The 4 structures are depicted in Figure 1. Helices II and III are antiparallel amphipathic helices (Qian et al 1996). The HPD motif is present in all known J domains, with the exception of the ring-infected erythrocyte surface antigen (RESA) proteins of Plasmodium falciparum (Bork et al 1992). Mutations of these residues abolish the stimulation of the Hsp70 ATPase activity (Cheetham and Caplan 1998).
Fig 1.
Ribbon representation of the structures of the DnaJ and HDJ1 J domains as determined using nuclear magnetic resonance. The PDB codes are 1XBL (Pellechia et al 1996) and 1BQ0 and 1BQZ (Huang et al 1999) for the J domain from E coli DnaJ and 1HDJ (Qian et al 1996) for the J domain from human HDJ1. The structures were visualized in Molscript (Kraulis 1991). The HPD motif is indicated as sticks. Helices II and III are in white, with the more mobile helices I and IV in black. The helices and HPD motif are labeled on the 1XBL structure
Recent attempts have been made to subclassify the many and varied DnaJ-like proteins into groups. E coli DnaJ contains 4 domains, namely, the J domain; a glycine-phenylalanine rich region, which is thought to act as a flexible linker region between the J domain and the rest of the protein but may also affect the interaction with Hsp70; a region containing 4 repeats of the form cysteine-XX-cysteine-X-glycine-X-glycine (CXXCXGXGX), which are thought to act as zinc finger–like motifs for interaction with proteins; and an uncharacterized C-terminal portion. Kelley (1998) proposed that DnaJ-like proteins should be divided into 2 groups. The first group would contain those DnaJ-like proteins that bore similarity to E coli DnaJ over the J domain and the glycine-phenylalanine rich region. The second group would contain those proteins that only bore resemblance to DnaJ over the J domain. An alternative division was proposed by Cheetham and Caplan (1998). They divided all DnaJ-like proteins into 3 groups. Type I DnaJ-like proteins contain similarity to DnaJ over all domains, namely, the J domain, the glycine-phenylalanine rich region, and the CXXCXGXG motifs. Type II proteins show similarity to DnaJ over the J domain and the glycine-phenylalanine rich region. Type III proteins, in contrast, have similarity only over the J domain to DnaJ (Cheetham and Caplan 1998). However, little analysis has been performed in the way of identifying residues critical to the J domain function, with the exception of the HPD motif. The J domains of the yeast endoplasmic reticulum proteins Scj1p and Sec63p have been swapped and have resulted in functional proteins (Schlenstedt et al 1995). However, substitution of the Sec3p J domain with the J domain of the cytosolic protein Sis1 resulted in a nonfunctional chimeric protein (Schlenstedt et al 1995). This indicates that there is a certain level of specialization within the J domain that is necessary to specify Hsp70-DnaJ-like partner proteins, with the possibility that variation in amino acid composition affects the specificity of the interaction. The alignments of DnaJ-like proteins have in general only used around 8 sequences (Bork et al 1992) and have not looked at sequence conservation, although the recent work by Ohtsuka and Hata (2000) showed an alignment of 23 sequences. It was also unclear as to whether the levels of sequence similarity remained constant across all 3 types of DnaJ-like proteins. Consequently, it was decided to align all known J domains and establish levels of sequence identity and similarity and compare these levels among the groups of type I, II, and III DnaJ-like proteins. It was theorized that this method would possibly provide an alternative method of distinguishing between groups of DnaJ-like proteins on the basis of the amino acid composition of their J domains and to identify residues important in specific Hsp70—DnaJ interaction.
MATERIALS AND METHODS
All proteins related to DnaJ were downloaded using the Entrez Browser (http://www.ncbi.nlm.nih.gov/Entrez/). Redundant sequences were removed, and a database containing all the proteins was created. The J domain sequences were then placed into the spreadsheet package Microsoft Excel and aligned manually. Altogether 223 sequences were used. A consensus sequence for all DnaJ-like proteins was then developed. The sequences were then divided into 3 groups according to the classification of Cheetham and Caplan (1998). Consensus sequences for these 3 groups were then developed, and the 4 sequences obtained were compared with each other. Checks of the consensus sequences were obtained by taking 8 random sequences from each group and aligning them against each other using freely available multiple alignment packages.
RESULTS
Trends in the level of conservation within and among J domains of types I, II, and III proteins
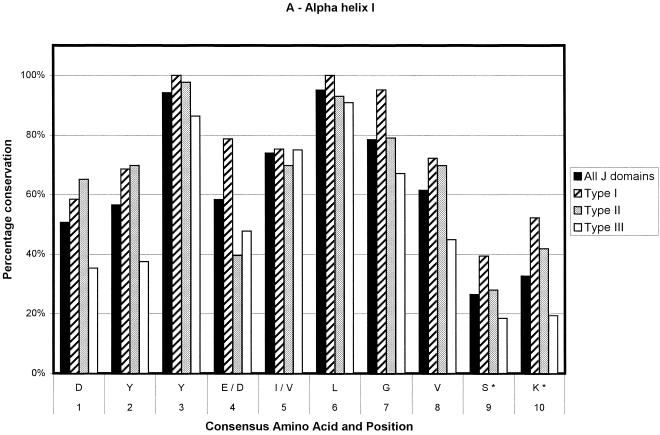
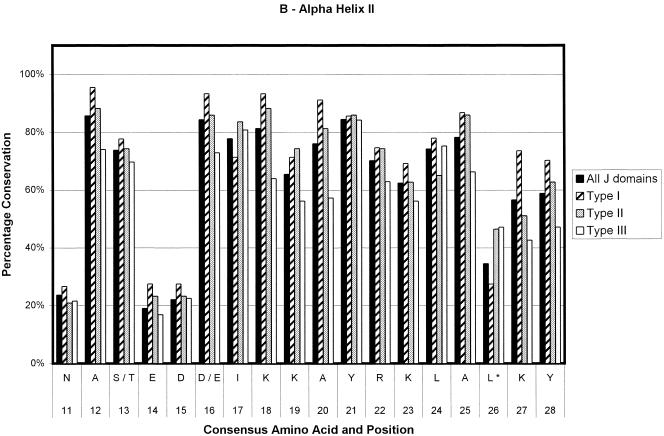
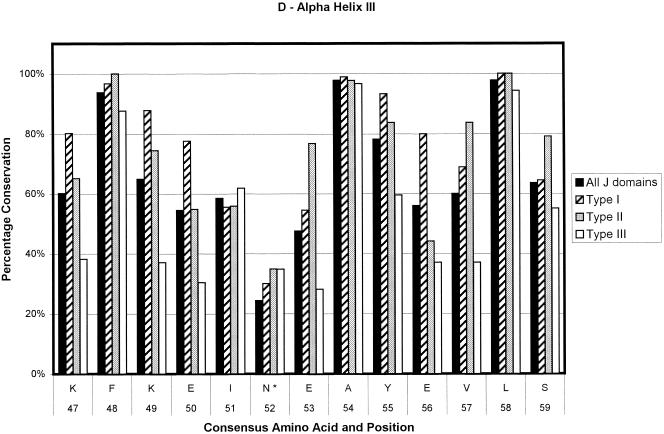
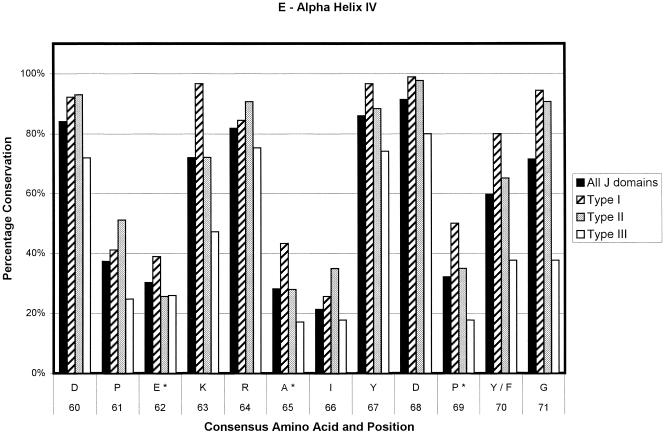
Ninety-one sequences were used in the generation of the type I consensus sequence, 43 for the type II consensus sequence, and 89 for the type III consensus sequence. In total, 223 sequences were used for the overall consensus sequence. The numbers of sequences contributing at any point could decrease slightly because of the insertion of gaps to optimize alignments and in the loop region. The results are depicted in Figure 2. The percentage conservation in the figure is a reflection of the level of sequence conservation of the given amino acid at a given position. Positions that are variable among the 4 derived consensus sequences are given in Table 1. As can be seen in Figure 2, type I J domains are highly conserved, type II slightly less so, and type III show the lowest level of conservation. Most of the loop region is not shown because of the low number of amino acids making up the alignment at this point and the low level of amino acid conservation. Most of the type I domains used in the alignments are from eubacteria. Hence, the evolutionary relationship among these proteins is likely to be fairly close. This could lead to an inherent bias in the determination of the consensus sequence of the type I proteins, with a possible overestimation of the levels of conservation of the residues at each position.
Fig 2.
Bar chart representations of the percentage conservation of each amino acid in the consensus sequences derived from an alignment of all J domains and the J domains of type I, II, and III DnaJ-like proteins. The data have been separated into helix I (a), helix II (b), the loop region (c), helix III (d), and helix IV (e). The single letter amino acid code has been used. The different consensus sequences were derived by determining the most highly conserved amino acid based on percentage identity at each position in all 223 J domain sequences and the J domains from 91 type I, 43 type II, and 89 type III DnaJ-like proteins. Only the consensus residues derived from the alignment of all J domain sequences are given. The positions where there is variation among the 4 derived consensus sequences are indicated with an asterisk and are detailed in Table 1
Table 1.
J domain positions that show residue variation among the 4 derived J domain consensus sequences
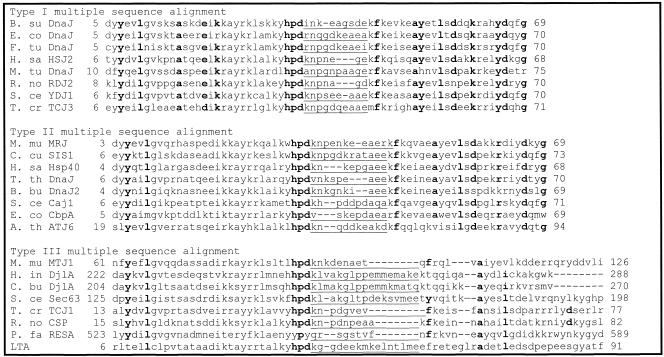
Figure 3 shows multiple sequence alignments done on 8 random J domain sequences for each of the 3 types of DnaJ-like proteins. Figure 3, similarly to Figure 2, illustrates that when using the Cheetham and Caplan (1998) classification to divide the proteins, the J domains of type I and II proteins have a far greater degree of conservation than those of type III proteins, particularly in the loop region. In addition, there is a decreasing level of conservation from type I to type III J domains, particularly with respect to the loop region. In general, the loop region is highly variable; however, in type I proteins, there appears to be a preference for certain amino acids (Table 1; Figs 2 and 3). There is a high level of identity in type I J domains, and these proteins have shorter loop regions, with a high percentage of glutamic acid and alanine residues in that region. Type II loop regions are slightly longer but retain the higher proportion of glutamic acid and alanine. Type III J domains are less conserved and have highly variable loop regions, both in terms of length and amino acid type with no bias toward glutamic acid and alanine. Intriguingly, the final amino acid of both type I and II domains is a highly conserved glycine (greater than 80%), which potentially reflects the J domain merging into the glycine-phenylalanine region.
Fig 3.
Multiple sequence alignments of selected J domain sequences. Bold letters indicate residues with levels of identity of greater than 90% in each consensus sequence. Underlining indicates the loop region. The position of the J domain in each protein is indicated. Organism names are abbreviated as follows: A. th, Arabidopsis thaliana (ATJ6—AAB91418); B. bu, Burrelia burgdorferi (DnaJ2—AAC66991); B. su, Bacillus subtilis (DnaJ—P17631); C. bu, Coxiella burnetii (DjlA—Q45885); C. cu, Cryptococcus curvatus (Sis1—CAA72798); E. co, Escherichia coli (DnaJ—P08622; CbpA—P36659); F. tu, F tularensis (DnaJ—P48207); H. in, Haemophilus influenzae (DjlA—P44607); H. sa, Homo sapiens (HSJ2—P31689; Hsp40—BAA08495); M. mu, Mus musculus (MRJ—O54946; MTJ1—Q61712); M tu, Mycobacterium tuberculosis (DnaJ—P07881); P. fa, P falciparum (RESA—Q26005); R. no, Rattus norvegicus (RDJ2—AAB64094; csp—I52655); S. ce, Saccharomyces cerevisiae (YDJ1—P25491; Caj1—P39101; Sec63—P14906); T. cr, Trypanosoma cruzi (TCJ3—AAC18896; TCJ1—AAC18894); and T. th, Thermus thermophilus (DnaJ—Q56237). LTA indicates large T antigen from the budgerigar fledgling disease virus (P13894)
Pairwise alignments were performed using the overall consensus sequence against 3 different J domains, where E coli DnaJ (Bardwell et al 1986) is a type I protein, the human protein HDJ1 (Raabe and Manley 1991) is a type II protein, and the murine DnaJ-like protein MTJ1 (Brightman et al 1995) is a type III protein. The results are shown in Table 2. The structure of the J domains from DnaJ and HDJ1 have both been determined, hence their use in these alignments (Pellechia et al 1996; Qian et al 1996). This alignment also reflects the greater degree of conservation between the J domains of type I and type II DnaJ-like proteins compared with the J domains from type III DnaJ-like proteins.
Table 2.
The percentage of identity and similarity for DnaJ HDJ1, and MTJ1 when aligned against the derived overall consensus sequence
Functional implications and the identification of an additional conserved motif
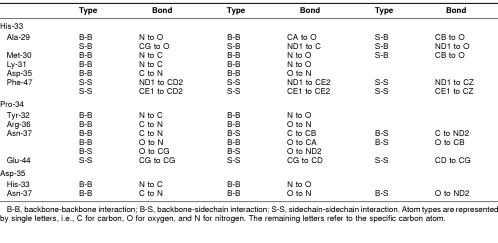
The Whatif program was used to establish residues potentially making contact with the highly conserved HPD tripeptide within the J domain (Chinea et al 1995; Vriend 1990). The E coli DnaJ J domain structure (1XBL) was used for this purpose. The results are summarized in Table 3 and pictured in Figure 4.
Table 3.
Potential interactions within 4 {Å} of the HPD tripeptide in the DnaJ domain
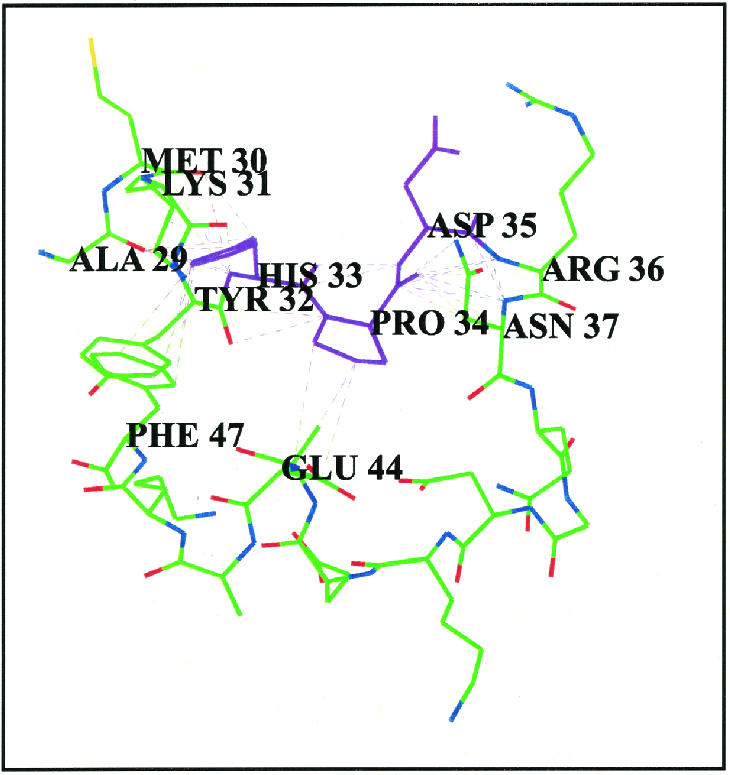
[om30p][bu401,16p,-6p]Fig 4. Residues that are potentially in contact with the conserved HPD tripeptide in E coli DnaJ. The HPD motif is in purple. Potential interactions are indicated as dotted lines. This model was developed using the Whatif program (Chinea et al 1995; Vriend 1990). A 4-Å cutoff limit was used
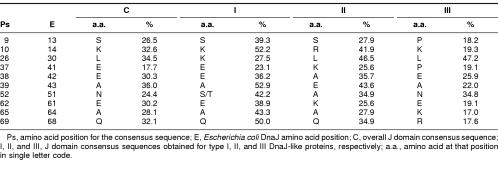
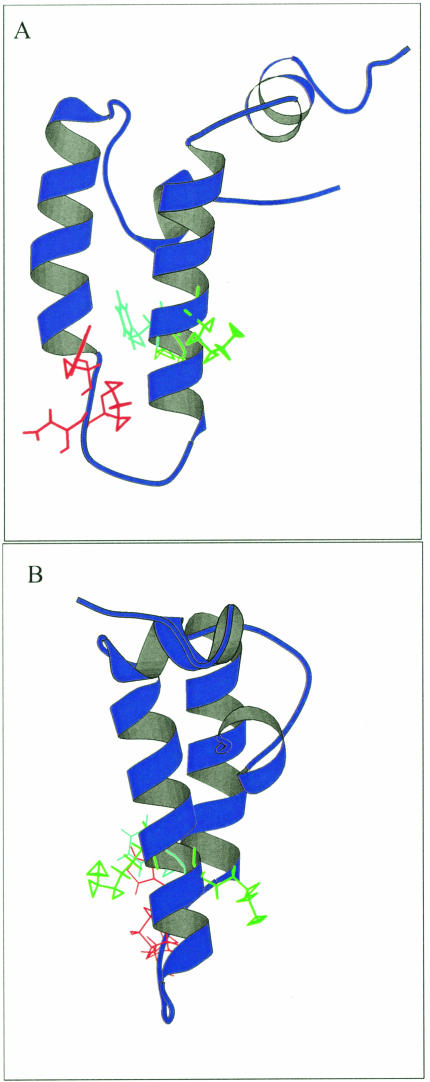
Many of the residues that have potential interactions with the HPD tripeptide are highly conserved, with the exception of Met-30. The consensus sequence contains a leucine residue at the position of the methionine residue (Leu-26 in the consensus sequence, Met-30 in E coli DnaJ). Intriguingly, this residue is not conserved across all 4 consensus sequences, with a lysine being more common than the leucine in type I domains (Table 1). Consequently, there is a high degree of likelihood that this residue is not important in a structural sense and may, therefore, be playing a role in specifying interactions with Hsp70 proteins. Other residues of interest include a phenylalanine at position 47 in E coli DnaJ. This residue is highly conserved and appears to be involved in many potential interactions with the histidine at position 33, generally for DnaJ-like proteins. Positive residues, often lysines that appear to flank this bulky, hydrophobic residue, give rise to what could be termed a KFK motif. Once again, the exception occurs in type III proteins, where there is just less than 40% lysine conservation at the 2 positions, although positively charged residues occur in half the type III sequences (data not shown). The phenylalanine-histidine interaction could be very important in terms of stabilizing the α-helices II and III of the J domain, in addition to the antiparallel bonding between the 2 helices. The flanking lysines generally protrude outward and could possibly play a role in interaction with Hsp70 or other regions of DnaJ-like proteins. The orientation of the phenylalanine and lysine residues in the structure of E coli DnaJ, with respect to the HPD motif, is depicted in Figure 5.
Fig 5.
Ribbon representation of the J domain in E coli DnaJ showing the KFK motif and the HPD motif. The structure used is 1XBL. The HPD motif is in red, the phenylalanine is in light blue, and the 2 lysines on either side of the phenylalanine are in green. The model was made using Molscript (Kraulis 1991). (A) Visualization of the phenylalanine residue with respect to the HPD motif. (B) Visualization of the orientation of the flanking lysine residues
DISCUSSION
In general, type I and II proteins show a greater degree of conservation in the J domain than do type III proteins. This could be a reflection of the fact that most of these proteins are present in prokaryotes where there are single Hsp70-DnaJ partner proteins. This could also be a result of the nearness of the evolutionary relation of type I proteins, as mentioned previously. Consequently, little variation is needed, since the DnaJ-like proteins will only have 1 Hsp70 partner to interact with. The converse of this is that in eukaryotes, where most type III proteins are located, the variation of the J domains is required to create specific DnaJ-like protein-Hsp70 partners. These J domains have possibly arisen as a result of gene duplication events. There might not have been the pressure on these type III J domains to retain the level of sequence identity that type I and type II J domains required, hence leading to a large level of sequence divergence. The evolutionary specialization of eukaryotic cells into functional compartments may have forced functional specialization of type III J domains. Further work could entail looking at the levels of nucleotide conservation, since this would give a better idea of the rate of evolutionary drift.
Presumably type I J domains, with their high level of similarity, can be freely interchanged in domain-swapping experiments, whereas type III J domains may not be able to interact nonspecifically with all Hsp70 proteins. The low level of conservation between type III proteins and the overall consensus sequence is further highlighted when levels of similarity and identity are calculated. A type III protein, such as the mouse protein MTJ1 (Brightman et al 1995), has a lower level of identity and similarity with respect to the J domain consensus sequence than does a type I protein such as DnaJ from E coli. The loop region is highly variable in type III J domains, both in type of amino acids and in length. This aspect could further contribute to specialization of the DnaJ-like protein in terms of facilitating the interaction with specific Hsp70s. There is generally a far lower level of variation in the loop region of type I J domains than in type III J domains. Generally, type I and II J domains have a higher level of alanine and glutamic acid residues in the loop region.
The J domain swapping experiment of Schlenstedt et al (1995), in which only the J domains of endoplasmic reticulum proteins were functionally equivalent, could be explained by the level of conservation and its relation to specialization. In other words, endoplasmic reticulum proteins could have conserved slightly different residues compared with proteins that would normally be located in the cytosol. Alternatively, DnaJ-like proteins could have altered their sequences in response to sequence changes in their partner Hsp70.
Ultimately, several amino acids have been identified that could be interesting in terms of mutagenesis studies. These can be divided into 3 categories: those that are purely structural, those that are important in terms of general interaction with Hsp70, and those that may provide specificity to the interaction with specific Hsp70 partner proteins.
In the first category, that of structurally important amino acids, there is a phenylalanine at consensus position 50 (position 47 in DnaJ), which is predicted to have several potential interactions with His-29 (33 in DnaJ). This phenylalanine is highly conserved, appears to be buried, and is found in another highly conserved tripeptide, KFK. Another residue of interest is the tyrosine located at position 3 on the consensus sequence in α-helix I (Fig 2). There are also 2 very highly conserved residues that are likely to play a structural role on α-helix III, namely, the alanine at consensus position 54 and the leucine at consensus position 58. All the highly conserved residues (>90% identity) appear to be important from a structural perspective. Interestingly, all these residues appear to form 2 clusters in the J domain, with the HPD residues and the phenylalanine forming the first cluster at the base of the J domain and the other residues at the top of the domain forming a second cluster. Hence, the 2 clusters could be stabilizing both the top and the base of the structure. A possible explanation for the deleterious nature of mutations in the HPD motif is the loss of the stabilizing nature of the tripeptide, in turn causing a deleterious effect on the structure.
The second group of residues are those that are likely to be important in terms of interacting with Hsp70. Into this group fall the histidine, proline, and aspartic acid that make up the HPD motif. Common mutations here include the histidine to glutamine mutation. An interesting alternative mutation would be to mutate the tripeptide to a tyrosine-proline-tyrosine, as is found in the RESA protein of Plasmodium falciparum. As far as is known, this double mutation has not been previously performed, and the functional significance of this putative P falciparum J domain is unknown. Other possible amino acids that could be important include a glutamic acid (which can be conservatively substituted to aspartic acid) at consensus position 16 and the aspartic acids at consensus positions 60 and 68. There are also conserved lysines at positions 18, 19, 23, 27, 47, 49, and 63 and an arginine residue at position 22. Recent work has indicated that positively charged residues, such as those mentioned above, could be interacting with negatively charged residues on Hsp70 (Gässler et al 1998).
The third group of residues are those that could potentially be involved in determining specificity. Any residue with a low percentage of occurrence could potentially be involved in determining specificity, since the structure of the J domains appears to be very conserved and any variation could be critical in terms of specificity. This is even more interesting when the generally lower level of conservation of type III J domains is taken into account. Table 1 shows the residues that vary among the 4 consensus sequences. An example is the asparagine residue at position 52 in the overall consensus sequence. This residue is replaced by a serine or a threonine in type I J domains and an alanine in type II. Only the type III sequence retains the asparagine. Another example is that of a leucine located at position 26 in the consensus sequence. There is a methionine in that position in E coli DnaJ, which potentially has an interaction with the HPD motif. This residue is also more likely to be a lysine in type I J domains but a leucine in type II and III J domains. The reason for the variation of these specific residues is unclear. In general, they are found at the end of the helices or in the loop region, with the exception of the asparagine 52 mentioned previously, which is located in the middle of helix III, and the glutamic acid located at consensus position 62 and the alanine located at consensus position 65, both in the middle of helix IV. These latter 2 residues are part of the region corresponding to the QKRAA motif (Auger and Roudier 1997; Suh et al 1999), with the residues EKRAI, that occurs at this point in the overall consensus sequence. The lysine and the arginine residues are significantly conserved, possibly implying that they are also important for the general binding of DnaJ-like proteins to Hsp70s, whereas the glutamic acid and the alanine show low levels of conservation and may be important in specificity.
The variation among the consensus sequences could also in part be ascribed to genetic drift and the differences that are seen are as a result of the phylogenetic distances. However, phylogenetic distance needs to be considered in a functional context. Functionality must affect the rate of evolution of a protein, resulting in different levels of conservation for different amino acids. Consequently, those residues that are important in terms of the maintenance of a correct structure or for generalized interaction with Hsp70s will be conserved in all J domains, whereas those residues important for determining specific DnaJ-like protein-Hsp70 partners will be less conserved and selected for, depending on the type of DnaJ-like protein.
Further work remains to be done with respect to comparing the conserved residues with conserved residues in the various domains of the Hsp70s. Work has recently been published postulating regions of potential interaction between DnaJ and DnaK, the E coli Hsp70 (Gässler et al 1998; Suh et al 1998). There is some evidence that DnaJ binds to 2 regions on DnaK (Suh et al 1998). The first is in a cleft formed on the underside of the ATPase domain of DnaK, and the second is near the substrate binding domain on DnaK. The critical interaction described in this work is the interaction at the ATPase domain. A mutation located in the cleft in the ATPase domain, namely, R167H, suppresses the mutation D35N, D35 being part of the HPD motif. There are also 2 residues that are located fairly close to Arg-165, namely, Asn-170 and Thr-173, that when mutated to alanines cause a decrease in DnaJ binding. Some work has indicated that the critical area of interaction between the J domain and a partner Hsp70 lies between residues 1 and 35, ie, helices I and II and part of the loop region (Greene et al 1998; Lu and Cyr 1998). There are several highly conserved residues in this region, which could then be critical in mediating this interaction. Consequently, there is a high likelihood that the J domain binds to the cleft in the ATPase domain, with the α-helices generally forming a structural scaffold, and several key residues mediate the general interaction and other residues mediate the specificity of the interaction. It remains to be seen if some of the hypothetical conclusions of this study could be extended to similar work on the Hsp70 family of proteins.
It appears that the presence of the glycine-phenylalanine rich region may be required for effective stimulation of an Hsp70's ATPase activity (Cheetham and Caplan 1998). This adds an interesting sidelight to the low level of conservation among type III J domains. Work has indicated that type I and II J domains cannot stimulate Hsp70 ATPase activity without the glycine-phenylalanine rich region (Szabo et al 1996). Type III J domains do, however, appear to be able to still stimulate ATPase activity. Several questions, therefore, need to be answered. First, does the lower level of sequence conservation in type III J domains indicate a change in the overall structure of the J domain, rendering the glycine-phenylalanine rich region unnecessary? Homology modeling does not show any major conformational changes; however, until the structure of more J domains is known, particularly that of the type III domains, this question cannot be answered unequivocally. Second, does the presence of the glycine-phenylalanine region contribute to the specificity of binding between type I and II DnaJ-like proteins and Hsp70s? Third, is the lower level of similarity of these type III domains due to additional interactions between the domain and the rest of the protein? Once again, in the absence of sufficient structural data, it is difficult to answer this question. It is also likely that there will be vast differences here with respect to type III proteins, since they have no other common domains.
In conclusion, the work presented herein identifies several residues in the conserved J domain that may be critical in terms of mediating the interaction with Hsp70. This may suggest a mechanism for determining the specificity of Hsp70—DnaJ-like protein interactions, since less conserved residues may come into play at this point, with the HPD motif being necessary for the general interaction mechanism and the stability of the J domain.
Fig 2.
Continued
Fig 2.
Continued
Fig 2.
Continued
Fig 2.
Continued
Acknowledgments
This work is supported by Wellcome grant 053027 to Drs Cheetham and Blatch. Ms Hennessy is supported by the South African National Research Foundation and the University of the Witwatersrand. Dr Warren Kaplan provided much initial help with Whatif and Molscript. We also appreciate the helpful comments made by the reviewers of the manuscript.
REFERENCES
- Auger I, Roudier J. A function for the QKRAA amino acid motif: mediating binding of DnaJ to DnaK. Implications for the association of rheumatoid arthritis with HLA-DR4. J Clin Invest. 1997;99:1818–1822. doi: 10.1172/JCI119348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell JCA, Tilly K, Craig E, King J, Zylicz M, Georgopoulos C. The nucleotide sequence of the Escherichia coli K12 dnaJ+ gene. J Biol Chem. 1986;261:1782–1785. [PubMed] [Google Scholar]
- Bork P, Sander C, Valencia A, Bukau B. A module of the DnaJ heat shock proteins found in malaria parasites. Trends Biochem Sci. 1992;17:129. doi: 10.1016/0968-0004(92)90319-5. [DOI] [PubMed] [Google Scholar]
- Brightman SE, Blatch GL, Zetter BR. Isolation of a mouse cDNA encoding MTJ1, a new murine member of the DnaJ family of proteins. Gene. 1995;153:249–254. doi: 10.1016/0378-1119(94)00741-a. [DOI] [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaption of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinea G, Padron G, Hooft RWW, Sander C, Vriend G. The use of position specific rotamers in model building by homology. Proteins. 1995;23:415–421. doi: 10.1002/prot.340230315. [DOI] [PubMed] [Google Scholar]
- Gässler CS, Buchberger A, Laufer T, Mayer MP, Schröder H, Valencia A, Bukau B. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc Natl Acad Sci U S A. 1998;95:15 229–15 234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene M, Maskos K, Landry SJ. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc Natl Acad Sci U S A. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Flanagan JM, Prestegard JH. The influence of C-terminal extension on the structure of the J domain in E coliDnaJ. Protein Sci. 1998;8:203–214. doi: 10.1110/ps.8.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley WL. The J-domain family and the recruitment of chaperone power. Trends Biochem Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. Molscript: a programme to produce both detailed and schematic plots of protein structures. J Appl Cryst. 1991;24:946–950. [Google Scholar]
- Lu Z, Cyr DM. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone YDJ1 assist Hsp70 in protein folding. J Biol Chem. 1998;273:5970–5978. doi: 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- Ohtsuka K, Hata M. Mammalian Hsp40/DnaJ homologs: cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones. 2000;5:98–112. doi: 10.1379/1466-1268(2000)005<0098:mhdhco>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellechia M, Szyperski T, Wall D, Georgopoulos C, Wüthrich K. NMR structure of the J-domain and the Gly/Phe-rich region of the Escherichia coli DnaJ chaperone. J Mol Biol. 1996;260:236–250. doi: 10.1006/jmbi.1996.0395. [DOI] [PubMed] [Google Scholar]
- Qian YQ, Patel D, Hartl F-U, McColl DJ. Nucleic magnetic resonance solution structure of the human Hsp40 (HDJ-1) J domain. J Mol Biol. 1996;260:224–235. doi: 10.1006/jmbi.1996.0394. [DOI] [PubMed] [Google Scholar]
- Raabe T, Manley JL. A human homologue of the Escherichia coli DnaJ heat-shock protein. Nucleic Acids Res. 1991;19:6645. doi: 10.1093/nar/19.23.6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Harris S, Risse B, Lill R, Silver PA. A yeast DnaJ homologue, Scj1p, can function in the endoplasmic reticulum via a conserved domain that specifies interactions with Hsp70s. J Cell Biol. 1995;129:979–988. doi: 10.1083/jcb.129.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh W-C, Burkholder WF, Lu CZ, Zhao X, Gottesman ME, Gross CA. Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc Natl Acad Sci U S A. 1998;95:15 223–15 228. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh W-C, Lu CZ, Gross CA. Structural features required for the interaction of the Hsp70 molecular chaperone DnaK with its cochaperone DnaJ. J Biol Chem. 1999;274:30534–30539. doi: 10.1074/jbc.274.43.30534. [DOI] [PubMed] [Google Scholar]
- Szabo A, Korszun R, Hartl F-U, Flanagan J. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured substrates. EMBO J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- Vriend G. Whatif: a molecular modelling and drug design programme. J Mol Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]