Abstract
Despite significant recent advances in molecular and structural studies of G protein-coupled receptors (GPCRs), understanding transmembrane signal transduction with chemical precision requires new approaches. Simple binary receptor-ligand or receptor-G protein complex models cannot describe adequately the relevant macromolecular signaling machines. GPCR “signalosomes” undergo complex dynamic assembly/disassembly reactions to create allosteric signaling conduits whose properties cannot necessarily be predicted from individual elements alone. The combinatorial possibilities inherent in a system with hundreds of potential components suggest that high-content miniaturized experimental platforms and computational approaches will be required. To study allosteric effects involved in signalosome reaction pathways, a bottom-up approach is proposed to use multicolor single-molecule detection fluorescence experiments in biochemically defined systems complemented by molecular dynamics models of macromolecular complexes. In bridging the gap between molecular and systems biology, this synthetic approach suggests a way forward from the flatlands to multi-dimensional data collection.
Keywords: G protein-coupled receptor, G protein, Allosterism, Signalosome, Single-molecule, Fluorescence
Do we need a new conceptual framework to study the relationship between structure and function in pharmacological receptors?
The past few years have witnessed remarkable progress in structural studies of heptahelical (7TM) receptors, which comprise a gene superfamily with more than 700 members in humans. The list of known crystal structures of GPCRs and their associated binding partners is steadily growing [1], and recently the first chemokine receptor was added, CXCR4 [2]. However, it remains an important and challenging problem to understand GPCR-mediated signaling. In addition to the large number of receptors, the number of possible combinations of ternary complexes between receptors and cellular proteins is almost astronomically large. For example, there are at least 21 Gα, 6 Gβ, and 12 Gγ subunits among heterotrimeric G proteins, 7 different GPCR kinases (GRKs), 4 arrestins and potentially other scaffold/adaptor proteins. We contend that to understand fully how GPCRs work in their native membrane environment will require new approaches to probe receptor structural dynamics in bilayers and to detect discrete receptor-ligand binding events and the formation of ternary complexes between activated receptors and their cellular protein partners. We propose a new strategy to determine the relationships between structural dynamics and biological function in macromolecular signaling complexes from mammalian cells, focusing on the 7TM GPCR “signalosome.”
GPCRs are not modular structures. For the most part, they cannot be dissected into fragments, expressed in E. coli and reconstituted in artificial environments. Furthermore, recent advances in cell biology and pharmacology prove that the canonical linear GPCR signaling pathway (receptor–G protein–effector), and the notion that receptors are merely two-state “off–on” switches, are gross oversimplifications. In fact, GPCRs are best thought of as exquisite allosteric machines with multiple “active” states that define state-specific ligand binding affinities/specificities and “signal outputs.” Our long-term strategy, therefore, is to develop a systematic approach and new tools to study the GPCR signalosome – the complex between receptor, ligand and accessory protein(s) in the membrane. Our tactic is to apply several new and exciting technologies to elucidate the relationships between receptor structural dynamics, the formation of supramolecular complexes and signal “output.” An interdisciplinary approach to this important problem will yield significant advances and provide a toolbox of enabling technologies that can be applied to other receptor systems as well. In bridging the gap between molecular and systems biology, this synthetic approach suggests a way forward from the flatlands1 to multi-dimensional data collection.
Signal processing by GPCR signalosomes
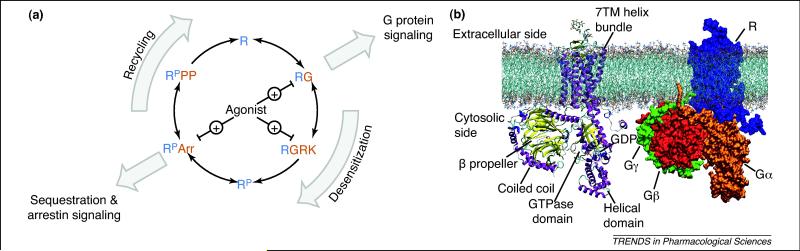
The operations of a GPCR might be analyzed in terms of the abstract concept of a probabilistic finite-state machine. Figure 1a illustrates the key states, transitions, and actions of this signal-processing unit. We can identify the agonist-dependent signaling switch and the phosphorylation-dependent desensitization/recycling switch. Moreover, additional compositional states that could be described as a four-way switch are characterized by the alternative macromolecular complexes (signalosomes) of receptor either with G protein, kinase, arrestin, or phosphatase. On another level of detail, each of these key states can be expanded to show how each of the proteins can be switched in a receptor-dependent manner between two or more states (Box 1, Figure I). With increasing level of detail (temporal and spatial), a given state will exhibit additional metastable configurational and/or conformational substates. Considering this nested hierarchy on the different levels of abstraction that are necessary to capture the reactive dynamics of the GPCR signal-processing unit, it becomes clear that novel experimental and computational methods are necessary to crack the code of how genetic information ultimately determines pharmacology.
Figure 1. Detection and amplification of signals: dynamic GPCR signalosomes as a macromolecular machine.
(a) Cyclic sequence (activation, desensitization, and recycling) of the key states in the GPCR signal transduction process. The corresponding characteristic macromolecular complexes or signalosomes are receptor–G protein (RG), receptor–GPCR kinase (RGRK), phosphorylated receptor–arrestin (RPArr), and phosphorylated receptor–protein phosphatase (RPPP). Note the quasi-irreversible reaction resulting in de-phosphorylated (R) and phosphorylated receptor (RP), respectively. (b) Molecular model of a representative GPCR signalosome, the receptor–G protein complex. Secondary structure cartoon (left) and space-filling model (right) illustrate the receptor–G protein complex in a phospholipid bilayer.
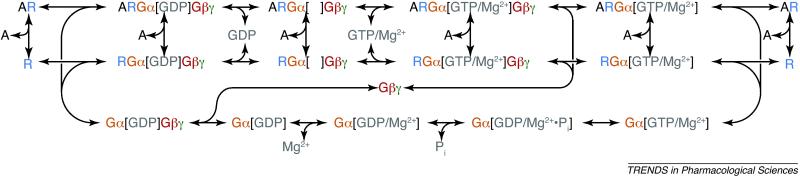
Figure I.
Examples of allosteric modulators (A) are pharmacological agents (agonists, partial agonists, inverse agonists, neutral antagonists, biased ligands), dimerizing receptors, chemical receptor modifications (side-chain protonation or phosphorylation), and physicochemical environmental effects (membrane curvature stress, lipid composition). Despite significant progress in the structural biology of G proteins, little is known about the precise interaction of a G protein with a GPCR (or GPCR dimer), and how this interaction leads to GDP release and the subsequent GTP uptake by the active complex [5]. Intermediate states of the receptor- and Gβγ-dependent nucleotide exchange process and GTPase cycle of Gα are shown. Note that for each state, multiple conformational substates of the participating proteins are possible. These conformational substates might be rapidly interconverting, or alternatively, they might correspond to metastable long-lived conformations that exhibit hysteretic behavior (conformational memory). A pharmacological agonist ligand functions as an allosteric modulator (A) that changes the distribution of conformational substates. The corresponding perturbation of the receptor free energy landscape will affect the binding of the inactive heterotrimer, Gα[GDP]Gβγ, and destabilization of its bound GDP. Structures for components of the lower part of the cycle have been reported.
The computational simulation of such systems requires a hierarchical approach that models each level from the atomistic to the macroscopic scale. Moreover, computational methods allow integration of diverse types of experimental information into comprehensive models of experimentally inaccessible structures. For example, no existing experimental method allows determination of the structure of a cell membrane in atomic detail. However, it is possible to obtain microsecond time scale, all-atom molecular dynamics (MD) simulation models of the receptor–G protein complex in a native-like membrane (Figure 1b). In this way, it might be possible to gain insight into the structural dynamics of this and related GPCR signalosomes and to describe the non-equilibrium thermodynamics of such functional modules. The long-term goal will be to reconstruct the atomistic mechanism that explains the flow of information (energy) through this signal-processing unit.
Multiscale modeling of macromolecular processes
To obtain useful computational models of biological systems is a central goal in science. One approach is to build larger systems bottom-up from smaller subsystems. Each subsystem itself is built from even smaller sub-subsystems. The nested hierarchy of systems has been described as a modular approach to understanding biological systems [3]. At the lowest level, there are atoms and bonds. Together these correspond to the familiar molecular graph, a network representation of atoms as nodes and bonds as edges, with characteristic length and time scales of nanometers and nanoseconds, respectively. Modeling of these systems generally involves quantum chemistry methods.
On the next level, there are molecules, intermolecular interactions and reactions. Here two types of network formulations are common. The first type is a network that shows intermolecular interactions analogous to the chemical bonds on the previous level, and this is typically found in proteomics studies of protein-protein interaction networks, in which the individual proteins are nodes and the interactions are edges. The second type is a network of reactions that represents the interconversion of molecules, a formulation that is reminiscent of the metabolic pathway charts of biochemistry. The protein interaction network encodes information (i.e., energy) in terms of the strength of interactions. The structures corresponding to the connected graphs are molecular complexes and functional modules of interacting molecules. By contrast, the metabolic pathways illustrate the interconversion of nodes corresponding to molecules (which are molecular graphs from the first layer) and the edges corresponding to reactions that are typically catalyzed by (enzymatic) protein functions. These two types of networks are functionally interrelated because proteins undergo reactions (posttranslational modifications such as phosphorylation and de-phosphorylation), and they are therefore also nodes (reactants/substrates or products) of metabolic networks. At the same time, enzymatic function of a protein is dependent on posttranslational modifications and binding partners (allosteric modulators). The enzymatic function itself has molecular complexes as structural correlates – the transient complexes of the enzyme with reactant(s), transition state, and product(s). Biomolecular simulations approximate the energetic interactions between atoms by classical mechanical force fields that are parameterized to fit the quantum chemical potential energy surface and condensed phase solvation properties of representative molecular fragments. Coarse-grained models simplify calculations of these networks by collecting groups of strongly interacting nodes into a single node; for example, one to five coarse-grained beads might represent an amino acid [4].
In metabolic networks, the nodes are molecules, and the nodes are connected by a directed graph of reactions. Connected sub-graphs represent reaction pathways. The molecules and reactions may be segregated in different cellular compartments, such as plasma membrane and cytoplasm. In this way, these networks encode localization (spatial information) and can be mathematically described by systems of ordinary differential equations (ODEs) with concentration terms encoding the size of nodes and rate constants determining the reactive flux between nodes. As biological systems operate out of thermodynamic equilibrium, the metabolic network is typically found in a quasi steady state. Perturbations of this steady state will lead to a characteristic temporal response profile of the nodal concentrations in the network.
From molecular to systems biology of GPCRs
Here we suggest how (in principle) the pharmacological action of drugs could be analyzed in this bottom-up approach to systems biology. We propose that a complete understanding of the 7TM receptor allosteric machinery on a structural and dynamical level might require rethinking of the existing terms of molecular biology and pharmacology. In particular, the analysis of structure-function relationships by biomolecular simulations will require new approaches to quantify function on an appropriate level. Biomolecular simulations are useful to compare and predict the relative stability of closely related structures. Therefore, they provide a platform to study models of homologous structures derived from experimentally determined template structures. It is important to analyze the reaction pathways in signal transduction networks to identify relevant targets for detailed analysis. For example, to compare two related pharmacological antagonists with different affinities for the receptor, one must perform free energy calculations with so-called “alchemical” transformations of one ligand into another. These calculations are once performed in the receptor-bound state and once in solution. The difference, together with the appropriate thermodynamic cycle, should correspond to the changes in dissociation constants and the corresponding difference in free energy. In this case, the existing set of crystal structures of antagonist-bound GPCRs is useful.
It is more complicated to compare two related agonists. The two ligands will differ in their affinity for the receptor, but agonist binding will lead to a shift in the equilibrium of metastable conformations of the receptors. Uncertainty about the structure of these metastable states is a problem because available crystal structures typically only give the inactive antagonist-bound ground state of the receptor and biomolecular simulations are currently not powerful enough to cover the time scales relevant to sample these slowly interconverting metastable states. Moreover, the pharmacological characterization of an agonist ligand will identify two different quantities, potency and efficacy. The efficacy reflects the extent of the pharmacological response at saturating concentrations relative to another (standard) agonist. Potency reflects the concentration necessary to elicit this response, which is typically determined as the concentration at half-maximal response (EC50).
To account for these two different qualities, one could consider the receptor as an enzyme that catalyzes the nucleotide exchange reaction of the G protein cycle. Note that the catalytic action of the receptor on the G protein cycle is not a classical enzymatic reaction because there is no change in the covalent bond structure; it is rather guanine nucleotide exchange factor (GEF) activity. Generally, the Michaelis constant (Km) and the maximum velocity (Vmax) characterize the enzymatic function. Allosteric modulation of enzymes results in changes of Km (K-type), Vmax (V-type), or both (mixed-type).
At a given concentration of G proteins, changes in Km and/or Vmax will both affect the rate of nucleotide exchange. In addition, the rate of desensitization (by receptor phosphorylation and arrestin binding) will limit the extent of G protein activation in vivo. Therefore, measurement of GTPγS uptake (a non-hydrolyzable GTP analogue) in plasma membrane preparations eliminates the contribution of desensitization due to absence of ATP in the reaction.
Calculations of reaction rates from biomolecular simulations are not trivial. If it is possible to identify a transition state then the rates can be related to its stability versus the reactants. As shown in Box 1, Figure I, the nucleotide exchange reaction can be broken down into several microscopically reversible reactions. The apparently irreversible GTP uptake reaction is the result of the large excess of GTP over GDP in live cells.
Allosteric modulation and mechanism of functional selectivity and conformational memory
The allosteric effect of the agonist on the catalysis of the nucleotide exchange reaction of the G protein cycle could be the result of changes in the stability of virtually any of these intermediates and transition states shown in Figure I. For example, the ability of an agonist-bound receptor to recruit G proteins might be different from its ability to induce nucleotide exchange and hence activation of the G protein. This idea stems from the observation that certain mutants of rhodopsin bind but fail to activate its G protein transducin [23]. This raises the question of whether or not a single active state of the agonist-bound receptor is responsible for binding and activation of the G protein.
Allosteric effects are bidirectional. The affinity of agonist ligands in presence and absence of G proteins allows measurement of the free energy contributions of allosteric coupling between the agonist and G protein binding sites. This G protein effect on agonist binding affinity is dependent on the concentration of GDP (which does not lead to G protein activation) as well as GTPγS (which irreversibly activates G proteins). In addition, we need to develop new methods (such as multicolor single-molecule detection (SMD) fluorescence methods) to measure the rate constants of the individual assembly and disassembly steps of the G protein activation reaction pathway. In this novel experimental paradigm, it is necessary to identify key states and corresponding molecular assemblies, and to characterize the transitions between these states. In this way, allosteric effects of agonists on transition states should be experimentally accessible.
The next open question, which is related to the issue of G protein specificity, is the mechanism of functional selectivity of different agonists. There is increasing evidence for ligand-specific signaling states of 7TM receptors [24]. Two related agonists on one receptor might exhibit different ratios of efficacies with respect to two downstream signaling pathways. For example, one ligand might preferentially activate Gq, whereas the other preferentially stimulates Gi. There might be preference within a single class, for example, Gi1 versus Gi2. Moreover, it might be possible that the receptor bound to one ligand or another prefers particular combinations of Gβ and Gγ subunits. Alternatively, functional selectivity might also arise as a switch from G protein signaling to arrestin signaling. In this case, phosphorylation of the receptor by GPCR kinases (GRKs) is responsible for the switch. Because Gβγ recruits (localizes) some GRKs to the plasma membrane, two ligands might have different ability to modulate allosterically the catalysis of this reaction. Moreover, functional selectivity could result from differential effects on G protein activation and desensitization. Alternatively, the functional selectivity might be explained by a preference of a ligand for the ternary complexes with G protein or arrestin (after phosphorylation). There is also a possibility of a phosphorylation “barcode” encoded by selective recruitment of various kinases.
Ligand binding/dissociation kinetics of orthosteric antagonists has been shown to play a role in receptor pharmacology resulting in non-competitive (insurmountable) antagonism versus competitive (surmountable) antagonism [25]. By analogy, different agonist ligands might affect the structural dynamics within the receptor in a way such that the distribution of states is largely unaffected, but instead the kinetics of exchange between them is affected. Simply stated, one ligand might “mobilize” the receptor more than another. The mobilized receptor might be relevant for activation of a large number of G proteins, whereas desensitization might be independent of receptor conformational mobilization. In that way, two ligands might induce the same percentage of high-affinity binding sites for the G proteins, but only one ligand will promote rapid equilibration with the low-affinity binding site. These are the types of issues that we intend to address.
G protein pre-coupling
Yet another related question is the possible role of G protein pre-coupling to the receptor. In one scenario, an un-liganded receptor might pre-select a specific G protein that it might be less likely to couple to after activation. Pre-coupling of receptor and G protein is unusual, but has been reported in some cases [26, 27]. For example, recent findings support a physiologically relevant pre-coupling mechanism in the case of pre-assembled, ligand-independent GPCR signalosomes of the relaxin receptor [28]. One the other hand, fluorescence recovery after photobleaching (FRAP) and SMD imaging experiments were used to study coupling of G proteins to cAMP receptor 1, a chemoattractant receptor, in Dictyostelium. The results support a ligand-induced coupling mechanism similar to that in rhodopsin and disfavor pre-coupling [29].
Moreover, pre-coupling might be required for initial recognition of agonists. In this case, the binding of the G protein would switch the conformation of the receptor to present a high-affinity binding site for the agonist ligand. The initial observation that purified CCR5 chemokine receptor cannot bind its natural ligands, MIP-1α, MIP-1β, or RANTES [30] at all, or with almost three orders of magnitude lower affinity in certain reconstituted systems as compared to cell membranes [31], may be explained by an unusually strong allosteric effect of G protein binding [32]. Here it will be interesting to observe subsequent rounds of G protein binding and activation. It is possible that after a single round the agonist dissociates. In this case the amplification of the “particle” detector would be “one” – one activated G protein for one agonist-binding event. Such self-limiting detection might have biological advantages since receptors would be instantaneously desensitized and recycled. Another alternative is that binding of the agonist would switch the conformation of the receptor that would then present a binding site for the G protein.
Early and relatively primitive SMD fluorescence experiments suggested conformational substates (“conformational complexity”) in the β2 adrenergic receptor [33], and this dynamic behavior was cited as one of the reasons that crystallization of GPCRs has been especially challenging for high-resolution structural analysis of GPCRs [34]. Such conformational complexity and the existence of multiple functional microdomains, switches, and ionic locks have also been demonstrated by mutagenesis and spectroscopy for rhodopsin [35-37]. However, while the “dynamic personality” of proteins [38] and the conformational complexity of GPCRs have gained acceptance, it is still unclear whether a single “on” state is responsible for G protein activation or not. In this case, all other conformational substates might be lumped into an “off” ensemble of states.
In addition, there is the possibility of a conformational “memory effect.” SMD fluorescence studies revealed evidence of molecular memory phenomena [39, 40]. It will be important to identify conformational memory effects in GPCRs, which might have a potential role in explaining the functional dynamics of GPCRs. For example, does the high-affinity ligand-binding state of the receptor, once formed by the action of a G protein, retain this state (at least transiently) after G protein dissociation? Relaxation of ligand-, or G protein-induced conformational states might be slow enough to allow the receptor to retain a pharmacologically relevant “active state” conformation even after dissociation of agonist or G protein subunit. In this way, the allosteric interaction between agonist and G protein would not require simultaneous occupancy of both binding sites. Related to the issue of G protein pre-coupling is a proposal to explain apparently allosteric interaction of different receptors by a G protein “steal” mechanism as an alternative to direct allosterism in a receptor heterodimer [41]. In this light, it will be important to characterize the ability of ligands to stabilize the receptor–G protein complex without the necessity to induce nucleotide exchange.
New experimental approaches: the chemokine receptor “signalosome”
We suggest that SMD fluorescence studies will shed light on the question of how to detect and define ligand-dependent signaling states of the receptor. Our own research program is focused on developing SMD fluorescence methods, namely SMD-TIRF (total-internal reflectance fluorescence) microscopy, which we will use to study chemokine receptors in bilayers. Chemokine receptors are involved in directed cell migration – or chemotaxis – of certain cell types during development, the immune response, inflammation, wound healing, stem cell maintenance/proliferation, and other physiological processes. Moreover, they play a role in the pathophysiology of tumor metastasis and retroviral infections. There are approximately 20 different chemokine receptors and 50 different types of chemokine ligands. There is significant “cross-talk” or “promiscuity” in chemokine receptor signaling as several chemokines can bind one receptor, and one chemokine might bind several receptors [42]. The pharmacology of chemokines is not well understood. A ligand bound to a receptor might act as an agonist, a partial agonist, a neutral antagonist, or an inverse agonist with respect to its ability to alter the equilibrium of inactive and active conformational state(s) of the receptor. The biology of chemokine receptors, with multiple cell-specific expression patterns, and how they function in a complex milieu of multiple chemokines at different concentrations that change rapidly over time is a complete mystery. CCR5 serves as the primary HIV-1 co-receptor and couples predominantly to Gαi2β1γ2, GRK2 and potentially GRK3, and arrestin-2 in their host cells [43, 44]. Besides studying these interactions that are part to the canonical signal transduction pathway of GPCRs, it will be interesting to develop methods observe the supramolecular structures involved in the early events in HIV-1 entry.
The interaction of the viral envelope protein (gp120-gp41) with the primary receptor CD4 on the target cells results in conformational changes within gp120. This new conformation of gp120 allows binding to the co-receptor (CCR5 or CXCR4) that triggers virus entry, which involves insertion of the fusion peptide gp41 in the cellular membrane. It has been shown that CD4 and CCR5 (or CXCR4) are each organized into homogeneous microclusters in the cell membrane. These CD4 clusters and chemokine receptor clusters frequently close enough to interact with a single HIV-1 virion [45]. It is thought that these microclusters are due to association of CCR5 with lipid rafts, which are important for HIV entry [46]. The lipid raft concept was highly controversial at first, but recent advances have demonstrated the present of nanoscale, dynamic, ordered assemblies of proteins and lipids, which are rich in cholesterol, saturated hydrocarbon chains, and sphingolipids [47-50]. We anticipate that SMD experiments with chemokine receptors reconstituted in a chemically defined lipid environment will give insights into the physicochemical driving forces involved in formation of these microclusters in live cells. These experiments will provide a platform for testing the role of other proteins, such as chaperones [51] and other scaffolding or adaptor proteins, in the formation of receptor oligomers. Moreover, it will be exciting to see whether the known lipid effects on rhodopsin can be generalized to chemokine receptors or not. For example, curvature and hydrophobic forces drive oligomerization and modulate activity of rhodopsin in the membrane [4, 52-54].
Understanding precise mechanisms of pharmacological allosteric GPCR modulators is another issue that can be addressed using the same technology that we propose to develop to study the chemokine receptor signalosome. Adding to the pharmacopoeia of drugs targeting the chemokine family of GPCRs is urgently needed [55]. For example, despite the lack of clinical efficacy of CCR1 antagonists in some trials [56], they have the exciting property of being able to suppress colon cancer liver metastasis [57]. Moreover, a CCR5 agonist drug might be expected to display adverse effects, but a drug that would selectively trigger CCR5 internalization would be clearly useful [58]. Defining the structural dynamics of chemokine receptor signaling might facilitate rational approaches to drug discovery. Although developing new approaches to GPCR-based drug discovery might be a long-term consequence of such work, we will focus here on developing and applying a useful toolkit and a conceptual framework for how to study the structural dynamics of chemokine receptor signalosomes.
Supported membranes for surface-selective spectroscopy and SMD-TIRF microscopy of CCR5
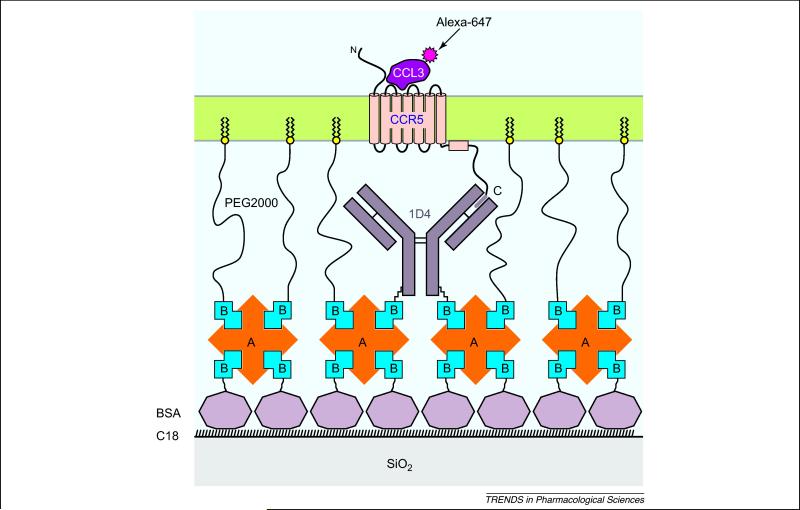
We have developed a strategy for functional reconstitution of CCR5 in solid-supported tethered phospholipid bilayer membranes for single-molecule fluorescence experiments (Figure 2). SMD-TIRF experiments allow selective excitation of fluorophores in the evanescent wave penetrating about 100-nm deep into the aqueous layer beyond the glass/water interface. Capturing a molecule of interest in this spatial region allows long-term observation of a single molecule. In this way, slow dissociation/association processes can be observed, and studies of conformational memory effects will be possible. Few general methods exist to orient proteins on surfaces, especially at high density [81]. Solid-supported bilayers are important models of biomembranes that enable certain classes of biophysical experiments [82]. One of the technical challenges is how to incorporate transmembrane proteins into these membranes. There are two lines of approaches successfully used for GPCRs.
Figure 2. Functional reconstitution of CCR5 in solid-supported tethered phospholipid bilayer membranes.
We developed the following method to self-assemble the depicted structure by sequential perfusion of the different protein solutions in a simple semi-microfluidics chamber [93]. The glass surface is first coated with octadecyl (C18) trialkoxy-silane. The binding capacity of the C18 modified glass for biotinylated BSA is substantially higher as compared to untreated glass [40]. The adsorbed layer of biotinylated BSA is further stabilized by neutravidin (A), a deglycosylated form of the tetrameric biotin-binding protein avidin with a more neutral isoelectric point. The anti-rhodopsin 1D4 mAb that recognizes the C-terminal nonapeptide of rhodopsin (“C9-tag”) is biotinylated specifically on the Fc stem. It is captured by avidin and the remaining biotin binding sites are saturated with biotin-PEG2000-DSPE. The mAb captures heterologously expressed CCR5 with an engineered C-terminal C9-tag from detergent solution. A lipid bilayer (light green) is formed by dilution of POPC/CHAPS bicelles. The fluorescently labeled chemokine (CCL3/MIP-1α-Alexa-647) is added in the last step, utilizing BSA as carriers.
In the first approach, detergent-solubilized receptor is rapidly diluted below the critical micellar concentration (CMC) in the presence of preformed phospholipid bilayer membranes. This method has been used to incorporate rhodopsin [83], the δ-opioid receptor [84], and the β2 adrenergic receptor into black lipid membranes for surface plasmon resonance (SPR) spectroscopy experiments, and the δ-opioid receptor into polyacrylamide-supported lipid bilayers on a fused silica cover slip for SMDTIRF experiments [85]. The drawback of this method is that the orientation of the membrane protein is difficult to control [86]. Moreover, protein aggregation competes with the process of incorporation and only a very small fraction of the protein eventually becomes incorporated into the membranes [83].
The second strategy to incorporate transmembrane proteins into membranes overcomes these shortcomings. It is based on capturing of receptors in defined orientation to the surface, followed by reconstitution of the supported lipid bilayer by micellar dilution. This method has been used to incorporate biotinylated rhodopsin (with carbohydrate-specific biotinylation of the receptor on the extracellular side facing the surface) captured by immobilized streptavidin for SPR spectroscopy [87], as well as the chemokine receptors CCR5 [30] and CXCR4 [88] for biochemical studies, in which both receptors were modified with a C-terminal C9 tag and captured using randomly oriented 1D4 mAb covalently immobilized on paramagnetic polystyrene beads. Moreover, N-terminal FLAG-tagged, fluorescently labeled β2 adrenergic receptor in detergent has been captured by biotinylated M1 anti-FLAG mAb immobilized to streptavidin on biotinylated BSA covered glass for TIRF experiments [89]. TIRF experiments have also been reported for plasma membrane fragments containing NK1 receptor biotinylated in vivo at its C-terminus, which was subsequently captured by streptavidin immobilized on quartz slides partially covered with biotinylated BSA [90].
Based on a combination of methods from these earlier studies, we designed a novel strategy suitable for SMD-TIRF experiments of CCR5. This new strategy should fulfill the following criteria: i) utilize glass or fused silica substrates; ii) allow for immunoaffinity-based capture of epitope-tagged receptors from detergent solution in a defined orientation; and iii) support formation of the bilayer membrane independently of the density of captured transmembrane proteins. Although one earlier study utilized lipid head-group biotinylated phospholipids bound to streptavidin as a membrane anchor [30], in our case it was necessary to introduce an additional spacer between the biotin-binding sites of the neutravidin layer and the bilayer membrane, because it was necessary to bridge the thickness of the IgG molecule that was also anchored to the neutravidin layer. A sufficiently long spacer was provided by biotinyl-PEG2000-DSPE, which can be stretched to about 10 nm [91]. The equilibrium separation depends on the lateral packing density and should be in the range of 4 to 7 nm [92], which is sufficient for globular proteins in the 100-kDa range. Moreover, we observed that the formation of the supported membranes upon dilution of a CHAPS/POPC bicelle system in some cases leads to the formation vesicles instead of a flat membrane. The desired formation of a flat membrane was critically dependent on the surface density of the biotinyl-PEG2000-DSPE anchor lipids, and hence the neutravidin surface density.
The number of available SMD fluorescence techniques is large and steadily growing, but for the study of GPCR signalosomes, a smaller number of methods appear to be especially well suited (Box 2). They are based on: i) intensity and residence time of fluorescence spots, ii) dual-color colocalization (>10 nm), and iii) FRET (3–10 nm). The unique feature of SMD-TIRF methods is that detailed kinetic information is accessible from observations of microscopic reversibility in a system in macroscopic equilibrium or under steady-state conditions. In this way, one can follow time-dependent pathways of reactions that are impossible to synchronize at the ensemble level [94] and use this property to determine the kinetics of transitions between assembly states. Obviously, conventional TIRF experiments may be performed in a complementary fashion.
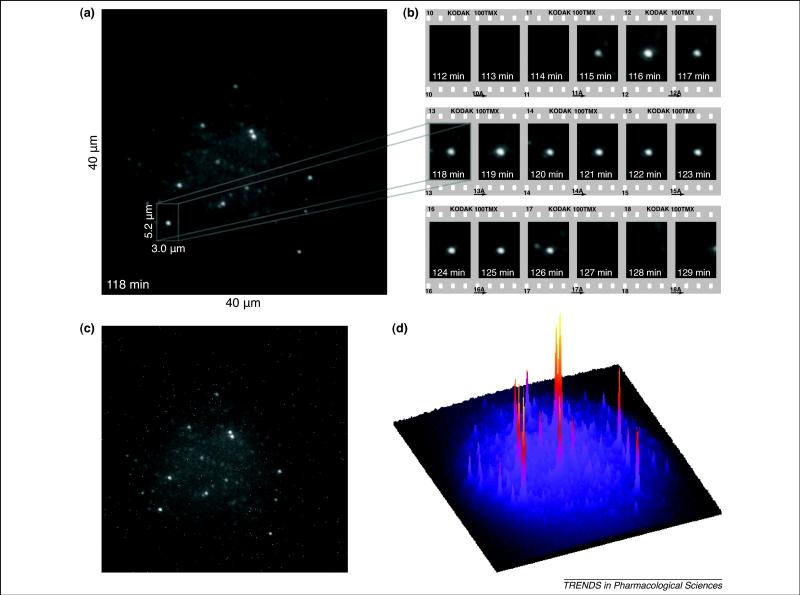
We obtained SMD images of single fluorescently-labeled chemokines (MIP-1α-Alexa-647) are shown bound to recombinant expressed CCR5 embedded in a bilayer membrane tethered to the surface of the chip (Figure 3). The fluorescence spots represent single fluorescent chemokines that have bound to CCR5. The binding event is visible since it occurs at a distance that is within the evanescent field of the TIRF illumination. The membrane-receptor assembly is stable for hours and can be interrogated repeatedly using the continuous flow cell strategy described above. For example, the MIP-1α-Alexa-647 can be washed out of the cell and a different concentration of MIP-1α-Alexa-647, or a different labeled chemokine, can be added in turn. Competition experiments with small molecules or with chemokine mixtures can be carried out as well.
Figure 3. Representative SMD-TIRF images of single chemokine molecules bound to CCR5 in tethered membranes.
(a) Image obtained with the SMD-TIRF microscope after subtraction of static background from ‘hot’ pixels of the CCD sensor and after application of a 2D convolution with a spot enhancing filter that was shown to be the optimal detector of a Gaussian-like spot in 1/ω2 noise [95]. It shows two classes of single-fluorophore spots. One side of the quadratic image corresponds to 40 μm in the sample plane. (b) Time series of the indicated region-of-interest from (a) with 10 seconds exposure time taken in one-minute intervals. The bright central spot corresponds to single fluorescently-labeled chemokines (CCL3/MIP-1α-Alexa-647) bound to CCR5 embedded in a bilayer membrane (see Figure 3) that is visible from 115 to 126 min. (c) Raw image corresponding to (a) before image processing. (d) A surface plot illustrating intensity of the spots as peaks. The tall signals are from single ligand-receptor complexes. The small signals (about 15% of the height of the Alexa-647 signals) are due to hydrophobic fluorescent impurities in a buffer to improve photostability and blinking of Alexa-647 [96]. We found that these impurities can efficiently be removed (not shown) by hydrophobic-affinity chromatography of the employed enzymes (glucose oxidase or catalase). This scheme allows acquisition of hundreds of images with sufficient signal-to-noise ratio to identify individual receptor-bound chemokine molecules before irreversible photobleaching.
Escaping the flatlands one molecule at a time: concluding remarks
To elucidate the structural dynamics of the GPCR signal-processing unit, it will be necessary to “color-code” different proteins with fluorescent probes. In biochemically-defined systems, the multicolor SMD-TIRF techniques will enable us to determine the molecular composition of individual protein complexes (assembly states) and to determine the equilibrium distribution of different assembly states. A time-ordered description of the assembly/disassembly processes of these complexes, which correspond to transitions between key states or functional intermediates, can be obtained. In this way, we will elucidate the potential roles of hysteresis (conformational memory), of sequential and/or cooperative processes, and of allosteric effects. A series of homologous components will be analyzed for the hypothetical role of these effects in the phenomenon of biased agonism (functional selectivity), if possible. We want to emphasize that this type of information is complementary both to structural work on static complexes, and to molecular pharmacology and cell biology.
Box 1. Allosteric modulation of the receptor–G protein interaction in the G protein cycle.
Recent reports of the structure of ligand-free opsin and opsin [6, 7] in complex with an active-state stabilizing peptide derived from a G protein provide additional clues about the conformation of the active receptor. For most GPCRs, a “fully-active” state might exist in a ternary complex of an agonist-bound receptor and a G protein. How the receptor switches to its active state to induce the release of bound GDP from the G protein α subunit is not known. In the G protein heterotrimer, the Gβγ dimer acts as a guanine nucleotide dissociation inhibitor (GDI), whereas the receptor functions as a GEF. At the same time, Gβγ enhances the receptor-dependent nucleotide exchange, possibly by restricting the orientation of Gα on the membrane. Experimentally, several discrete steps in the G protein-activation pathway have been defined (Figure I): the receptor–G protein complex first exhibits reduced affinity for the bound GDP nucleotide [9], the GDP consequently dissociates [10], and finally GTP/Mg2+ binds to the empty nucleotide-binding pocket and causes G protein release [11]. In fact, in rod outer segment disc membranes, affinities of the receptor-G complex for GDP and GTP were measured 25 years ago [9]. Crystal structures are available for the isolated Gαt[GTPγS] [12] and the Gβγ heterodimer [13]. The interaction of Gα[GTP] with its effector, for example, phospholipase C (PLC), or with a regulator of G protein signaling (RGS) protein results in increased GTPase activity. These proteins therefore act as GTPase-activating proteins (GAPs) that accelerate the decay of the active G protein. Moreover, a transition state analogue of the GTPase reaction, Gαt[GDP-AlF4-/Ca2+] [14], and the hydrolysis product Gαt[GDP/Mg2+] [15] are known in high-resolution atomic detail. In the GTP-bound form, and in the transition state of GTP hydrolysis with GDP and monophosphate still bound, Mg2+ is bound with high affinity [14, 16]; whereas, in the GDP-bound Gα the presence of Mg2+ is controversial [15, 17]. In crystal structures of the GDP-bound heterotrimeric G protein, no bound Mg2+ has been observed [18, 19]. Consequently, it is likely that the GDP-bound receptor–G protein complex might lack Mg2+ as well. Mechanistically, this would make sense, especially because Mg2+ was found to inhibit GDP release in a small G protein [20]. However, Gαi mutagenesis experiments suggested a sequential release mechanism in which Mg2+ release facilitated subsequent GDP release in engineered G proteins with high basal nucleotide exchange rates [21]. There is the possibility that after GTP-uptake Gα dissociates first, followed by Gβγ. Moreover, it is possible that the GTP-loaded heterotrimer dissociates from the receptor before separation of the subunits. Furthermore, the standard model of dissociation of the heterotrimeric G proteins after receptor activation was challenged by in vivo FRET (Förster resonance energy transfer) experiments, which suggested that heterotrimeric Gi protein apparently undergoes subunit rearrangement instead of dissociation upon activation [22].
Box 2. Enabling and emerging technologies for single-molecule studies of GPCRs and higher order complexes.
Fluorescence correlation spectroscopy (FCS) has been applied to ligand-binding studies with fluorophore-labeled ligands in cells [59] or membrane preparations [60].
Fluorescence intensity distribution analysis (FIDA) is a SMD method sensitive to brightness. It has been applied as ultra high-throughput screening (uHTS) [61] to monitor binding of fluorophore-labeled ligands to receptors in plasma membrane preparations [62].
Single-particle tracking (SPT) methods have been used to study the mobility of GPCR fusion proteins of with fluorescent proteins [63, 64], enzyme tags (HaloTag or ACP tag) that allow specific conjugation with fluorophores [65, 66], or epitope tags that allow visualization with gold-tagged Fab antibody fragments [63]. SPT has been used to infer receptor binding of fluorescent protein fusion constructs of heterotrimeric G proteins [67, 68]. SPT has been applied to monitor the interaction of a Gα peptide with photoactivated rhodopsin [69]. SPT of fluorescently labeled antagonist ligands bound to receptors have demonstrated short-lived receptor dimers in living cells [64].
Single-molecule detection (SMD) has been used to visualize quantum dot-conjugated peptide antagonists bound to receptors embedded in solid-supported membranes [70].
High-density lipoprotein (HDL) particles known as nanodiscs [71] and nanoscale apolipoprotein-bound bilayers (NABBs) [72] have been used to reconstitute receptors in a native-like phospholipid matrix. SMD-TIRF microscopy has been used to visualize fluorophore-conjugated receptors reconstituted in nanodiscs [73, 74].
Single-particle electron microscopy (sp-EM) was used to quantify monomeric and dimeric receptors labeled with nanogold-maleimide site-specifically at a single reactive cysteine and reconstituted in NABBs under cryo conditions [72]. Alternatively, mass tagging of the receptor with a mAb Fab fragment bound to a C-terminal epitope allows determination of the orientation of receptor dimers by sp-EM in negatively stained samples [72].
Modular assembly of detergent-solubilized ternary complexes on beads and rapid mix flow cytometry was employed to measure the dissociation kinetics these complexes using several alternative capturing and labeling schemes [75]. While these experiments are limited to measurements of larger ensembles, they demonstrate the feasibility to reconstitute functional ternary complexes from purified components.
Atomic-force microscopy (AFM) has been used to visualize the supramolecular (lateral) organization of receptors in membranes from retinal photoreceptor cells [76].
Single-molecule force spectroscopy is an AFM technique that has been applied to study unfolding of rhodopsin [77].
Site-specific unnatural amino acid (UAA) mutagenesis can introduce novel chemical functionalities in correctly folded, post-translationally modified proteins expressed in mammalian cell culture. Receptors modified with keto and azide groups [78, 79] enable bioorthogonal labeling reactions to introduce fluorophores suitable for SMD.
Homogeneous time resolved fluorescence resonance energy transfer using a conformationally sensitive antibody to CCR5 allows quantification of femtomole amounts of receptors [80]. This auxiliary technology is relevant for monitoring folded receptor during purification
Funding information
This work was supported by NIH Grant EY12049 and the Crowley Family Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The term “escaping the flatlands” was inspired by the work of American information designer Edward Tufte.
References
- 1.Hanson MA, Stevens RC. Discovery of new GPCR biology: one receptor structure at a time. Structure. 2009;17:8–14. doi: 10.1016/j.str.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu B, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;7:7. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartwell LH, et al. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 4.Periole X, et al. G protein-coupled receptors self-assemble in dynamics simulations of model bilayers. J. Am. Chem. Soc. 2007;129:10126–10132. doi: 10.1021/ja0706246. [DOI] [PubMed] [Google Scholar]
- 5.Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat. Rev. Mol. Cell Bio. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- 6.Park JH, et al. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 7.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–U430. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 8.Sprang SR, et al. Structural basis of effector regulation and signal termination in heterotrimeric G alpha proteins. Adv. Protein Chem. 2007;74:1–65. doi: 10.1016/S0065-3233(07)74001-9. [DOI] [PubMed] [Google Scholar]
- 9.Bennett N, Dupont Y. The G-protein of retinal rod outer segments (transducin) - mechanism of interaction with rhodopsin and nucleotides. J. Biol. Chem. 1985;260:4156–4168. [PubMed] [Google Scholar]
- 10.Ernst OP, et al. Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit. Proc. Natl. Acad. Sci. USA. 2007;104:10859–10864. doi: 10.1073/pnas.0701967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn H. Light-regulated and GTP-regulated interaction of GTPase and other proteins with bovine photoreceptor-membranes. Nature. 1980;283:587–589. doi: 10.1038/283587a0. [DOI] [PubMed] [Google Scholar]
- 12.Noel JP, et al. The 2.2 Å crystal structure of transducin-α complexed with GTPγS. Nature. 1993;366:654–663. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- 13.Sondek J, et al. Crystal structure of a GA protein βγ dimer at 2.1 Å resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 14.Sondek J, et al. GTPase mechanism of G proteins from the 1.7-Å crystal-structure of transducin α· GDP· AlF4-. Nature. 1994;372:276–279. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]
- 15.Lambright DG, et al. Structural determinants for activation of the α-subunit of a heterotrimeric G protein. Nature. 1994;369:621–628. doi: 10.1038/369621a0. [DOI] [PubMed] [Google Scholar]
- 16.Coleman DE, et al. Structures of active conformations of Giα1 and the mechanism of GTP hydrolysis. Science. 1994;265:1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 17.Coleman DE, Sprang SR. Crystal structures of the G protein Giα1 complexed with GDP and Mg2+: A crystallographic titration experiment. Biochemistry. 1998;37:14376–14385. doi: 10.1021/bi9810306. [DOI] [PubMed] [Google Scholar]
- 18.Wall MA, et al. The structure of the G-protein heterotrimer Giα1β1γ2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 19.Lambright DG, et al. The 2.0 Å crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 20.Pan JY, et al. Influence of Mg2+ on the structure and function of Rab5. J. Biol. Chem. 1996;271:1322–1328. doi: 10.1074/jbc.271.3.1322. [DOI] [PubMed] [Google Scholar]
- 21.Kapoor N, et al. Structural evidence for a sequential release mechanism for activation of heterotrimeric G proteins. J. Mol. Biol. 2009;393:882–897. doi: 10.1016/j.jmb.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 22.Frank M, et al. G protein activation without subunit dissociation depends on a Gαi-specific region. J. Biol. Chem. 2005;280:24584–24590. doi: 10.1074/jbc.M414630200. [DOI] [PubMed] [Google Scholar]
- 23.Franke RR, et al. Rhodopsin mutants that bind but fail to activate transducin. Science. 1990;250:123–125. doi: 10.1126/science.2218504. [DOI] [PubMed] [Google Scholar]
- 24.Kenakin T. Ligand-selective receptor conformations revisited: the promise and the problem. Trends Pharmacol. Sci. 2003;24:346–354. doi: 10.1016/S0165-6147(03)00167-6. [DOI] [PubMed] [Google Scholar]
- 25.Kenakin T, et al. Determining the potency and molecular mechanism of action of insurmountable antagonists. J. Pharmacol. Exp. Ther. 2006;319:710–723. doi: 10.1124/jpet.106.107375. [DOI] [PubMed] [Google Scholar]
- 26.Siciliano SJ, et al. Interaction between the C5a receptor and Gi in both the membrane-bound and detergent-solubilized states. J. Biol. Chem. 1990;265:19568–19574. [PubMed] [Google Scholar]
- 27.Nobles M, et al. Heterotrimeric G proteins precouple with G protein-coupled receptors in living cells. Proc. Natl. Acad. Sci. USA. 2005;102:18706–18711. doi: 10.1073/pnas.0504778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halls ML, Cooper DM. Sub-picomolar relaxin signalling by a pre-assembled RXFP1, AKAP79, AC2, β-arrestin 2, PDE4D3 complex. EMBO J. 2010;29:2772–2787. doi: 10.1038/emboj.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu XH, et al. Coupling mechanism of a GPCR and a heterotrimeric G protein during chemoattractant gradient sensing in Dictyostelium. Sci. Signal. 2010;3:ra71. doi: 10.1126/scisignal.2000980. [DOI] [PubMed] [Google Scholar]
- 30.Mirzabekov T, et al. Paramagnetic proteoliposomes containing a pure, native, and oriented seven-transmembrane segment protein, CCR5. Nat. Biotechnol. 2000;18:649–654. doi: 10.1038/76501. [DOI] [PubMed] [Google Scholar]
- 31.Nisius L, et al. Large-scale expression and purification of the major HIV-1 coreceptor CCR5 and characterization of its interaction with RANTES. Protein Express. Purif. 2008;61:155–162. doi: 10.1016/j.pep.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Staudinger R, et al. Allosteric regulation of CCR5 by guanine nucleotides and HIV-1 envelope. Biochem. Biophys. Res. Commun. 2001;286:41–47. doi: 10.1006/bbrc.2001.5345. [DOI] [PubMed] [Google Scholar]
- 33.Peleg G, et al. Single-molecule spectroscopy of the β2 adrenergic receptor: Observation of conformational substates in a membrane protein. Proc. Natl. Acad. Sci. USA. 2001;98:8469–8474. doi: 10.1073/pnas.151239698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kobilka BK. G protein coupled receptor structure and activation. Biochim. Biophys. Acta. 2007;1768:794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakmar TP, et al. Rhodopsin: Insights from recent structural studies. Annual Review of Biophysics and Biomolecular Structure. 2002;31:443–484. doi: 10.1146/annurev.biophys.31.082901.134348. [DOI] [PubMed] [Google Scholar]
- 36.Kim JM, et al. Structural origins of constitutive activation in rhodopsin: Role of the K296/E113 salt bridge. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12508–12513. doi: 10.1073/pnas.0404519101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogel R, et al. Functional role of the “Ionic Lock” - An interhelical hydrogen-bond network in family a heptahelical receptors. Journal of Molecular Biology. 2008;380:648–655. doi: 10.1016/j.jmb.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 38.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 39.Lu HP, et al. Single-molecule enzymatic dynamics. Science. 1998;282:1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 40.Zhuang XW, et al. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 41.Birdsall NJM. Class A GPCR heterodimers: evidence from binding studies. Trends Pharmacol. Sci. 2010;31:499–508. doi: 10.1016/j.tips.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 42.Allen SJ, et al. Chemokine: Receptor structure, interactions, and antagonism. Annual Review of Immunology. 2007;25:787–820. doi: 10.1146/annurev.immunol.24.021605.090529. [DOI] [PubMed] [Google Scholar]
- 43.Oppermann M, et al. Differential effects of CC chemokines on CC chemokine receptor 5 (CCR5) phosphorylation and identification of phosphorylation sites on the CCR5 carboxyl terminus. J. Biol. Chem. 1999;274:8875–8885. doi: 10.1074/jbc.274.13.8875. [DOI] [PubMed] [Google Scholar]
- 44.Vila-Coro AJ, et al. Characterization of RANTES- and aminooxypentane-RANTES-triggered desensitization signals reveals differences in recruitment of the G protein-coupled receptor complex. J. Immunol. 1999;163:3037–3044. [PubMed] [Google Scholar]
- 45.Singer II, et al. CCR5, CXCR4, and CD4 are clustered and closely apposed on microvilli of human macrophages and T cells. Journal of Virology. 2001;75:3779–3790. doi: 10.1128/JVI.75.8.3779-3790.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carter GC, et al. HIV entry in macrophages is dependent on intact lipid rafts. Virology. 2009;386:192–202. doi: 10.1016/j.virol.2008.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elson EL, et al. Phase Separation in Biological Membranes: Integration of Theory and Experiment. Annual Review of Biophysics, Vol 39. 2010;39:207–226. doi: 10.1146/annurev.biophys.093008.131238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lingwood D, Simons K. Lipid Rafts As a Membrane-Organizing Principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 49.Simons K, Gerl MJ. Revitalizing membrane rafts: new tools and insights. Nature Reviews Molecular Cell Biology. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 50.Kusumi A, et al. Hierarchical organization of the plasma membrane: Investigations by single-molecule tracking vs. fluorescence correlation spectroscopy. Febs Letters. 2010;584:1814–1823. doi: 10.1016/j.febslet.2010.02.047. [DOI] [PubMed] [Google Scholar]
- 51.Hammad MM, Dupre DJ. Chaperones contribute to G protein coupled receptor oligomerization, but do not participate in assembly of the G protein with the receptor signaling complex. J Mol Signal. 2010;5:16. doi: 10.1186/1750-2187-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Botelho AV, et al. Curvature and hydrophobic forces drive oligomerization and modulate activity of rhodopsin in membranes. Biophysical Journal. 2006;91:4464–4477. doi: 10.1529/biophysj.106.082776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Madathil S, Fahmy K. Lipid Protein Interactions Couple Protonation to Conformation in a Conserved Cytosolic Domain of G Protein-coupled Receptors. Journal of Biological Chemistry. 2009;284:28801–28809. doi: 10.1074/jbc.M109.002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soubias O, et al. Contribution of Membrane Elastic Energy to Rhodopsin Function. Biophysical Journal. 2010;99:817–824. doi: 10.1016/j.bpj.2010.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horuk R. OPINION Chemokine receptor antagonists: overcoming developmental hurdles. Nat. Rev. Drug Discov. 2009;8:23–33. doi: 10.1038/nrd2734. [DOI] [PubMed] [Google Scholar]
- 56.Gladue RP, Brown MF. Current status of CCR1 antagonists in clinical trials. In: Neote K, et al., editors. Chemokine Biology: Basic Research and Clinical Application. II. Birkha□user; 2007. [Google Scholar]
- 57.Kitamura T, et al. Inactivation of chemokine (C-C motif) receptor 1 (CCR1) suppresses colon cancer liver metastasis by blocking accumulation of immature myeloid cells in a mouse model. Proc. Natl. Acad. Sci. USA. 2010;107:13063–13068. doi: 10.1073/pnas.1002372107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leach K, et al. Allosteric GPCR modulators: taking advantage of permissive receptor pharmacology. Trends Pharmacol. Sci. 2007;28:382–389. doi: 10.1016/j.tips.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 59.Pramanik A, et al. Fluorescence correlation spectroscopy detects galanin receptor diversity on insulinoma cells. Biochemistry. 2001;40:10839–10845. doi: 10.1021/bi010514q. [DOI] [PubMed] [Google Scholar]
- 60.Middleton RJ, Kellam B. Fluorophore-tagged GPCR ligands. Curr. Opin. Chem. Biol. 2005;9:517–525. doi: 10.1016/j.cbpa.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 61.Rudiger M, et al. Single-molecule detection technologies in miniaturized high throughput screening: Binding assays for G protein-coupled receptors using fluorescence intensity distribution analysis and fluorescence anisotropy. Journal of Biomolecular Screening. 2001;6:29–37. doi: 10.1177/108705710100600105. [DOI] [PubMed] [Google Scholar]
- 62.Scheel AA, et al. Receptor-ligand interactions studied with homogeneous fluorescence-based assays suitable for miniaturized screening. J. Biomol. Screen. 2001;6:11–18. doi: 10.1177/108705710100600103. [DOI] [PubMed] [Google Scholar]
- 63.Suzuki K, et al. Rapid hop diffusion of a G-protein-coupled receptor in the plasma membrane as revealed by single-molecule techniques. Biophys. J. 2005;88:3659–3680. doi: 10.1529/biophysj.104.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hern JA, et al. Formation and dissociation of M-1 muscarinic receptor dimers seen by total internal reflection fluorescence imaging of single molecules. Proc. Natl. Acad. Sci. USA. 2010;107:2693–2698. doi: 10.1073/pnas.0907915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prummer M, et al. Post-translational covalent labeling reveals heterogeneous mobility of individual G protein-coupled receptors in living cells. ChemBioChem. 2006;7:908–911. doi: 10.1002/cbic.200500477. [DOI] [PubMed] [Google Scholar]
- 66.Miyanaga Y, et al. Single-molecule imaging techniques to visualize chemotactic signaling events on the membrane of living Dictyostelium cells. Methods Mol. Biol. 2009;571:417–435. doi: 10.1007/978-1-60761-198-1_28. [DOI] [PubMed] [Google Scholar]
- 67.Perez JB, et al. Monitoring the diffusion of single heterotrimeric G proteins in supported cell-membrane sheets reveals their partitioning into microdomains. J. Mol. Biol. 2006;363:918–930. doi: 10.1016/j.jmb.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 68.van Hemert F, et al. Mobility of G proteins is heterogeneous and polarized during chemotaxis. J. Cell Sci. 2010;123:2922–2930. doi: 10.1242/jcs.063990. [DOI] [PubMed] [Google Scholar]
- 69.Kim TY, et al. Monitoring the Interaction of a Single G-Protein Key Binding Site with Rhodopsin Disk Membranes upon Light Activation. Biochemistry. 2009;48:3801–3803. doi: 10.1021/bi900308c. [DOI] [PubMed] [Google Scholar]
- 70.Zhou M, et al. Peptide-labeled quantum dots for imaging GPCRs in whole cells and as single molecules. Bioconjugate Chem. 2007;18:323–332. doi: 10.1021/bc0601929. [DOI] [PubMed] [Google Scholar]
- 71.Bayburt TH, et al. Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2002;2:853–856. [Google Scholar]
- 72.Banerjee S, et al. Rapid incorporation of functional rhodopsin into nanoscale apolipoprotein bound bilayer (NABB) particles. J. Mol. Biol. 2008;377:1067–1081. doi: 10.1016/j.jmb.2008.01.066. [DOI] [PubMed] [Google Scholar]
- 73.Whorton MR, et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl. Acad. Sci. USA. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuszak AJ, et al. Purification and functional reconstitution of monomeric μ-opioid receptors - allosteric modulation of agonist binding by Gi2. J. Biol. Chem. 2009;284:26732–26741. doi: 10.1074/jbc.M109.026922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buranda T, et al. Some mechanistic insights into GPCR activation from detergent-solubilized ternary complexes on beads. Mechanisms and Pathways of Heterotrimeric G Protein Signaling. 2007;74:95–135. doi: 10.1016/S0065-3233(07)74003-2. [DOI] [PubMed] [Google Scholar]
- 76.Fotiadis D, et al. Atomic-force microscopy: Rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- 77.Sapra KT, et al. Detecting molecular interactions that stabilize native bovine rhodopsin. J. Mol. Biol. 2006;358:255–269. doi: 10.1016/j.jmb.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 78.Ye S, et al. Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. J. Biol. Chem. 2008;283:1525–1533. doi: 10.1074/jbc.M707355200. [DOI] [PubMed] [Google Scholar]
- 79.Ye S, et al. FTIR analysis of GPCR activation using azido probes. Nat. Chem. Biol. 2009;5:397–399. doi: 10.1038/nchembio.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Knepp AM, et al. Direct Measurement of Thermal Stability of Expressed CCR5 and Stabilization by Small Molecule Ligands. Biochemistry. 2011;50:502–511. doi: 10.1021/bi101059w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin PC, et al. Site-specific protein modification through Cu-I-catalyzed 1,2,3-triazole formation and its implementation in protein microarray fabrication. Angew. Chem. Int. Ed. 2006;45:4286–4290. doi: 10.1002/anie.200600756. [DOI] [PubMed] [Google Scholar]
- 82.Tanaka M, Sackmann E. Polymer-supported membranes as models of the cell surface. Nature. 2005;437:656–663. doi: 10.1038/nature04164. [DOI] [PubMed] [Google Scholar]
- 83.Salamon Z, et al. Conformational-Changes in Rhodopsin Probed by Surface-Plasmon Resonance Spectroscopy. Biochemistry. 1994;33:13706–13711. doi: 10.1021/bi00250a022. [DOI] [PubMed] [Google Scholar]
- 84.Alves ID, et al. Direct observation of G-protein binding to the human delta-opioid receptor using plasmon-waveguide resonance spectroscopy. J. Biol. Chem. 2003;278:48890–48897. doi: 10.1074/jbc.M306866200. [DOI] [PubMed] [Google Scholar]
- 85.Tokimoto T, et al. Probing orientations of single fluorescent labels on a peptide reversibly binding to the human delta-opioid receptor. Appl. Spectrosc. 2007;61:130–137. doi: 10.1366/000370207779947512. [DOI] [PubMed] [Google Scholar]
- 86.Rigaud JL, et al. Reconstitution of membrane-proteins into liposomes - application to energy-transducing membrane-proteins. Biochim. Biophys. Acta. 1995;1231:223–246. doi: 10.1016/0005-2728(95)00091-v. [DOI] [PubMed] [Google Scholar]
- 87.Bieri C, et al. Micropatterned immobilization of a G protein-coupled receptor and direct detection of G protein activation. Nat. Biotechnol. 1999;17:1105–1108. doi: 10.1038/15090. [DOI] [PubMed] [Google Scholar]
- 88.Babcock GJ, et al. Ligand binding characteristics of CXCR4 incorporated into paramagnetic proteoliposomes. J. Biol. Chem. 2001;276:38433–38440. doi: 10.1074/jbc.M106229200. [DOI] [PubMed] [Google Scholar]
- 89.Neumann L, et al. Functional immobilization of a ligand-activated G-protein-coupled receptor. ChemBioChem. 2002;3:993–998. doi: 10.1002/1439-7633(20021004)3:10<993::AID-CBIC993>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 90.Martinez KL, et al. Ligand binding to G protein-coupled receptors in tethered cell membranes. Langmuir. 2003;19:10925–10929. [Google Scholar]
- 91.Sheth SR, Leckband D. Measurements of attractive forces between proteins and end-grafted poly(ethylene glycol) chains. Proc. Natl. Acad. Sci. USA. 1997;94:8399–8404. doi: 10.1073/pnas.94.16.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kuhl TL, et al. Modulation of interaction forces between bilayers exposing short-chained ethylene-oxide headgroups. Biophys. J. 1994;66:1479–1488. doi: 10.1016/S0006-3495(94)80938-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ross JL, Fygenson DK. Mobility of taxol in microtubule bundles. Biophys. J. 2003;84:3959–3967. doi: 10.1016/S0006-3495(03)75123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weiss S. Fluorescence spectroscopy of single biomolecules. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 95.Sage D, et al. Automatic tracking of individual fluorescence particles: Application to the study of chromosome dynamics. IEEE Trans. Image Process. 2005;14:1372–1383. doi: 10.1109/tip.2005.852787. [DOI] [PubMed] [Google Scholar]
- 96.Rasnik I, et al. Nonblinking and longlasting single-molecule fluorescence imaging. Nat. Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]