Abstract
Pili pickup sticks: Rodlike type 1 bacterial pili particles (see picture, red) self-assemble into highly ordered nanostructures through molecular recognition in the presence of suitable inducers. 1D bundles, 2D double-layer lattices, and 3D multilayer lattices were produced by varying the nature and concentration of the inducers. The self-assembled pili serve as templates for nucleating and organizing inorganic nanomaterials such as silica.
Keywords: bacterial pili, nanostructures, self-assembly, silica, template synthesis
The assembly of nanomaterials into ordered hierarchical structures is a very important step towards the fabrication of supramolecular structures and nanodevices. A promising strategy is to employ biomolecules with well-defined nanostructures as templates for directing the assembly of nanomaterials.[1] Towards this end, some filamentous biomacromolecules, such as bacteriophage, flagella, and tobacco mosaic virus, have been used to organize or nucleate inorganic nanomaterials.[2] However, they normally do not self-assemble into a 3D crystal lattice. To date, most nanostructures generated on the filamentous biotemplates are single nanotubes or nanowires. However, an ordered organization of nanotubes or nanowires is needed to satisfy the development of nanodevices. Thus, there is a need for a filamentous biotemplate to be assembled into a crystal-like lattice that can be further used as a template for directing nanomaterials synthesis. To our knowledge, the most common higher-order nanostructures assembled from filamentous biomacromolecules are limited to bundle structures through electrostatic interactions, liquid-crystalline assembly, or capillary forces.[2b, 3] Herein, we report the fabrication of highly ordered 3D lattice structures from rodlike type 1 bacterial pili particles in the presence of inducer molecules through molecular-recognition-based self-assembly and identify the proper inducer molecules that can tune the self-assembled structures of pili.
Type 1 bacterial pilus is a rigid, straight, naturally occurring protein nanorod (6–7 nm wide and 1–2 µm long) that can be detached from bacterial cells (Figure 1a,b and Figure S1 in the Supporting Information).[4] It is helically assembled from more than 3000 copies of a protein subunit called fimA (Figure 1g) with 27 subunits in eight turns.[4] It has an anionic surface with an isoelectric point of 3.92.[5] Its biological function is to assist the adhesion of bacteria to solid surfaces.[6] Although self-assembly behavior of pili was observed occasionally a long time ago,[5] the reproducible production of 3D hierarchical nanostructures made of multiple layers of pili has never been reported.
Figure 1.
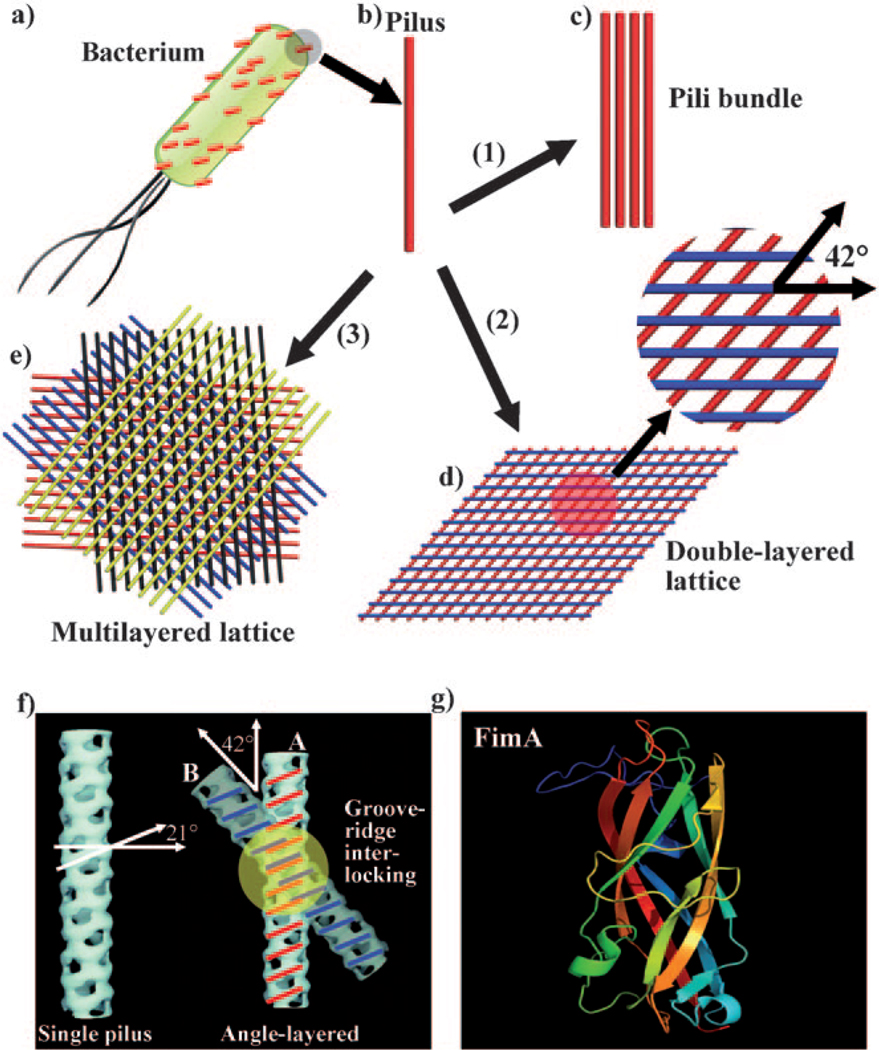
Self-assembly of pili into 1D bundles, 2D lattices, and 3D lattices. a) Illustration of a bacterial cell with many pili protruding from its surface. b) An individual pilus. c) Pili bundles. d) Double-layer pili lattices. The two neighboring layers of pili have a fixed angle of 42°. e) Four-layer pili lattices. A red layer of parallel pili was formed, with subsequent assembly of a blue layer of pili on top of the red layer with a twist angle of 42°. After that, a black and a yellow layer are deposited in turn on top of the blue layer and black layer, with the neighboring layer twisted at the same angle of 42°, resulting in the formation of a four-layer pili lattice. f) Molecular recognition between two pili particles. A single pilus has a set of alternating ridges and grooves with a pitch angle of 21°. When the crossing angle between pili A and B is 42°, the ridges of pilus A can make a reasonable interlocking fit into the corresponding grooves of pilus B. g) The 3D structure of the subunit of pili FimA (PDB ID: 2JTY). Inducing agents include: 1) more than 160 mm hexamethylenediamine; 2) 80 mm hexamethylenediamine; and 3) 80 mm pimelic acid or 1,3-propanedisulfonic acid. A high-resolution version of (e) is shown in Figure S3 in the Supporting Information.
We discovered the formation of 3D ordered pili lattices in the presence of pimelic acid. This fact encouraged us to hypothesize that linear molecules structurally similar to pimelic acid can influence the assembly of pili into lattices. To identify the molecules that can induce the formation of ordered pili lattices, we employed linear molecules with either backbones between the two distal functional groups or functional groups at the two distal ends different from those of the pimelic acid molecule and systemically studied the assembly of pili particles in the presence of these molecules (Table S1 in the Supporting Information). We found a reproducible approach to the formation of highly ordered nanostructures, including 1D bundles, 2D double-layer lattices, and 3D multilayer lattices from the rodlike type 1 bacterial pili particles by varying the inducer molecules and their concentrations in the aqueous phase (Figure 1). Specifically, we found that linear molecules with identical cationic functional groups (NH2) at the two distal ends tend to induce the formation of double-layer lattices (Figure 1d and Table S1 and Figure S2 in the Supporting Information), whereas those with identical anionic functional groups (SO3H or COOH) at the two distal ends tend to induce the formation of multilayer lattices (Table S1 in the Supporting Information, Figure 1e). Furthermore, the self-assembled pili could be used as templates to nucleate and organize inorganic nanomaterials, such as silica.
Type 1 pili particles used herein were detached and purified from E. coli K12 ER2738 (Figure S1 in the Supporting Information). To induce self-assembly, the pili solution was diluted to a concentration of OD280nm = 0.25 (OD280nm = optical density at 280 nm) in specific aqueous solutions (Table S1 in the Supporting Information) and incubated at 4°C. In the presence of a high concentration of hexamethylenediamine molecules (above 160 mm), the pili solution became cloudy within 1 h, because pili were self-assembled into bundle structures in which pili aligned in parallel (Figure 1c and Figure 2a). After pili were incubated in a lower concentration (80 mm) of hexamethylenediamine, the solution was clear for the first two days and became cloudy after three days, thus suggesting that pili slowly self-assembled into large aggregates. On day seven, precipitates appeared in the solution, thus indicating a continuous growth of pili assemblies. TEM examination revealed the formation of double-layer pili lattices that were made up of two twisted individual layers. Each layer is made of parallel pili, and pili from the two adjacent layers are at a fixed angle of 42° (Figure 1d, Figure 2b, and Figure S2 in the Supporting Information). The center-to-center distance between the neighboring pili in the same layer was about 13 nm.
Figure 2.
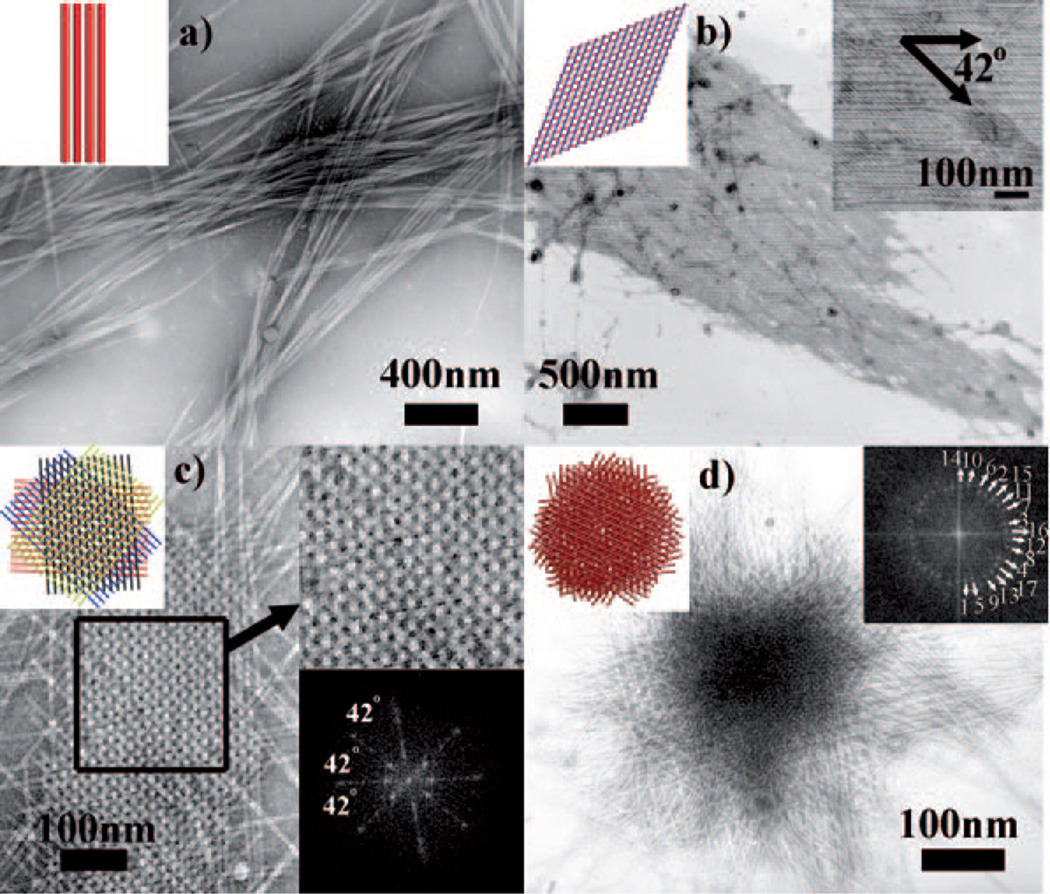
TEM images of pili assemblies. a) Bundles. b) A double-layer lattice with a fixed twist angle of 42° between two layers. c) A four-layer pili lattice. FFT (inset, lower right) of selected area confirms the highly ordered structure and the twist angle between neighboring layers. d) A 17-layer pili lattice. FFT image (inset, upper right) confirms there are 17 layers of pili in the lattice with a fixed twist angle of 42° between adjacent layers. The spot corresponding to each layer is marked in the 2D-FFT image. The inset in the upper left corner of each TEM image illustrates the corresponding pili assemblies.
In the presence of pimelic acid with a proper concentration (80 mm), the pili solution stayed clear until day six. After day six, the solution became cloudy, and small precipitates appeared, owing to the formation of highly ordered multilayer pili lattices (Figure 1e and Figure 2 c,d). The pili lattices were made up of at least four alternately twisted layers of parallel pili, with the neighboring layers having a fixed angle of 42°. This highly ordered lattice structure, made of vertically stacked layers of pili with a twist angle of 42° between neighboring layers, could be verified by the 2D fast Fourier transform (2D-FFT) image (Figure 2c,d, inset). In a five-layer lattice, the angle between the direction of pili in layer 1 and the direction of pili in layer 5 is only about 12°, which makes them almost overlapped in TEM images. Therefore, when the number of layers is more than four in pili crystals, it is difficult to identify the number of layers by simply using 2D TEM imaging (Figure 2d). But the FFT image can be used to identify the number of pili layers. The FFT image (inset in Figure 2d) shows a total of 17 layers of pili in the crystal with a fixed twist angle of 42° between neighboring layers.
Our work shows that the careful choice of inducers in the pili solution can control the self-assembled nanostructures (Table S1 in the Supporting Information). First, we found that pili could self-assemble only in the presence of hexamethylenediamine, pimelic acid, or 1,3-propanedisulfonic acid, but could not in the presence of other species we tested (Table S1 in the Supporting Information). Those successful assembly-inducers have either positively or negatively charged distal ends connected by a long central carbon chain. Molecules with two neutral ends (1,4-bis(2-hydroxyethyl)piperazine) or with one neutral end and one negative end (HEPES; 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid), or even with one positive end and one negative end (6-aminocaproic acid) could not induce the assembly of pili. These facts suggest that only molecules with the same charges on both sides could induce the assembly of pili. Second, the concentrations of the inducers in pili solutions could control whether the self-assembled nanostructures were 1D bundles, 2D double-layer lattices, or 3D multilayer lattices. High concentrations of positively charged inducers (hexamethylenediamine) could induce the self-assembly of pili into bundles through electrostatic interactions (within 1 h). Double-layer pili lattices were formed in an 80 mm hexamethylenediamine solution after a longer incubation time (three days). In this case, only a proper concentration of hexamethylenediamine could induce the formation of double-layer pili lattices. If the concentration was too high, only bundles were produced; if the concentration was too low, no assemblies were observed.
However, 3D multilayer pili lattices could never be induced by hexamethylenediamine solution, no matter how long we incubated the system or what concentration we used. They were formed in the presence of either 80 mm pimelic acid (seven days) or 80 mm 1,3-propanedisulfonic acid (20 days, Table S1 in the Supporting Information), both of which have negative charges (COOH or SO3H) at two distal ends. These inducer molecules could not electrostatically attract pili like hexamethylenediamine. Since they could not interact with pili directly, the nucleation and growth of pili lattices were slower. Compared to 1,3-propanedisulfonic acid, pimelic acid has a better precipitating efficiency, so the growth rate of pili in its solution (seven days) was faster than in 1,3-propanedisulfonic acid solution (20 days).
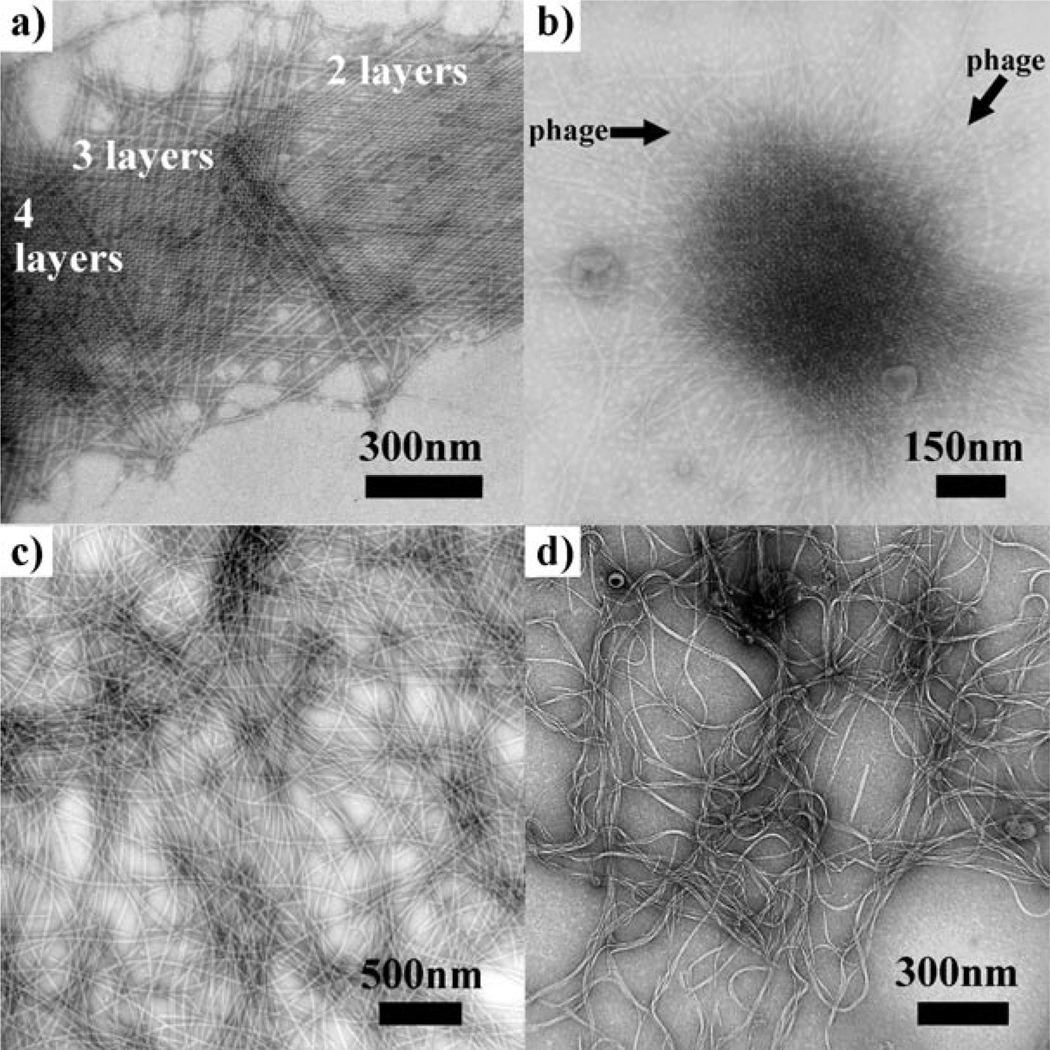
If we regard the multilayer pili lattice as a crystal formed from rodlike colloidal particles, the growth of pili lattices followed a layer-by-layer fashion[7] through 2D nucleation on surfaces (Figure 3a). This 2D nucleation strongly slowed the rate at which crystal growth occurred,[8] consistent with our finding that multilayer pili lattices were formed slowly (seven days). Furthermore, pili could be crystallized into lattices even in the presence of M13 phage as a contaminant (Figure 3b), which has a similar isoelectric point (4.2), diameter (7 nm), and length (1 µm) to the pili particles. This fact suggested that there were specific molecular recognition interactions between neighboring pili layers that make them recognizable, and the presence of M13 phage could not disturb the assembly of pili into 3D lattices. To test the importance of molecular recognition between pili in the assembly process, bacterial flagella and M13 phages, which do not have recognition between themselves, were incubated under the same conditions. As expected, no lattices were formed from flagella or M13 phages (Figure 3c, d).
Figure 3.
Mechanism study of the formation of 3D lattices from pili particles. a) Evidence showing that the crystallization of pili took place in a layer-by-layer fashion. b) Pili crystallized into lattices with M13 phage as a contaminant. c) Bacterial flagella after incubation in 80 mm pimelic acid. d) Phages after incubation in 80 mm pimelic acid.
We believe the molecular recognition driving the formation of the pili lattices arises from the molecular structures of pili. On each pilus, there is a set of alternating ridges and grooves with a pitch angle of 21° (Figure 1 f).[4] When pili self-assemble, the ridges of pilus A must make an interlocking fit into corresponding grooves of pilus B, which can happen only when pili A and B are at an angle of 2 × 21° = 42° (Figure 1g). This angle is same as the twist angle of 42° found in double- or multilayer lattices. Moreover, the measured center-to-center distance between the neighboring pili in the same layer was about 13 nm, and the length of each side of the unit rhombus in the lattice was about 19 nm (Figure S2 in the Supporting Information). This value matches the reported helical repeat of type 1 pilus (19.31 nm),[4] which again confirmed the existence of molecular recognition in the lattice. This molecular-structure-based recognition favors the nucleation and growth of pili lattices. Such molecular-structure-directed self-assembly was also observed in F-actin.[9]
There might be two competing driving forces for the assembly of pili into 3D lattices. One is the molecular recognition (Figure 1 f) between pili in the two adjacent pili layers, which enables the pili in the upper layer to form a 42° angle with the pili in the bottom layer and favors the layer-by-layer assembly (Figure 1e); another is the like-charge attraction[10] (Figure S4 in the Supporting Information) between anionic pili in the same layer modulated by the counterions,[11] which enables the pili to be parallel to each other in the same layer and favors the layer expansion. Therefore, in order to form a multilayer lattice, the molecular recognition driving force between neighboring layers should surpass the like-charge interaction between the parallel neighboring pili in the same layer. Linear molecules with two anionic groups at the ends tend to disfavor the like-charge interaction between neighboring anionic pili in the same layer (Figure S4c in the Supporting Information) and instead favor the vertical assembly of additional layers through molecular recognition and the concomitant formation of multilayer lattices. However, linear molecules with two cationic groups at the ends will tend to favor the like-charge interaction between parallel pili in the same layer (Figure S4b in the Supporting Information) more than the molecular recognition between adjacent pili layers, and thus the formation of either bundles or double-layer lattices is more favored. This rationale is consistent with our findings (Table S1 in the Supporting Information and Figure 2) and also explains why a proper inducer molecule is required to promote the assembly of pili into multilayer 3D lattices. Furthermore, linear molecules are needed for lattice formation, because such molecules will tend to make pili parallel to each other and not touching each other in the same layer. This conclusion is also in agreement with our finding that small cations with higher concentration (Mg2+, 50mm) could also induce the formation of bundles (Figure S4a in the Supporting Information). However, the inorganic cations could never induce the formation of ordered lattices, which further confirms the necessity of using linear molecules as inducers.
Pili bundles, double-layer lattices, and multilayer lattices are well-organized biotemplates (Figure 2) for nanosynthesis. We selected silica as a model inorganic nanomaterial for this purpose. Specifically, all pili assemblies were fixed in 1% glutaraldehyde solution, and then 3-aminopropyltriethoxysilane (APTES) and tetraethyl orthosilicate (TEOS) were added in turn under stirring so that silica could be uniformly coated onto them.[2c] Silica–pili hybrid nanorods with multichannels, nanorhombuses, and nanoflowers were produced by using pili bundles, double-layer pili lattices, and multilayer pili lattices as templates, respectively (Figure 4). Silica coatings were confirmed by energy-dispersive X-ray (EDX) spectroscopy analysis (Figure 4d). The fabricated 2D silica nanorhombuses and 3D silica nanoflowers have ordered pili lattices inside, which could be easily removed by calcinations for further applications.
Figure 4.
Silica–pili composites formed by templating silica nucleation on pili assemblies. a) Silica-coated pili bundles. b) A fragment of a silica-coated double-layer pili lattice. c) A silica-coated multilayer pili lattice. d) SEM image and EDX analysis of composites shown in (c). EDX analysis (inset) shows elemental composition of Si and O with a proper weight percentage. The cartoon of each mineralized pili assembly structure is shown as the insets in (a)–(c).
Before silica precursors were added, pili assemblies were first fixed and cross-linked by glutaraldehyde so that glutaraldehyde molecules reacted with the amino groups from pili but not with carboxy groups. Therefore, the remaining carboxy groups made the pili surface highly negative so that APTES could easily be absorbed onto pili assemblies. The close contact of APTES with the pili surface enhances the hydrolysis of APTES to form silicic acid, which functions as a nucleation site for subsequent silica growth.[2c] After the addition of TEOS, polycondensation of TEOS and the growth of silica will be based on the silica nuclei on the pili surface, ultimately resulting in the formation of pili–silica hybrids.
In summary, for the first time, we reproducibly generated controlled nanoarchitectures of 1D, 2D, and 3D pili assemblies and demonstrated their use as novel biotemplates for the synthesis of inorganic nanomaterials. Pili lattices were composed of multilayers of pili particles through the molecular recognition between neighboring pili layers. The free-standing pili bundles, double-layer lattices, and multilayer lattices could serve as biotemplates for the synthesis and assembly of inorganic nanomaterials. A foreign peptide can be genetically inserted into each subunit constituting the pili, thus leading to the display of the peptide on the side wall of the pili. Therefore, the lattices assembled from peptide-displayed pili can enable many possible applications in materials science, photonic and electronic device fabrication, bioengineering, and nanomedicine.
Experimental Section
Assembly of pili into lattices: The purified pili solution was diluted to a concentration of OD280nm = 0.5, mixed with a specific aqueous solution (Table S1 in the Supporting Information) with a 1:1 ratio, and incubated at 4°C for different times. After incubation, pili assemblies were negatively stained with 1% uranyl acetate and characterized under transmission electron microscopy (TEM).
Footnotes
We thank the National Science Foundation (DMR-0847758, CBET-0854414, CBET-0854465), National Institutes of Health (R21EB009909-01A1, R01L092526-01A2, R03AR056848-01), and Oklahoma Center for the Advancement of Science and Technology for financial support.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201102052.
References
- 1.a) Ma N, Sargent EH, Kelley SO. J. Mater. Chem. 2008;18:954–964. [Google Scholar]; b) Sotiropoulou S, Sierra-Sastre Y, Mark SS, Batt CA. Chem. Mater. 2008;20:821–834. [Google Scholar]; c) van Bommel KJC, Friggeri A, Shinkai S. Angew. Chem. 2003;115:1010–1030. doi: 10.1002/anie.200390284. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2003;42:980–999. doi: 10.1002/anie.200390284. [DOI] [PubMed] [Google Scholar]; d) Wang Q, Lin TW, Tang L, Johnson JE, Finn MG. Angew. Chem. 2002;114:477–480. doi: 10.1002/1521-3773(20020201)41:3<459::aid-anie459>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2002;41:459–462. [Google Scholar]
- 2.a) Cao BR, Mao CB. Langmuir. 2007;23:10701–10705. doi: 10.1021/la7014435. [DOI] [PubMed] [Google Scholar]; b) Wang FK, Cao BR, Mao CB. Chem. Mater. 2010;22:3630–3636. doi: 10.1021/cm902727s. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang FK, Li D, Newton SM, Klebba PE, Mao CB. Adv. Funct. Mater. 2009;19:3355–3355. [Google Scholar]; d) Balci S, Bittner AM, Hahn K, Scheu C, Knez M, Kadri A, Wege C, Jeske H, Kern K. Electrochim. Acta. 2006;51:6251–6257. [Google Scholar]; e) Bromley KM, Patil AJ, Perriman AW, Stubbs G, Mann S. J. Mater. Chem. 2008;18:4796–4801. [Google Scholar]; f) He T, Abbineni G, Cao BR, Mao CB. Small. 2010;6:2230–2235. doi: 10.1002/smll.201001108. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Mao CB, Liu AH, Cao BR. Angew. Chem. 2009;121:6922–6943. [Google Scholar]; Angew. Chem. Int. Ed. 2009;48:6790–6810. doi: 10.1002/anie.200900231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Lin Y, Balizan E, Lee LA, Niu ZW, Wang Q. Angew. Chem. 2010;122:880–884. doi: 10.1002/anie.200904993. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2010;49:868–872. doi: 10.1002/anie.200904993. [DOI] [PubMed] [Google Scholar]; b) Lee SW, Mao CB, Flynn CE, Belcher AM. Science. 2002;296:892–895. doi: 10.1126/science.1068054. [DOI] [PubMed] [Google Scholar]; c) Hamley IW. Soft Matter. 2010;6:1863–1871. [Google Scholar]
- 4.Hahn E, Wild P, Hermanns U, Sebbel P, Glockshuber R, Haner M, Taschner N, Burkhard P, Aebi U, Muller SA. J. Mol. Biol. 2002;323:845–857. doi: 10.1016/s0022-2836(02)01005-7. [DOI] [PubMed] [Google Scholar]
- 5.Brinton CC. Trans. N. Y. Acad. Sci. 1965;27:1003–1022. doi: 10.1111/j.2164-0947.1965.tb02342.x. [DOI] [PubMed] [Google Scholar]
- 6.Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, Hultgren SJ. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 7.Malkin AJ, Kuznetsov YG, Land TA, Deyoreo JJ, Mcpherson A. Nat. Struct. Biol. 1995;2:956–959. doi: 10.1038/nsb1195-956. [DOI] [PubMed] [Google Scholar]
- 8.Boistelle R, Astier JP. J. Cryst. Growth. 1988;90:14–30. [Google Scholar]
- 9.Yamamoto K, Yanagida M, Kawamura M, Maruyama K, Noda H. J. Mol. Biol. 1975;91:463–466. doi: 10.1016/0022-2836(75)90272-7. [DOI] [PubMed] [Google Scholar]
- 10.a) Marvin DA. Curr. Opin. Struct. Biol. 1998;8:150–158. doi: 10.1016/s0959-440x(98)80032-8. [DOI] [PubMed] [Google Scholar]; b) Tang JX, Janmey PA, Lyubartsev A, Nordenskiold L. Biophys. J. 2002;83:566–581. doi: 10.1016/S0006-3495(02)75192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zimmermann K, Hagedorn H, Heuck CC, Hinrichsen M, Ludwig H. J. Biol. Chem. 1986;261:1653–1655. [PubMed] [Google Scholar]; d) Purdy KR, Fraden S. Phys. Rev. E. 2004;70 doi: 10.1103/PhysRevE.70.061703. 061703. [DOI] [PubMed] [Google Scholar]; e) Butler JC, Angelini T, Tang JX, Wong GCL. Phys. Rev. Lett. 2003;91 doi: 10.1103/PhysRevLett.91.028301. 028301. [DOI] [PubMed] [Google Scholar]
- 11.a) Souza GR, Christianson DR, Staquicini FI, Ozawa MG, Snyder EY, Sidman RL, Miller JH, Arap W, Pasqualini R. Proc. Natl. Acad. Sci. USA. 2006;103:1215–1220. doi: 10.1073/pnas.0509739103. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Souza GR, Molina JR, Raphael RM, Ozawa MG, Stark DJ, Levin CS, Bronk LF, Ananta JS, Mandelin J, Georgescu MM, Bankson JA, Gelovani JG, Killian TC, Arap W, Pasqualini R. Nat. Nanotechnol. 2010;5:291–296. doi: 10.1038/nnano.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]